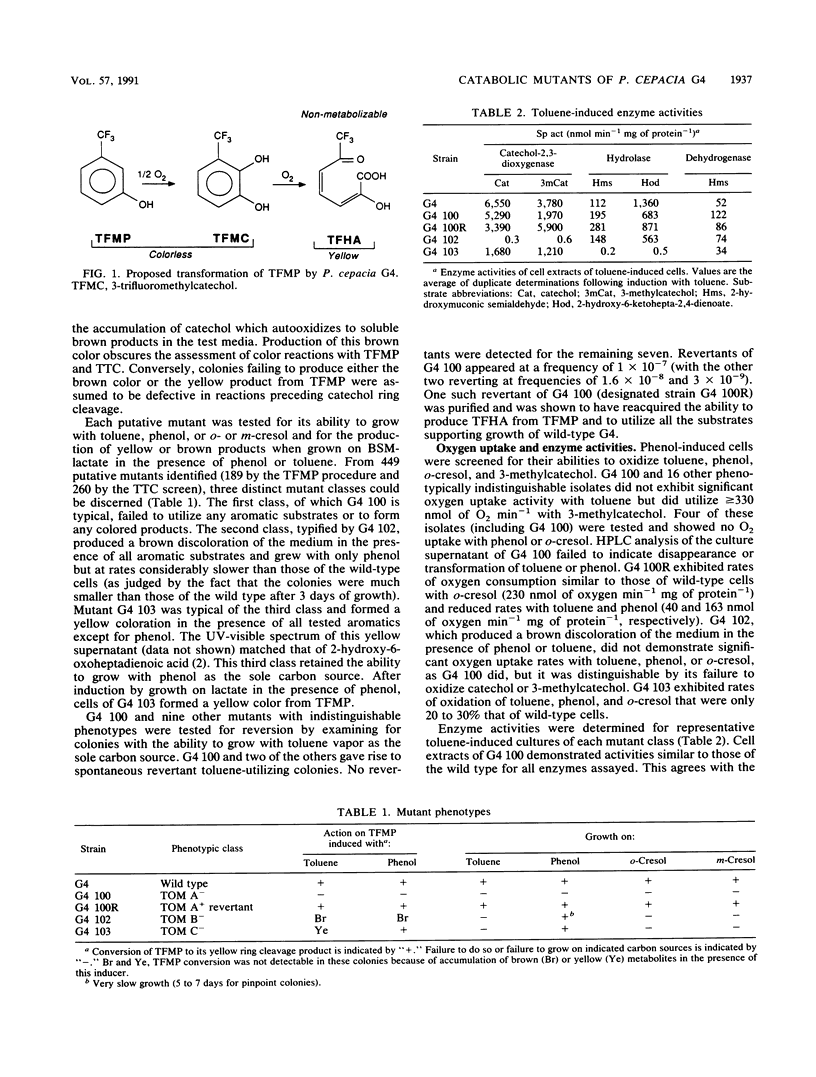
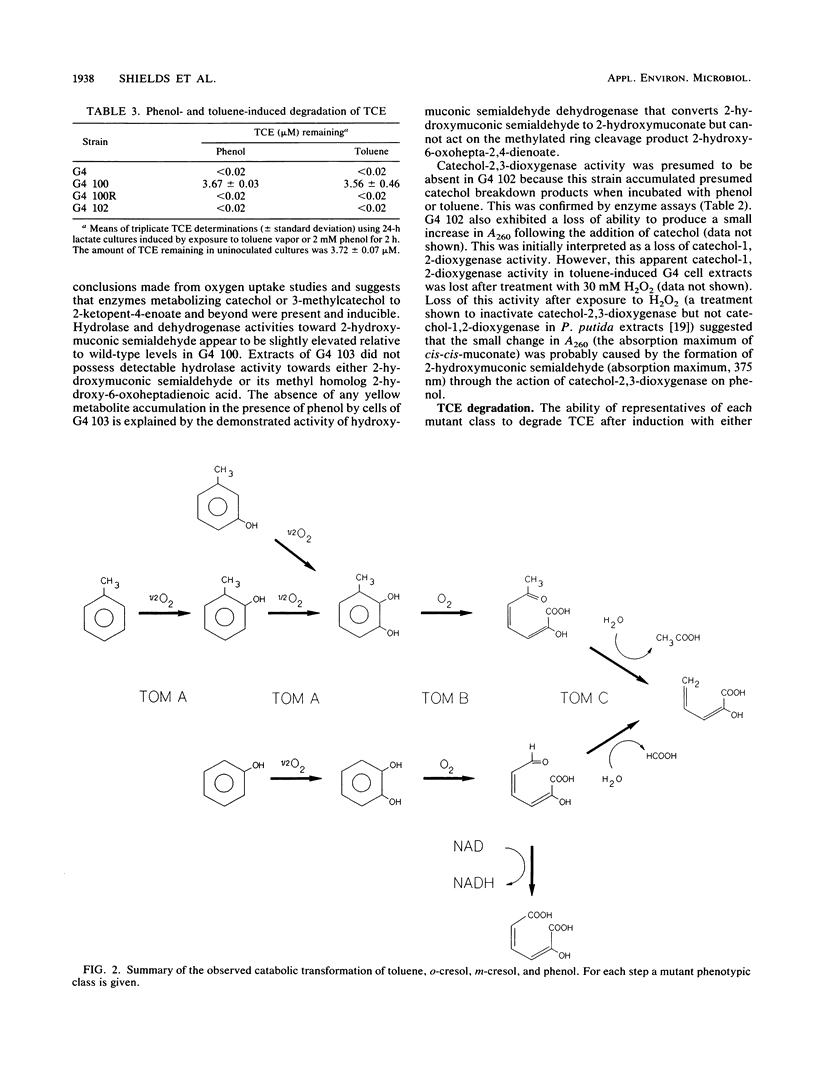
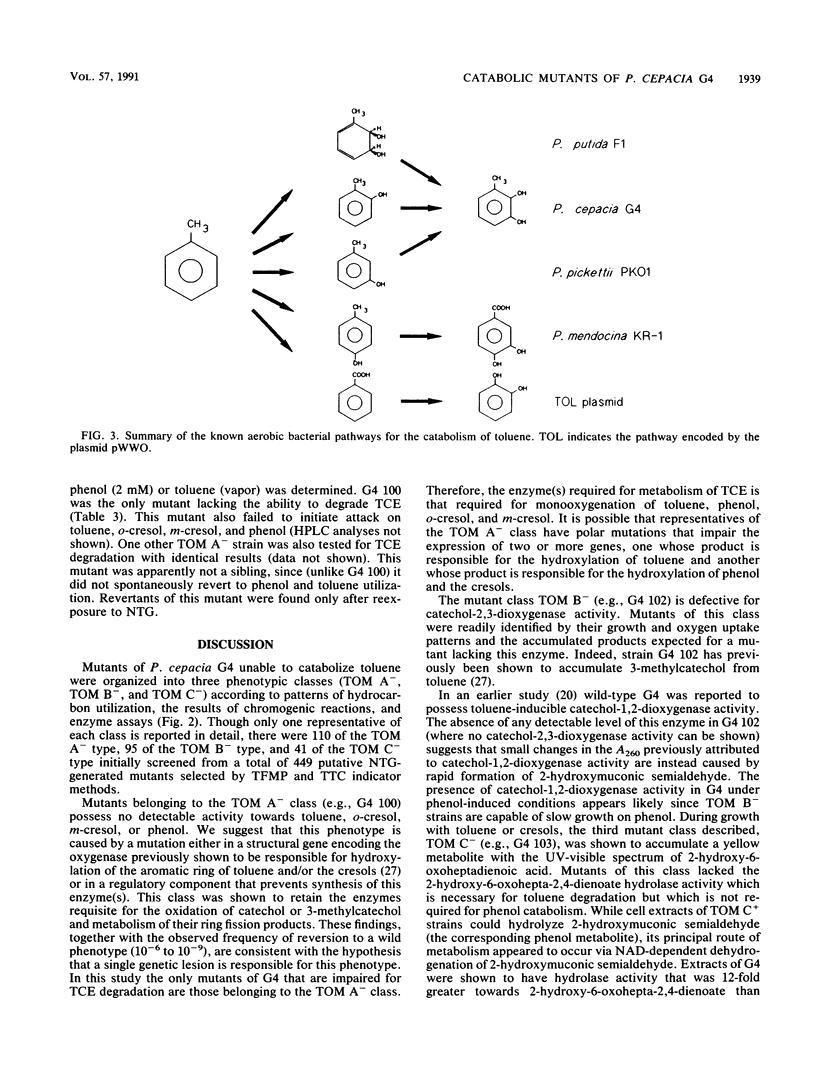
Abstract
Pseudomonas cepacia G4 possesses a novel pathway of toluene catabolism that is shown to be responsible for the degradation of trichloroethylene (TCE). This pathway involves conversion of toluene via o-cresol to 3-methylcatechol. In order to determine the enzyme of toluene degradation that is responsible for TCE degradation, chemically induced mutants, blocked in the toluene ortho-monooxygenase (TOM) pathway of G4, were examined. Mutants of the phenotypic class designated TOM A- were all defective in their ability to oxidize toluene, o-cresol, m-cresol, and phenol, suggesting that a single enzyme is responsible for conversion of these compounds to their hydroxylated products (3-methylcatechol from toluene, o-cresol, and m-cresol and catechol from phenol) in the wild type. Mutants of this class did not degrade TCE. Two other mutant classes which were blocked in toluene catabolism, TOM B-, which lacked catechol-2,3-dioxygenase, and TOM C-, which lacked 2-hydroxy-6-oxoheptadienoic acid hydrolase activity, were fully capable of TCE degradation. Therefore, TCE degradation is directly associated with the monooxygenation capability responsible for toluene, cresol, and phenol hydroxylation.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arciero D., Vannelli T., Logan M., Hooper A. B. Degradation of trichloroethylene by the ammonia-oxidizing bacterium Nitrosomonas europaea. Biochem Biophys Res Commun. 1989 Mar 15;159(2):640–643. doi: 10.1016/0006-291x(89)90042-9. [DOI] [PubMed] [Google Scholar]
- Bayly R. C., Dagley S., Gibson D. T. The metabolism of cresols by species of Pseudomonas. Biochem J. 1966 Nov;101(2):293–301. doi: 10.1042/bj1010293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayly R. C., Wigmore G. J. Metabolism of phenol and cresols by mutants of Pseudomonas putida. J Bacteriol. 1973 Mar;113(3):1112–1120. doi: 10.1128/jb.113.3.1112-1120.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochner B. R., Savageau M. A. Generalized indicator plate for genetic, metabolic, and taxonomic studies with microorganisms. Appl Environ Microbiol. 