Abstract
Cladistic analysis of cranial and dental evidence has been widely used to generate phylogenetic hypotheses about humans and their fossil relatives. However, the reliability of these hypotheses has never been subjected to external validation. To rectify this, we applied identical methods to equivalent evidence from two groups of extant higher primates for whom reliable molecular phylogenies are available, the hominoids and papionins. We found that the phylogenetic hypotheses based on the craniodental data were incompatible with the molecular phylogenies for the groups. Given the robustness of the molecular phylogenies, these results indicate that little confidence can be placed in phylogenies generated solely from higher primate craniodental evidence. The corollary of this is that existing phylogenetic hypotheses about human evolution are unlikely to be reliable. Accordingly, new approaches are required to address the problem of hominin phylogeny.
The upsurge in paleoanthropological field research over the past quarter century has resulted in the recognition of many new hominin species, including Australopithecus afarensis (1), Paranthropus aethiopicus (2), Ardipithecus ramidus (3, 4), Australopithecus anamensis (5), Australopithecus bahrelghazali (6), Homo antecessor (7), and Australopithecus garhi (8). This has led to commensurate interest in the generation of reliable hypotheses about human phylogeny (8–14). Without a reliable phylogeny, little confidence can be placed in hypotheses of ancestry, or in scenarios linking events in human evolution with environmental and ecological influences. However, the phylogenetic relationships of the dozen, or so, species whose remains comprise the hominin fossil record are far from certain. Despite, in paleontological terms, a relative abundance of fossil evidence, cladistic analyses of the hominins have so far yielded conflicting and weakly supported hypotheses of relationships (9–14, 15, 16). Conventionally, this state of affairs has been attributed to poor character choice, taxonomic disagreements, or flaws in the available analytical methods (10, 12, 14). A fourth possibility, namely, that the type of qualitative and quantitative craniodental characters normally used to reconstruct the phylogenetic relationships of hominin species and genera are not reliable for this purpose, has only recently been entertained (8, 13, 17–21).
To assess the likely reliability of standard hominin cranial and dental characters for interspecific and intergeneric phylogenetic reconstruction, we used hominin cladistic methods to analyze comparable characters from two groups of extant primates: the hominoids, the higher primate group most closely related to the fossil hominins, and the papionins, the Old World monkey tribe comprising the baboons, mangabeys, and macaques. We then judged the resulting cladograms against the groups' consensus molecular phylogenies (22, 23). This approach, which is similar to Hartman's (24), assumes that congruence between the morphological and molecular phylogenies indicates that equivalent hominin fossil evidence yields reliable phylogenies, whereas incongruence indicates the converse. The assumption that any conflict between the molecular and morphological cladograms results from limitations of the morphological evidence can be justified on several counts. First, phylogenetic relationships are genetic relationships. Thus, in phylogenetics, morphology can never be more than a proxy for molecular data. Second, because osseous and other morphological characters can be highly influenced by external stimuli, such as the forces generated by habitual activities (13, 19–21), they can be expected to provide misleading information about phylogeny more frequently than genetical characters. Third, both molecular cladograms used in this study are supported by multiple lines of independent biomolecular and karyological evidence (22, 23). Congruence among multiple lines of independent evidence is the strongest support possible for a phylogenetic hypothesis. Lastly, molecular phylogenetic methods have been successfully tested on taxa of known phylogeny whereas comparable tests of morphological phylogenetic methods have not been successful (25–27).
In the present study, we concentrated on testing the phylogenetic utility of quantitative rather than qualitative craniodental data. Although more hominin cladistic analyses have been based on qualitative characters than on quantitative characters, there is in fact no intrinsic difference between qualitative and quantitative characters as far as the cladistic methodology is concerned (28). Moreover, qualitatively scored characters are necessarily more subjective than measurements, and the few studies that have examined the reproducibility of qualitative assessments of morphology have shown that many of them are impossible to replicate (14, 29). Furthermore, unlike discrete character states, morphometric data can be adjusted to account for any intertaxon body size differences, thereby reducing a further potential source of confusion. Nonetheless, to relate our results to all of the published cladistic analyses of the hominins, we also included an assessment of the phylogenetic utility of qualitatively derived character states.
