Abstract
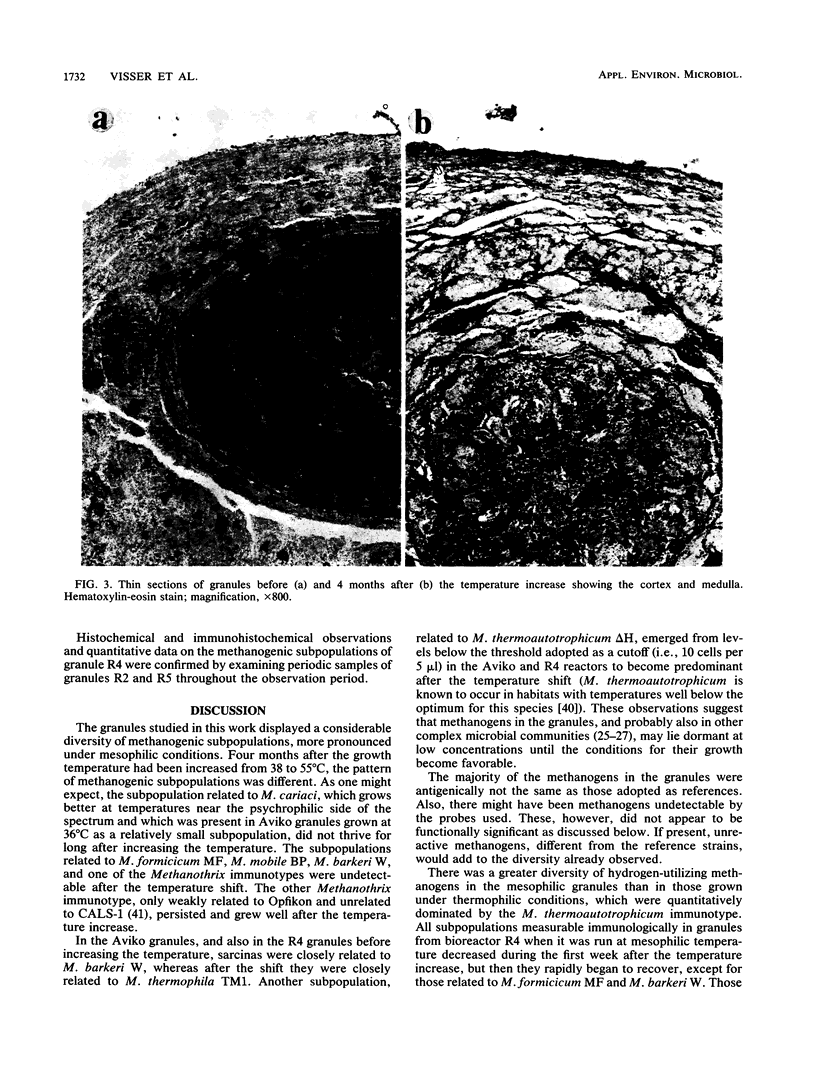
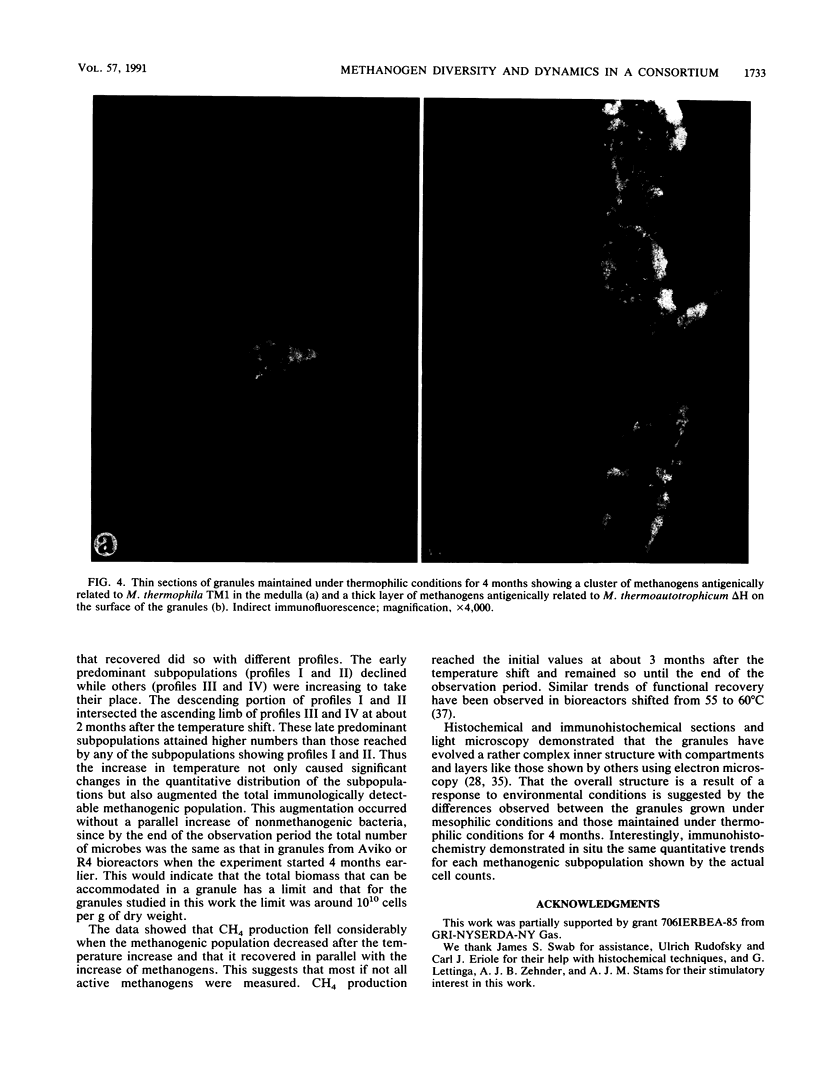
Upflow anaerobic sludge blanket bioreactor granules were used as an experimental model microbial consortium to study the dynamics and distribution of methanogens. Immunologic methods revealed a considerable diversity of methanogens that was greater in mesophilic granules than in the same granules 4 months after a temperature shift from 38 to 55°C. During this period, the sizes of the methanogenic subpopulations changed with distinctive profiles after the initial reduction caused by the shift. Methanogens antigenically related to Methanobrevibacter smithii PS and ALI, Methanobacterium hungatei JF1, and Methanosarcina thermophila TM1 increased rapidly, reached a short plateau, and then fell to lower concentrations that persisted for the duration of the experiment. A methanogen related to Methanogenium cariaci JR1 followed a similar profile at the beginning, but it soon diminished below detection levels. Methanothrix rods weakly related to the strain Opfikon increased rapidly, reaching a high-level, long-lasting plateau. Two methanogens related to Methanobrevibacter arboriphilus AZ and Methanobacterium thermoautotrophicum ΔH emerged from very low levels before the temperature shift and multiplied to attain their highest numbers 4 months after the shift. Histochemistry and immunohistochemistry revealed thick layers, globular clusters, and lawns of variable density which were distinctive of the methanogens related to M. thermoautotrophicum ΔH, M. thermophila TM1, and M. arboriphilus AZ and M. soehngenii Opfikon, respectively, in thin sections of granules grown at 55°C for 4 months. Mesophilic granules showed a different pattern of methanogenic subpopulations.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Balch W. E., Fox G. E., Magrum L. J., Woese C. R., Wolfe R. S. Methanogens: reevaluation of a unique biological group. Microbiol Rev. 1979 Jun;43(2):260–296. doi: 10.1128/mr.43.2.260-296.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartrain M., Zeikus J. G. Microbial ecophysiology of whey biomethanation: characterization of bacterial trophic populations and prevalent species in continuous culture. Appl Environ Microbiol. 1986 Jan;51(1):188–196. doi: 10.1128/aem.51.1.188-196.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway de Macario E., Macario A. J., Jovell R. J. Slide immunoenzymatic assay (SIA) in hybridoma technology. Methods Enzymol. 1986;121:509–525. doi: 10.1016/0076-6879(86)21051-4. [DOI] [PubMed] [Google Scholar]