1977 Feb;33(2):434–444. doi: 10.1128/aem.33.2.434-444.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- DAGLEY S., GIBSON D. T. THE BACTERIAL DEGRADATION OF CATECHOL. Biochem J. 1965 May;95:466–474. doi: 10.1042/bj0950466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day D. L., Yasinow D., McDonough J., Shuster C. W. Use of ureidopenicillins for selection of plasmid vector transformants in Pseudomonas aeruginosa and Pseudomonas putida. J Bacteriol. 1984 Mar;157(3):937–939. doi: 10.1128/jb.157.3.937-939.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engesser K. H., Cain R. B., Knackmuss H. J. Bacterial metabolism of side chain fluorinated aromatics: cometabolism of 3-trifluoromethyl(TFM)-benzoate by Pseudomonas putida (arvilla) mt-2 and Rhodococcus rubropertinctus N657. Arch Microbiol. 1988 Jan;149(3):188–197. doi: 10.1007/BF00422004. [DOI] [PubMed] [Google Scholar]
- Ewers J., Freier-Schröder D., Knackmuss H. J. Selection of trichloroethene (TCE) degrading bacteria that resist inactivation by TCE. Arch Microbiol. 1990;154(4):410–413. doi: 10.1007/BF00276540. [DOI] [PubMed] [Google Scholar]
- Folsom B. R., Chapman P. J., Pritchard P. H. Phenol and trichloroethylene degradation by Pseudomonas cepacia G4: kinetics and interactions between substrates. Appl Environ Microbiol. 1990 May;56(5):1279–1285. doi: 10.1128/aem.56.5.1279-1285.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox B. G., Borneman J. G., Wackett L. P., Lipscomb J. D. Haloalkene oxidation by the soluble methane monooxygenase from Methylosinus trichosporium OB3b: mechanistic and environmental implications. Biochemistry. 1990 Jul 10;29(27):6419–6427. doi: 10.1021/bi00479a013. [DOI] [PubMed] [Google Scholar]
- HUGHES D. E. A press for disrupting bacteria and other micro-organisms. Br J Exp Pathol. 1951 Apr;32(2):97–109. [PMC free article] [PubMed] [Google Scholar]
