Abstract
Transcriptional activation often requires the rapid assembly of complexes between dimeric transcription factors and specific DNA sites. Here we show that members of the basic region leucine zipper and basic region helix–loop–helix zipper transcription factor families follow an assembly pathway in which two protein monomers bind DNA sequentially and form their dimerization interface while bound to DNA. Nonspecific protein or DNA competitors have little effect on the rate of assembly along this pathway, but slow a competing pathway in which preformed dimers bind DNA. The sequential monomer-binding pathway allows the protein to search for and locate a specific DNA site more quickly, resulting in greater specificity prior to equilibrium.
Activation of transcription requires the formation of large, multicomponent protein–DNA complexes (1). These complexes choose their route of assembly from many potential pathways. In vivo, the pathway chosen will depend on the availability of components and the relative rate constants of individual steps along each pathway, as well as on the propensities of intermediates to engage in alternative interactions with other nuclear components (2). Some of these alternative interactions can be nonproductive or even detrimental, whereas others might allow productive retargeting of a transcription factor in response to cellular stimuli.
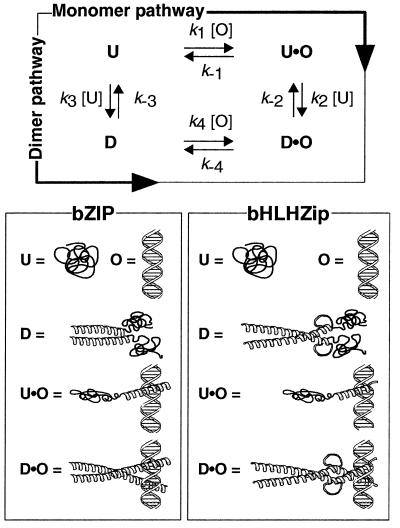
Many transcription factors bind DNA to form dimeric (2:1) protein–DNA complexes. Examples include basic region leucine zipper (bZIP) proteins and basic region helix–loop–helix zipper (bHLHZip) proteins. For these proteins, there exist two limiting pathways that may describe the route of complex assembly (Fig. 1). The protein can dimerize first, then associate with DNA (dimer pathway), or can follow a pathway in which two monomers bind DNA sequentially and assemble their dimerization interface while bound to DNA (monomer pathway) (3–10). Because these transcription factors (or their derivatives) often have been observed in dimeric form in the absence of DNA, complex assembly has been assumed to proceed via the dimer pathway. Here we show that, for members of the bZIP and bHLHZip families, the rate of DNA binding is rapid and cannot be accounted for by assembly through the dimer pathway. For these proteins, we propose that DNA binding occurs via the monomer pathway. Furthermore, we show that excess nonspecific protein or DNA competitors, even at low concentrations, slow the dimer pathway but not the monomer pathway. By minimizing apportionment of a transcription factor into incorrect, nonspecific complexes prior to equilibrium, the monomer pathway allows for rapid identification of a specific DNA site in response to cellular stimuli. Therefore, not only does the monomer pathway allow for faster assembly of certain bZIP and bHLHZip transcription factor dimer–DNA complexes, it also provides an efficient means of discriminating between specific and nonspecific DNA target sites. This method of kinetic discrimination has implications for the regulation of transcription by both cellular and viral proteins.
Figure 1.
Two pathways for binding bZIP and bHLHZip proteins to DNA.
Materials and Methods
Peptides and DNA.
Activating transcription factor 2 (ATF-2)350–505 (ref. 11) and Max22–113 (ref. 12) have been reported. Max22–105SH (ref. 12) contained residues 22–105 of Max with the amino acid sequence Gly-Gly-Cys-Asp appended to the C terminus. Max22–105SFlu was produced by alkylation of the unique cysteine in Max22–105SH with 5-iodoacetamidofluorescein (Molecular Probes) and was purified by reverse-phase HPLC. Max22–105SS was produced by oxidation of Max22–105SH and was purified by reverse-phase HPLC. DNA sequences employed were as follows: CRE24, d(AGTGGAGATGACGTCATCTCGTGC), Ebox22, d(GTGTAGGCCACGTGACCGGGTG), and hsEbox22, d(GTGTAGGCCTAGTGACCGGGTG).
CD Experiments.
