Abstract
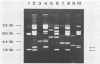
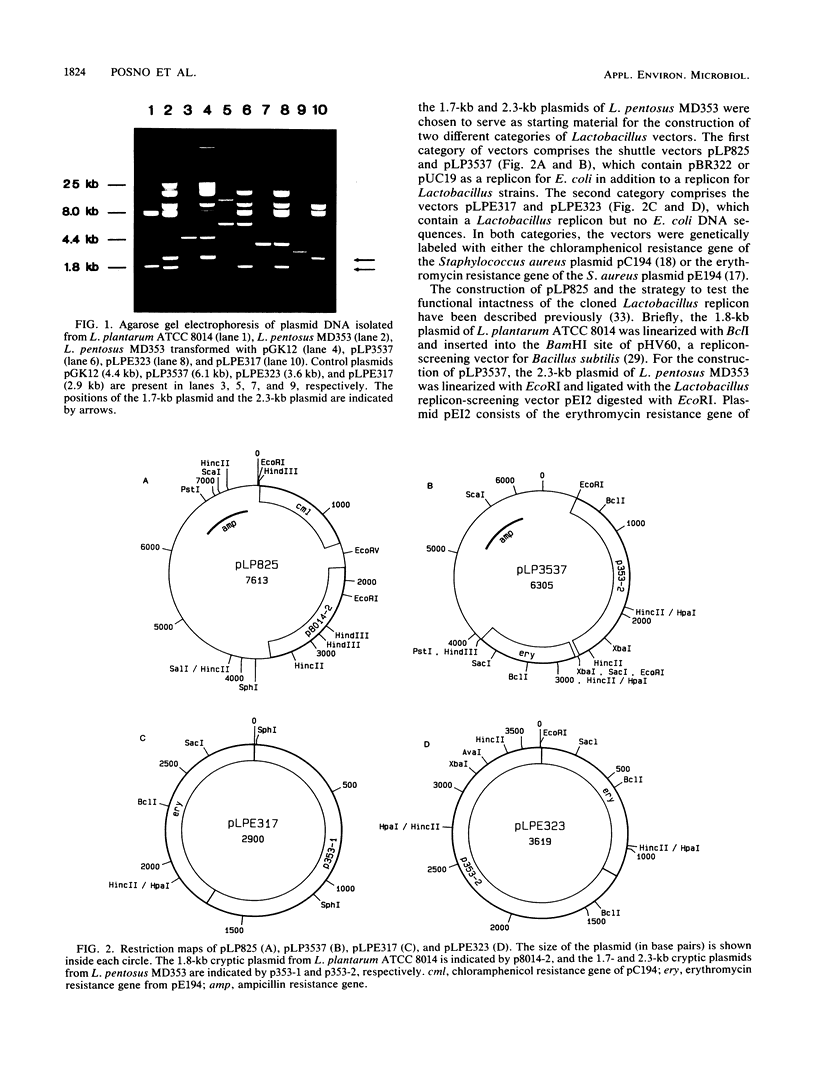
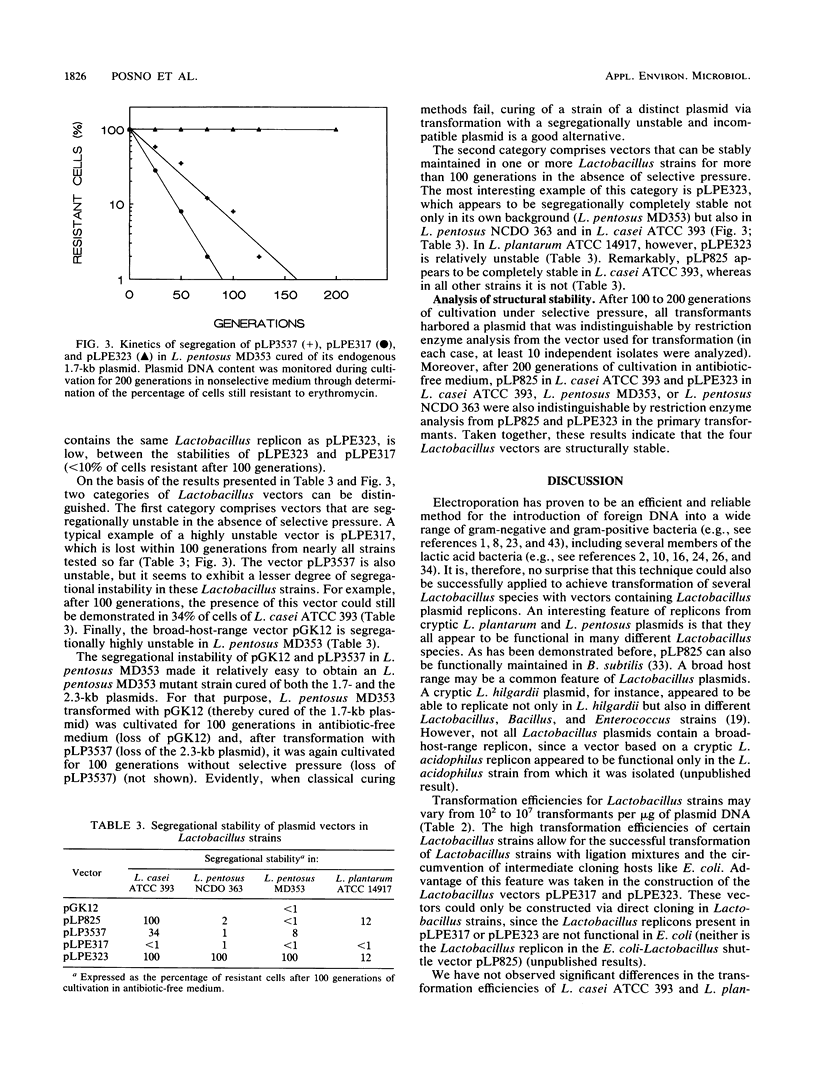
Three new Lactobacillus vectors based on cryptic Lactobacillus plasmids were constructed. The shuttle vector pLP3537 consists of a 2.3-kb plasmid from Lactobacillus pentosus MD353, an erythromycin resistance gene from Staphylococcus aureus plasmid pE194, and pUC19 as a replicon for Escherichia coli. The vectors pLPE317 and pLPE323, which do not contain E. coli sequences, were generated by introducing the erythromycin resistance gene of pE194 into a 1.7- and a 2.3-kb plasmid from L. pentosus MD353, respectively. These vectors and the shuttle vector pLP825 (M. Posno, R. J. Leer, J. M. M. van Rijn, B. C. Lokman, and P. H. Pouwels, p. 397-401, in A. T. Ganesan and J. A. Hoch, ed., Genetics and biotechnology of bacilli, vol. 2, 1988) could be introduced by electroporation into Lactobacillus casei, L. pentosus, L. plantarum, L. acidophilus, L. fermentum, and L. brevis strains with similar efficiencies. Transformation efficiencies were strain dependent and varied from 102 to 107 transformants per μg of DNA. Plasmid DNA analysis of L. pentosus MD353 transformants revealed that the introduction of pLP3537 or pLPE323 was invariably accompanied by loss of the endogenous 2.3-kb plasmid. Remarkably, pLPE317 could only be introduced into an L. pentosus MD353 strain that had been previously cured of its endogenous 1.7-kb plasmid. The curing phenomena are most likely to be explained by the incompatibility of the vectors and resident plasmids. Lactobacillus vectors are generally rapidly lost when cells are cultivated in the absence of selective pressure. However, pLPE323 is stable in three of four Lactobacillus strains tested so far.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Augustin J., Götz F. Transformation of Staphylococcus epidermidis and other staphylococcal species with plasmid DNA by electroporation. FEMS Microbiol Lett. 1990 Jan 1;54(1-3):203–207. doi: 10.1016/0378-1097(90)90283-v. [DOI] [PubMed] [Google Scholar]
- Bates E. E., Gilbert H. J. Characterization of a cryptic plasmid from Lactobacillus plantarum. Gene. 1989 Dec 21;85(1):253–258. doi: 10.1016/0378-1119(89)90491-5. [DOI] [PubMed] [Google Scholar]