Materials and Methods
Standard paleoanthropological measurements of the cranium, mandible, and dentition were used to compile quantitative data sets for the extant hominoids and papionins. The hominoid data set comprised values for 129 measurements recorded on males and females of Gorilla, Homo, Pan, Pongo, and an Old World monkey, Colobus, which was included as an outgroup. Seventy-seven of the one hundred and twenty-nine measurements were recorded on 37 Gorilla gorilla, 75 Homo sapiens, 35 Pan troglodytes, 41 Pongo pygmaeus, and 24 Colobus guereza (30). The other 52 measurements were taken on 20 G. gorilla, 20 H. sapiens, 20 P. troglodytes, 20 P. pygmaeus and 20 C. guereza (31).
The papionin data set consisted of values for 62 measurements recorded on mixed sex samples of Macaca, Cercocebus, Lophocebus, Mandrillus, Papio, Theropithecus, and an outgroup, Pan. All 62 measurements were recorded on 26 Cercocebus galeritus/torquatus, 40 Lophocebus albigena/atterimus, 40 Macaca fascicularis/mulatta, 62 Mandrillus leucophaeus/sphinx, 39 Papio hamadryas, 44 Theropithecus gelada, and 17 P. troglodytes (32). Fifty-five of the measurements were taken on a further 14 C. torquatus and 12 P. troglodytes. These data were kindly provided by A. T. Chamberlain (University of Sheffield).
A character state data matrix was derived from each data set. The confounding effects of the body size differences between the taxa were minimized by dividing the values for each specimen by their geometric mean (33). The size-adjusted data were converted into discrete character states using divergence coding (34). Alternative size-adjustment methods and coding procedures either give similar results to the techniques used here (35) or are inappropriate for analyses of fossil taxa (36).
A qualitative data matrix was also generated for the hominoids. This consisted of the states of 96 craniodental characters recorded on specimens of Gorilla, Homo, Hylobates, Pan, Pongo, and an outgroup. Sixty-two characters were taken from Shoshani et al. (37), two from Braga (38), six from Andrews (39), four from Schwartz (40), and two from Delson and Andrews (41). The remainder were the cranial and dental characters from Groves (42) that were excluded, without explanation, from the data set of Shoshani et al. (37). (This dataset is published as supplemental data on the PNAS web site, www.pnas.org.)
The matrices were used to perform two tests of the hypothesis that craniodental characters are reliable for reconstructing the phylogenetic relationships of fossil primate species and genera. The first test was based on parsimony analysis, which identifies the cladogram/s requiring the smallest number of ad hoc hypotheses of character state change to account for the distribution of character states among the taxa. Each matrix was subjected to parsimony analysis by using paup 3.0s (43), and the shortest cladogram compared with the appropriate consensus molecular cladogram (Figs. 1 and 2). We reasoned that, for the hypothesis to be supported, the favored cladogram should not contain any “false” (i.e., nonmolecular) clades, since it has been standard practice in hominin cladistic analyses to assume that all clades of a cladogram are equally reliable (9–14). Thus, the hypothesis would have been supported if an analysis favored a fully resolved cladogram that matched the molecular cladogram, or a partially resolved cladogram that comprised only molecular clades. Alternatively, the hypothesis would have been supported had the analysis yielded multiple equally parsimonious cladograms whose strict consensus comprised only clades that were compatible with the molecular cladogram.