- Hareland W. A., Crawford R. L., Chapman P. J., Dagley S. Metabolic function and properties of 4-hydroxyphenylacetic acid 1-hydroxylase from Pseudomonas acidovorans. J Bacteriol. 1975 Jan;121(1):272–285. doi: 10.1128/jb.121.1.272-285.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukor J. J., Olsen R. H. Molecular cloning, characterization, and regulation of a Pseudomonas pickettii PKO1 gene encoding phenol hydroxylase and expression of the gene in Pseudomonas aeruginosa PAO1c. J Bacteriol. 1990 Aug;172(8):4624–4630. doi: 10.1128/jb.172.8.4624-4630.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little C. D., Palumbo A. V., Herbes S. E., Lidstrom M. E., Tyndall R. L., Gilmer P. J. Trichloroethylene biodegradation by a methane-oxidizing bacterium. Appl Environ Microbiol. 1988 Apr;54(4):951–956. doi: 10.1128/aem.54.4.951-956.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray K., Williams P. A. Role of catechol and the methylcatechols as inducers of aromatic metabolism in Pseudomonas putida. J Bacteriol. 1974 Mar;117(3):1153–1157. doi: 10.1128/jb.117.3.1153-1157.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NOZAKI M., KAGAMIYAMA H., HAYAISHI O. METAPYROCATECHASE. I. PURIFICATION, CRYSTALLIZATION AND SOME PROPERTIES. Biochem Z. 1963;338:582–590. [PubMed] [Google Scholar]
- Nelson M. J., Montgomery S. O., Mahaffey W. R., Pritchard P. H. Biodegradation of trichloroethylene and involvement of an aromatic biodegradative pathway. Appl Environ Microbiol. 1987 May;53(5):949–954. doi: 10.1128/aem.53.5.949-954.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson M. J., Montgomery S. O., O'neill E. J., Pritchard P. H. Aerobic metabolism of trichloroethylene by a bacterial isolate. Appl Environ Microbiol. 1986 Aug;52(2):383–384. doi: 10.1128/aem.52.2.383-384.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson M. J., Montgomery S. O., Pritchard P. H. Trichloroethylene metabolism by microorganisms that degrade aromatic compounds. Appl Environ Microbiol. 1988 Feb;54(2):604–606. doi: 10.1128/aem.54.2.604-606.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldenhuis R., Vink R. L., Janssen D. B., Witholt B. Degradation of chlorinated aliphatic hydrocarbons by Methylosinus trichosporium OB3b expressing soluble methane monooxygenase. Appl Environ Microbiol. 1989 Nov;55(11):2819–2826. doi: 10.1128/aem.55.11.2819-2826.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ornston L. N., Ornston M. K., Chou G. Isolation of spontaneous mutant strains of Pseudomonas putida. Biochem Biophys Res Commun. 1969 Jul 7;36(1):179–184. doi: 10.1016/0006-291x(69)90666-4. [DOI] [PubMed] [Google Scholar]