- Bouia A., Bringel F., Frey L., Kammerer B., Belarbi A., Guyonvarch A., Hubert J. C. Structural organization of pLP1, a cryptic plasmid from Lactobacillus plantarum CCM 1904. Plasmid. 1989 Nov;22(3):185–192. doi: 10.1016/0147-619x(89)90001-2. [DOI] [PubMed] [Google Scholar]
- Bringel F., Frey L., Hubert J. C. Characterization, cloning, curing, and distribution in lactic acid bacteria of pLP1, a plasmid from Lactobacillus plantarum CCM 1904 and its use in shuttle vector construction. Plasmid. 1989 Nov;22(3):193–202. doi: 10.1016/0147-619x(89)90002-4. [DOI] [PubMed] [Google Scholar]
- Bron S., Luxen E. Segregational instability of pUB110-derived recombinant plasmids in Bacillus subtilis. Plasmid. 1985 Nov;14(3):235–244. doi: 10.1016/0147-619x(85)90007-1. [DOI] [PubMed] [Google Scholar]
- Bron S., Luxen E., Swart P. Instability of recombinant pUB110 plasmids in Bacillus subtilis: plasmid-encoded stability function and effects of DNA inserts. Plasmid. 1988 May;19(3):231–241. doi: 10.1016/0147-619x(88)90041-8. [DOI] [PubMed] [Google Scholar]
- Calvin N. M., Hanawalt P. C. High-efficiency transformation of bacterial cells by electroporation. J Bacteriol. 1988 Jun;170(6):2796–2801. doi: 10.1128/jb.170.6.2796-2801.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currier T. C., Nester E. W. Isolation of covalently closed circular DNA of high molecular weight from bacteria. Anal Biochem. 1976 Dec;76(2):431–441. doi: 10.1016/0003-2697(76)90338-9. [DOI] [PubMed] [Google Scholar]
- Gros M. F., te Riele H., Ehrlich S. D. Rolling circle replication of single-stranded DNA plasmid pC194. EMBO J. 1987 Dec 1;6(12):3863–3869. doi: 10.1002/j.1460-2075.1987.tb02724.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruss A., Ehrlich S. D. The family of highly interrelated single-stranded deoxyribonucleic acid plasmids. Microbiol Rev. 1989 Jun;53(2):231–241. doi: 10.1128/mr.53.2.231-241.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashiba H., Takiguchi R., Ishii S., Aoyama K. Transformation of Lactobacillus helveticus subsp. jugurti with plasmid pLHR by electroporation. Agric Biol Chem. 1990 Jun;54(6):1537–1541. [PubMed] [Google Scholar]