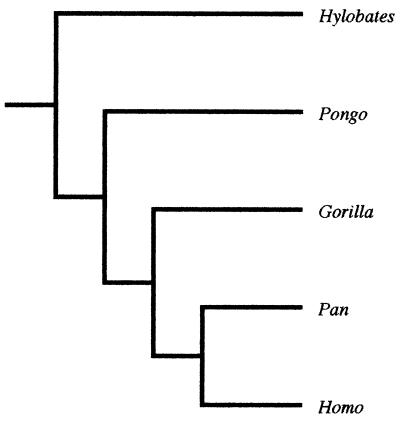
Figure 1.
Hominoid molecular relationships.
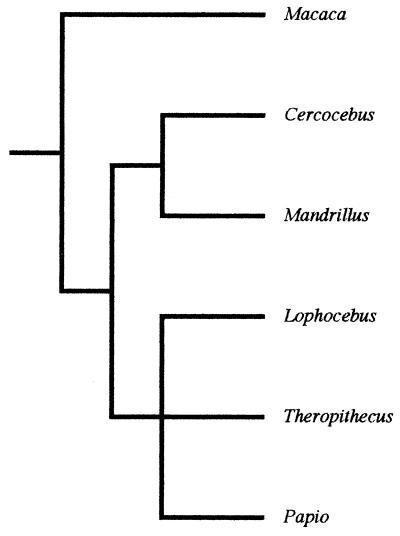
Figure 2.
Papionin molecular relationships.
The second test of the hypothesis was based on the phylogenetic bootstrap, which is a method of assessing the confidence interval associated with a given clade (44, 45). Using paup 3.0s, 10,000 matrices were derived from each matrix by sampling with replacement. The new matrices were subjected to parsimony analysis, and a consensus cladogram of the most parsimonious cladograms was computed using a confidence region of 70% (46). Thereafter, the clades of the consensus cladogram were compared with the appropriate molecular cladogram. In this test, it was reasoned that, for the hypothesis to be supported, the best supported clades should not be false clades, since it is commonly assumed in hominin analyses that the better the bootstrap support for a clade, the more likely the clade is to be “true” (15, 16).
In both the parsimony and bootstrap analyses, the quantitative characters were treated as linearly ordered and freely reversing variables (47). Twenty-seven of the qualitative characters were also treated as linearly ordered, freely reversing variables, because their states clearly formed linear transformation series. The other 69 qualitative characters were treated as unordered variables. Cladograms were obtained using paup's branch-and-bound search routine.
Results
The hypothesis was not supported by the parsimony analyses of the two quantitative data sets. The hominoid analysis yielded a most parsimonious cladogram whose branching pattern differed from that of the hominoid consensus molecular cladogram. It suggested that Homo is the sister taxon of a (Gorilla, Pan, Pongo) clade, and that Pan is the sister taxon of a (Gorilla, Pongo) clade. The papionin analysis also favored a single most parsimonious cladogram that was incompatible with the group's molecular cladogram. It suggested that the first branching event in papionin evolution separated the Lophocebus lineage from the common ancestor of Cercocebus, Macaca, and the baboons, while the second branching event separated the Cercocebus lineage from the common ancestor of Macaca and the baboons. The third branching event, according to this cladogram, separated the macaque lineage from the common ancestor of the baboons, while the final phylogenetically informative branching event separated the Theropithecus lineage from the common ancestor of Papio and Mandrillus. The hypothesis was also not supported by the parsimony analysis of the hominoid qualitative matrix. The cladogram agreed with the hominoid molecular cladogram in locating Hylobates as the basal hominoid. However, it differed from the molecular cladogram in grouping Homo and Pongo in one clade and Pan and Gorilla in a second, the latter being supported by four more putative synapomorphies than the Pan-Homo clade of the molecular cladogram.