- Shields M. S., Montgomery S. O., Chapman P. J., Cuskey S. M., Pritchard P. H. Novel pathway of toluene catabolism in the trichloroethylene-degrading bacterium g4. Appl Environ Microbiol. 1989 Jun;55(6):1624–1629. doi: 10.1128/aem.55.6.1624-1629.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsien H. C., Brusseau G. A., Hanson R. S., Waclett L. P. Biodegradation of trichloroethylene by Methylosinus trichosporium OB3b. Appl Environ Microbiol. 1989 Dec;55(12):3155–3161. doi: 10.1128/aem.55.12.3155-3161.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wackett L. P., Brusseau G. A., Householder S. R., Hanson R. S. Survey of microbial oxygenases: trichloroethylene degradation by propane-oxidizing bacteria. Appl Environ Microbiol. 1989 Nov;55(11):2960–2964. doi: 10.1128/aem.55.11.2960-2964.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wackett L. P., Gibson D. T. Degradation of trichloroethylene by toluene dioxygenase in whole-cell studies with Pseudomonas putida F1. Appl Environ Microbiol. 1988 Jul;54(7):1703–1708. doi: 10.1128/aem.54.7.1703-1708.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wackett L. P., Householder S. R. Toxicity of Trichloroethylene to Pseudomonas putida F1 Is Mediated by Toluene Dioxygenase. Appl Environ Microbiol. 1989 Oct;55(10):2723–2725. doi: 10.1128/aem.55.10.2723-2725.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worsey M. J., Williams P. A. Characterization of a spontaneously occurring mutant of the TOL20 plasmid in Pseudomonas putida MT20: possible regulatory implications. J Bacteriol. 1977 Jun;130(3):1149–1158. doi: 10.1128/jb.130.3.1149-1158.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zylstra G. J., McCombie W. R., Gibson D. T., Finette B. A. Toluene degradation by Pseudomonas putida F1: genetic organization of the tod operon. Appl Environ Microbiol. 1988 Jun;54(6):1498–1503. doi: 10.1128/aem.54.6.1498-1503.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]