- Horinouchi S., Weisblum B. Nucleotide sequence and functional map of pC194, a plasmid that specifies inducible chloramphenicol resistance. J Bacteriol. 1982 May;150(2):815–825. doi: 10.1128/jb.150.2.815-825.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horinouchi S., Weisblum B. Nucleotide sequence and functional map of pE194, a plasmid that specifies inducible resistance to macrolide, lincosamide, and streptogramin type B antibodies. J Bacteriol. 1982 May;150(2):804–814. doi: 10.1128/jb.150.2.804-814.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josson K., Scheirlinck T., Michiels F., Platteeuw C., Stanssens P., Joos H., Dhaese P., Zabeau M., Mahillon J. Characterization of a gram-positive broad-host-range plasmid isolated from Lactobacillus hilgardii. Plasmid. 1989 Jan;21(1):9–20. doi: 10.1016/0147-619x(89)90082-6. [DOI] [PubMed] [Google Scholar]
- Kok J., van der Vossen J. M., Venema G. Construction of plasmid cloning vectors for lactic streptococci which also replicate in Bacillus subtilis and Escherichia coli. Appl Environ Microbiol. 1984 Oct;48(4):726–731. doi: 10.1128/aem.48.4.726-731.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebl W., Bayerl A., Schein B., Stillner U., Schleifer K. H. High efficiency electroporation of intact Corynebacterium glutamicum cells. FEMS Microbiol Lett. 1989 Dec;53(3):299–303. doi: 10.1016/0378-1097(89)90234-6. [DOI] [PubMed] [Google Scholar]
- Luchansky J. B., Muriana P. M., Klaenhammer T. R. Application of electroporation for transfer of plasmid DNA to Lactobacillus, Lactococcus, Leuconostoc, Listeria, Pediococcus, Bacillus, Staphylococcus, Enterococcus and Propionibacterium. Mol Microbiol. 1988 Sep;2(5):637–646. doi: 10.1111/j.1365-2958.1988.tb00072.x. [DOI] [PubMed] [Google Scholar]
- Matsushima P., Cox K. L., Baltz R. H. Highly transformable mutants of Streptomyces fradiae defective in several restriction systems. Mol Gen Genet. 1987 Mar;206(3):393–400. doi: 10.1007/BF00428877. [DOI] [PubMed] [Google Scholar]
- McIntyre D. A., Harlander S. K. Improved electroporation efficiency of intact Lactococcus lactis subsp. lactis cells grown in defined media. Appl Environ Microbiol. 1989 Oct;55(10):2621–2626. doi: 10.1128/aem.55.10.2621-2626.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay L. L., Baldwin K. A. Applications for biotechnology: present and future improvements in lactic acid bacteria. FEMS Microbiol Rev. 1990 Sep;7(1-2):3–14. doi: 10.1111/j.1574-6968.1990.tb04876.x. [DOI] [PubMed] [Google Scholar]
- Mercenier A., Chassy B. M. Strategies for the development of bacterial transformation systems. Biochimie. 1988 Apr;70(4):503–517. doi: 10.1016/0300-9084(88)90086-7. [DOI] [PubMed] [Google Scholar]