The bootstrap analyses similarly failed to uphold the hypothesis. None of the clades supported by 70% or more of the bootstrap samples were compatible with the consensus molecular cladograms. The hominoid quantitative analysis supported a (Gorilla, Pan, Pongo) clade at 95%, and a (Gorilla, Pongo) clade at 73%. The papionin quantitative analysis supported a (Cercocebus, Macaca, baboon) clade at 98%, a (Macaca, baboon) clade at 78%, a baboon clade at 97%, and a (Mandrillus, Papio) clade at 73%. The analysis of the hominoid qualitative data yielded one clade, which incorrectly linked Gorilla and Pan to the exclusion of the other taxa (92%).
Discussion
The results of the parsimony and bootstrap tests indicate that cladistic analyses based on standard craniodental characters cannot be relied on to reconstruct the phylogenetic relationships of the hominoids, papionins, and, by extension, the fossil hominins. More problematically, the tests suggest that such analyses can strongly support phylogenetic hypotheses that are misleading. For example, the bootstrap-based tests indicate that craniodental data can return impressive levels of statistical support (e.g., 97%) for patterns of phylogenetic relationship that are most likely incorrect. In other words, cladistic analyses of higher primate craniodental morphology may yield not only “false-positive” results, but false-positive results that pass, by a substantial margin, the statistical test favored by many researchers. It should be noted that the strong bootstrap support for the clades undermines the objection that, because they were designed to recover hominin phylogeny, standard paleoanthropological measurements cannot be expected to recover the phylogenetic relationships of other taxa. Even if this were true, which we dispute, the logic of cladistics is such that the measurements should be ambiguous with regard to the relationships of the extant hominoids and papionins; they most certainly should not provide strong support for “false” clades. It should also be noted that the results of the quantitative analyses cannot be dismissed as artifacts of size-related shape change, since the most parsimonious cladograms do not group the taxa in the manner that would be expected if skull size is the principal signal in the transformed data.
The results of our analyses are in line with Hartman's (24) finding that hominoid dental characters are not reliable for phylogenetic reconstruction. More importantly, they indicate that the problem is not confined to the dentition of the hominoids. It afflicts other groups of higher primates, and extends to quantitative and qualitative cranial characters. Our results suggest that paleoanthropologists should not rely on phylogenetic hypotheses for fossil hominins that are based solely on craniodental evidence. Most likely, these hypotheses reflect a mixture of the true phylogeny and the phylogenetically misleading effects of convergence, parallelism, reversal, and/or behaviorally induced morphogenesis. If anything, it is likely that extrapolating from the results of the present study to the results of hominin cladistic studies overestimates the reliability of the latter, since we did not account for two factors that routinely complicate analyses of the hominin fossil record: namely, uncertain species identification and intraspecific morphological change through time. We stress that these results do not indicate that the cladistic methodology is flawed, or that primate craniodental data are problematic for phylogenetic reconstruction at all taxonomic levels. Rather, our results show that the type of craniodental characters that have hitherto been used in hominin phylogenetics are probably not reliable for reconstructing the phylogenetic relationships of higher primate species and genera, including those among the hominins. Although this study addressed the reliability of hominin phylogenetic hypotheses, it is noteworthy that our results are in line with recent observations regarding the use of osteological data to reconstruct the phylogenetic relationships of fossil catarrhines (48), Miocene hominoids (49), and fossil and extant colobines (50). There is, we suggest, a pressing need to undertake comparable analyses to the one reported here for other mammalian groups whose fossil members are also known principally from craniodental evidence.
How can the reliability of human phylogenetic hypotheses be improved? One strategy is to devise techniques for analyzing hominin craniodental morphology that are more sensitive to any phylogenetic signal than the methods presently in use (51, 52). Another approach is to develop rigorous comparative methods for discriminating between phylogenetically informative and phylogenetically misleading craniodental similarities. For example, the pursuit of detailed information about the ontogeny of characters may help distinguish homologous and homoplastic resemblances (53–56). Likewise, functional analyses may enable researchers to predict where similarities resulting from behavior-induced morphogenesis are likely to occur in the hominin cranium (13, 19–21). A third strategy is to develop techniques for assigning postcranial specimens to taxa in the absence of associated skeletons, thereby overcoming the taphonomy-imposed focus on craniodental morphology and enabling hominin cladistic analyses to be based on a wider sample of the phenotype (57). Significantly, recent studies suggest that data sets based on cranial, postcranial, and soft-tissue characters can yield relationships that are compatible with low-level primate molecular phylogenies (37, 58–60). Last, we suggest that more attention should be paid to nonmorphological lines of evidence that may have a bearing on hominin phylogenetic relationships, such as the temporal distribution of the species (61).