- Costerton J. W., Cheng K. J., Geesey G. G., Ladd T. I., Nickel J. C., Dasgupta M., Marrie T. J. Bacterial biofilms in nature and disease. Annu Rev Microbiol. 1987;41:435–464. doi: 10.1146/annurev.mi.41.100187.002251. [DOI] [PubMed] [Google Scholar]
- Kemp H. A., Archer D. B., Morgan M. R. Enzyme-Linked Immunosorbent Assays for the Specific and Sensitive Quantification of Methanosarcina mazei and Methanobacterium bryantii. Appl Environ Microbiol. 1988 Apr;54(4):1003–1008. doi: 10.1128/aem.54.4.1003-1008.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi H. A., Conway de Macario E., Williams R. S., Macario A. J. Direct characterization of methanogens in two high-rate anaerobic biological reactors. Appl Environ Microbiol. 1988 Mar;54(3):693–698. doi: 10.1128/aem.54.3.693-698.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLeod F. A., Guiot S. R., Costerton J. W. Layered structure of bacterial aggregates produced in an upflow anaerobic sludge bed and filter reactor. Appl Environ Microbiol. 1990 Jun;56(6):1598–1607. doi: 10.1128/aem.56.6.1598-1607.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macario A. J., Conway de Macario E., Ney U., Schoberth S. M., Sahm H. Shifts in methanogenic subpopulations measured with antibody probes in a fixed-bed loop anaerobic bioreactor treating sulfite evaporator condensate. Appl Environ Microbiol. 1989 Aug;55(8):1996–2001. doi: 10.1128/aem.55.8.1996-2001.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macario A. J., Conway de Macario E. Quantitative immunologic analysis of the methanogenic flora of digestors reveals a considerable diversity. Appl Environ Microbiol. 1988 Jan;54(1):79–86. doi: 10.1128/aem.54.1.79-86.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulis L., Chase D., Guerrero R. Microbial communities. Bioscience. 1986 Mar;36(3):160–170. [PubMed] [Google Scholar]
- Varel V. H., Isaacson H. R., Bryant M. P. Thermophilic methane production from cattle waste. Appl Environ Microbiol. 1977 Feb;33(2):298–307. doi: 10.1128/aem.33.2.298-307.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R. Bacterial evolution. Microbiol Rev. 1987 Jun;51(2):221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinder S. H., Anguish T., Cardwell S. C. Effects of Temperature on Methanogenesis in a Thermophilic (58 degrees C) Anaerobic Digestor. Appl Environ Microbiol. 1984 Apr;47(4):808–813. doi: 10.1128/aem.47.4.808-813.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]