- Michel B., Niaudet B., Ehrlich S. D. Intermolecular recombination during transformation of Bacillus subtilis competent cells by monomeric and dimeric plasmids. Plasmid. 1983 Jul;10(1):1–10. doi: 10.1016/0147-619x(83)90052-5. [DOI] [PubMed] [Google Scholar]
- Muriana P. M., Klaenhammer T. R. Conjugal Transfer of Plasmid-Encoded Determinants for Bacteriocin Production and Immunity in Lactobacillus acidophilus 88. Appl Environ Microbiol. 1987 Mar;53(3):553–560. doi: 10.1128/aem.53.3.553-560.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick R. P. Plasmid incompatibility. Microbiol Rev. 1987 Dec;51(4):381–395. doi: 10.1128/mr.51.4.381-395.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell Ian B., Achen Marc G., Hillier Alan J., Davidson Barrie E. A Simple and Rapid Method for Genetic Transformation of Lactic Streptococci by Electroporation. Appl Environ Microbiol. 1988 Mar;54(3):655–660. doi: 10.1128/aem.54.3.655-660.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaugen M. The complete nucleotide sequence of a small cryptic plasmid from Lactobacillus plantarum. Plasmid. 1989 Sep;22(2):175–179. doi: 10.1016/0147-619x(89)90028-0. [DOI] [PubMed] [Google Scholar]
- Vescovo M., Morelli L., Bottazzi V. Drug resistance plasmids in Lactobacillus acidophilus and Lactobacillus reuteri. Appl Environ Microbiol. 1982 Jan;43(1):50–56. doi: 10.1128/aem.43.1.50-56.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth R., Friesenegger A., Fiedler S. Transformation of various species of gram-negative bacteria belonging to 11 different genera by electroporation. Mol Gen Genet. 1989 Mar;216(1):175–177. doi: 10.1007/BF00332248. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- te Riele H., Michel B., Ehrlich S. D. Are single-stranded circles intermediates in plasmid DNA replication? EMBO J. 1986 Mar;5(3):631–637. doi: 10.1002/j.1460-2075.1986.tb04257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- te Riele H., Michel B., Ehrlich S. D. Single-stranded plasmid DNA in Bacillus subtilis and Staphylococcus aureus. Proc Natl Acad Sci U S A. 1986 Apr;83(8):2541–2545. doi: 10.1073/pnas.83.8.2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Lelie D., van der Vossen J. M., Venema G. Effect of Plasmid Incompatibility on DNA Transfer to Streptococcus cremoris. Appl Environ Microbiol. 1988 Apr;54(4):865–871. doi: 10.1128/aem.54.4.865-871.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]