Supplementary Material
Acknowledgments
We thank the following institutions for access to specimens in their care: the Natural History Museum (London), the Anthropologisches Institut und Museum (Universität Zürich-Irchel, Zurich), the Muséum d'Histoire Naturelle (Geneva), the Muséum d'Histoire Naturelle (Paris), and the Museum für Naturkunde (Humboldt-Universitat zu Berlin, Berlin). We also thank A. T. Chamberlain for allowing us to use his data. This study benefited from the advice of L. C. Aiello, A. Bilsborough, A. T. Chamberlain, M. C. Dean, D. E. Lieberman, P. O'Higgins, C. R. C. Paul, D. R. Pilbeam, M. Ruta, D. S. Strait, and several anonymous reviewers. M.C. was funded by a Natural Environment Research Council studentship and is now funded by a Wellcome Trust Bioarchaeology Fellowship. B.W. was funded by The Leverhulme Trust and is now funded by The Henry Luce Foundation.
References
- 1.Johanson D C, White T D, Coppens Y. Kirtlandia. 1978;28:1–14. [Google Scholar]
- 2.Walker A C, Leakey R E F, Harris J M, Brown F H. Nature (London) 1986;322:517–522. [Google Scholar]
- 3.White T D, Suwa G, Asfaw B. Nature (London) 1994;371:306–312. doi: 10.1038/371306a0. [DOI] [PubMed] [Google Scholar]
- 4.White T D, Suwa G, Asfaw B. Nature (London) 1995;375:88. doi: 10.1038/375088a0. [DOI] [PubMed] [Google Scholar]
- 5.Leakey M G, Feibel C S, McDougall I, Walker A C. Nature (London) 1995;376:565–571. doi: 10.1038/376565a0. [DOI] [PubMed] [Google Scholar]
- 6.Brunet M, Beauvilian A, Coppens Y, Heintz E, Moutaye A H E, Pilbeam D R. C R Acad Sci Paris. 1996;322:907–913. [Google Scholar]
- 7.Bermudez de Castro J M, Arsuaga J L, Carbonell E, Rosas A, Martinez I, Mosquera M. Science. 1997;276:1392–1395. doi: 10.1126/science.276.5317.1392. [DOI] [PubMed] [Google Scholar]
- 8.Asfaw B, White T D, Lovejoy O, Latimer B, Simpson S, Suwa G. Science. 1999;284:629–635. doi: 10.1126/science.284.5414.629. [DOI] [PubMed] [Google Scholar]
- 9.Stringer C B. J Hum Evol. 1987;16:135–146. [Google Scholar]
- 10.Chamberlain A T, Wood B A. J Hum Evol. 1987;16:119–133. [Google Scholar]
- 11.Wood B A. Koobi Fora Research Project, Volume 4: Hominid Cranial Remains. Oxford: Oxford Univ. Press; 1991. [Google Scholar]
- 12.Skelton R R, McHenry H M. J Hum Evol. 1992;23:309–349. [Google Scholar]
- 13.Lieberman D E, Pilbeam D R, Wood B A. J Hum Evol. 1996;17:503–511. [Google Scholar]
- 14.Strait D S, Grine F E, Moniz M A. J Hum Evol. 1997;32:17–82. doi: 10.1006/jhev.1996.0097. [DOI] [PubMed] [Google Scholar]
- 15.Corruccini R S. In: Integrative Approaches to the Past: Paleoanthropological Advances in Honor of F. Clark Howell. Corruccini R S, Ciochon R L, editors. Englewood Cliffs, NJ: Prentice–Hall; 1994. pp. 167–183. [Google Scholar]
- 16.Wood B A, Collard M. Science. 1999;284:65–71. doi: 10.1126/science.284.5411.65. [DOI] [PubMed] [Google Scholar]
- 17.Trinkaus E. Am J Phys Anthropol. 1990;83:1–11. doi: 10.1002/ajpa.1330830102. [DOI] [PubMed] [Google Scholar]
- 18.Trinkaus E. In: Continuity or Replacement: Controversies in Homo sapiens Evolution. Bräuer G, Smith F H, editors. Rotterdam, the Netherlands: Balkema; 1992. pp. 1–7. [Google Scholar]
- 19.Lieberman D E. Curr Anthropol. 1995;36:159–197. [Google Scholar]
- 20.Lieberman D E. Annu Rev Anthropol. 1997;26:185–210. [Google Scholar]
- 21.Lieberman D E. Evol Anthropol. 1999;7:142–151. [Google Scholar]
- 22.Ruvolo M. Mol Biol Evol. 1997;14:248–265. doi: 10.1093/oxfordjournals.molbev.a025761. [DOI] [PubMed] [Google Scholar]
- 23.Harris E E, Disotell T R. Mol Biol Evol. 1998;15:892–900. doi: 10.1093/oxfordjournals.molbev.a025993. [DOI] [PubMed] [Google Scholar]
- 24.Hartman S E. J Hum Evol. 1988;17:489–502. [Google Scholar]
- 25.Fitch W M, Atchley W R. In: Molecules and Morphology in Evolution: Conflict or Compromise? Patterson C, editor. Cambridge, U.K.: Cambridge Univ. Press; 1987. pp. pp.203–216. [Google Scholar]
- 26.Atchley W R, Fitch W M. Science. 1991;254:554–558. doi: 10.1126/science.1948030. [DOI] [PubMed] [Google Scholar]
- 27.Hillis D M, Bull J J, White M E, Badgett M R, Molineux I J. Science. 1992;255:589–592. doi: 10.1126/science.1736360. [DOI] [PubMed] [Google Scholar]
- 28.Rae T C. Cladistics. 1998;14:221–228. doi: 10.1111/j.1096-0031.1998.tb00335.x. [DOI] [PubMed] [Google Scholar]
- 29.Ahern J H. Am J Phys Anthropol. 1998;105:461–480. doi: 10.1002/(SICI)1096-8644(199804)105:4<461::AID-AJPA5>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 30.Wood B A, Li Y, Willoughby C. J Anat. 1991;174:185–205. [PMC free article] [PubMed] [Google Scholar]
- 31.Chamberlain A T. Ph.D. thesis. Liverpool, U.K.: Univ. of Liverpool; 1987. [Google Scholar]
- 32.Collard M. Ph.D. thesis. Liverpool, U.K.: Univ. of Liverpool; 1998. [Google Scholar]
- 33.Jungers W L, Falsetti A B, Wall C E. Yearbook Phys Anthropol. 1995;38:137–161. [Google Scholar]
- 34.Thorpe R S. Evolution (Lawrence, Kans) 1984;38:244–255. doi: 10.1111/j.1558-5646.1984.tb00282.x. [DOI] [PubMed] [Google Scholar]
- 35.Creel N. Syst Zool. 1986;35:81–99. [Google Scholar]
- 36.Strait D S, Moniz M, Strait P. Syst Biol. 1996;45:67–78. [Google Scholar]
- 37.Shoshani J, Groves C P, Simons E L, Gunnell G F. Mol Phyl Evol. 1996;5:102–154. doi: 10.1006/mpev.1996.0009. [DOI] [PubMed] [Google Scholar]
- 38.Braga J. Folia Primatol. 1995;65:144–153. doi: 10.1159/000156880. [DOI] [PubMed] [Google Scholar]
- 39.Andrews P. In: Molecules and Morphology in Evolution: Conflict or Compromise? Patterson C, editor. Cambridge, U.K.: Cambridge Univ. Press; 1987. pp. 23–53. [Google Scholar]
- 40.Schwartz J H. Curr Anthropol. 1984;25:655–672. [Google Scholar]
- 41.Delson E, Andrews P. In: Phylogeny of the Primates: A Multidisciplinary Approach. Luckett W P, Szalay F S, editors. New York: Plenum; 1975. pp. 405–446. [Google Scholar]
- 42.Groves C P. In: Comparative Primate Biology, Volume 1: Systematics, Evolution and Anatomy. Swindler D R, Erwin J, editors. New York: Liss; 1986. pp. 187–217. [Google Scholar]
- 43.Swofford D L. Phylogenetic Analysis Using Parsimony Version 3.0s. Champaign, IL: Illinois Natural History Survey; 1991. [Google Scholar]
- 44.Felsenstein J. Evolution (Lawrence, Kans) 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 45.Sanderson M J. Syst Biol. 1995;44:299–320. [Google Scholar]
- 46.Hillis D M, Bull J J. Syst Biol. 1993;42:182–192. [Google Scholar]
- 47.Slowinski J. Syst Biol. 1993;42:155–165. [Google Scholar]
- 48.Harrison T. In: Species, Species Concepts and Primate Evolution. Kimbel W H, Martin L B, editors. New York: Plenum; 1993. pp. 345–371. [Google Scholar]
- 49.Pilbeam D R. Mol Phylogenet Evol. 1996;5:155–168. doi: 10.1006/mpev.1996.0010. [DOI] [PubMed] [Google Scholar]
- 50.Jablonski N. Curr Biol. 1999;9:122. doi: 10.1016/s0960-9822(99)80077-3. [DOI] [PubMed] [Google Scholar]
- 51.Cheetham A H. Paleobiology. 1987;13:286–296. [Google Scholar]
- 52.Budd A F, Coates A G. Paleobiology. 1992;18:425–446. [Google Scholar]
- 53.Wood B A. In: Evolutionary History of the “Robust” Australopithecines. Grine F E, editor. New York: Aldine de Gruyter; 1988. pp. 269–284. [Google Scholar]
- 54.Bromage T G. J Hum Evol. 1989;18:751–773. [Google Scholar]
- 55.McCollum M. Science. 1999;284:301–305. doi: 10.1126/science.284.5412.301. [DOI] [PubMed] [Google Scholar]
- 56.Lovejoy C O, Cohn M J, White T D. Proc Natl Acad Sci USA. 1999;96:13247–13252. doi: 10.1073/pnas.96.23.13247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Aiello L C, Wood B A. Am J Phys Anthropol. 1994;95:409–426. doi: 10.1002/ajpa.1330950405. [DOI] [PubMed] [Google Scholar]
- 58.Begun D R, Ward C V, Rose M D. In: Function, Phylogeny, and Fossils: Miocene Hominoid Evolution and Adaptations. Begun D R, Ward C V, Rose M D, editors. New York: Plenum; 1997. pp. 389–415. [Google Scholar]
- 59.Groves C P. In: Old World Monkeys. Whitehead P F, Jolly C J, editors. Cambridge, U.K.: Cambridge Univ. Press; 2000. , in press. [Google Scholar]
- 60.Gibbs S. Ph.D. thesis. Liverpool, U.K.: Univ. of Liverpool; 1999. [Google Scholar]
- 61.Paul C R C. In: Problems of Phylogenetic Reconstruction. Joysey K A, Friday A E, editors. London: Academic; 1982. pp. 75–117. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.