Equilibrium CD spectra were acquired at 25°C on an Aviv 62DS spectrophotometer, and ellipticity at 222 nm was plotted as a function of protein concentration. Each data point represented the average of three independent trials. The data were fit to the equation (13): 2M0fm2 + fmKdim − Kdim = 0, where M0 = [protein]Total, Kdim = the equilibrium dissociation constant of the protein dimer, and fm = the fraction of the total CD signal observed [(Θobserved − Θmax)/(Θmin − Θmax)]. For both ATF-2350–505 and Max22–113, equivalent Kdim values were also determined by fluorescence polarization. ATF-2350–505 CD spectra were acquired in 1× PBS (1.4 mM KH2PO4/4.3 mM Na2HPO4/2.7 mM KCl/137 mM NaCl, pH 7.4) containing 1 mM DTT. Max22–113 spectra were acquired in 1× PBS. CD experiments performed in the presence of 1× PBS containing 5% glycerol and 1 mM EDTA demonstrated that these two components did not increase the amount of α-helical structure within Max22–113.
To monitor the Max22–113 monomer–dimer transition by CD, a solution of [Max22–113]Total = 50 μM was diluted 50-fold into PBS buffer at 25°C and the negative ellipticity at 222 nm was monitored as a function of time. Each point was averaged over 20 sec and corrected for the signal arising from buffer.
Stopped-Flow Fluorescence.
To monitor the kinetics of the ATF-2350–505 monomer–dimer transition, a solution of ATF-2350–505 in 1× PBS containing 1 mM DTT was diluted 10-fold and the intrinsic tryptophan fluorescence was monitored as a function of time. Each point represented the average of data accumulated over 0.2 sec. Kinetic traces were acquired at 25°C on a KinTek SF-2001 stopped-flow spectrophotometer with an excitation wavelength of 280 nm, and emission was monitored at 340 nm with a 12-nm bandpass filter (Corion, Holliston, MA). Relaxation rates (1/τ) were determined from the fit to a single exponential and represented the average of at least 50 independent experiments at each concentration between 10 and 30 μM ATF-2350–505. Data were fit to the linearized rate equation 1/τ = 4k3(ATF-2350–505)monomer + k−3 (14) to determine k3 and k−3.
To monitor [ATF-2350–505]2⋅CRE24 association kinetics, a 200-nM solution of ATF-2350–505 was mixed rapidly with 100 nM CRE24 and the fluorescence was monitored as a function of time. Each point represented the average of data accumulated over 0.2 sec. The relaxation rate (1/τ) was determined from the fit to a single exponential and represented the average of 50 independent experiments. Simulations of the monomer and dimer pathways were performed by using kinteksim software (15). The starting dimer concentration used to simulate the dimer pathway was defined by either k−3/k3 or Kdim.
Stopped-Flow Polarization.
A solution of 5 μM Max22–105SFlu was diluted 10-fold into 1× PBS at 25°C, and the fluorescence anisotropy was monitored as a function of time (16). Each point represented the average of data accumulated over 0.5 sec.
Electrophoretic Mobility-Shift Experiments.
Max22–113 was incubated in binding buffer (1× PBS containing 0.1% Nonidet P-40, 400 μg/ml BSA, 5% glycerol, and 1 mM EDTA) for 30 min at 25°C. [32P]Ebox22 was added to a concentration of 50 pM, and the reaction was followed by loading aliquots on a nondenaturing 8% (49:1) polyacrylamide gel. Radioactivity was quantified by using a STORM 840 PhosphorImager (Molecular Dynamics). Simulations of the monomer and dimer pathways were performed by using kinteksim software (15). For experiments comparing the rate of binding through the monomer and dimer pathways, a mixture of 8 nM Max22–113 and 1.9 nM Max22–105SS was incubated for 30 min at 25°C in binding buffer in the absence or presence of 10 nM hsEbox22. To each reaction was added 2 nM [32P]Ebox22, and the partitioning of [32P]Ebox22 into complexes with Max22–113 and Max22–105SS was monitored as a function of time. The reactions were followed by electrophoresis as described above. Analogous experiments were performed with either 0.57 μg/ml calf thymus DNA or 22.7 μg/ml HeLa cell nuclear extracts (Promega) as the competitor.
Results and Discussion
The bZIP Protein ATF-2350–505 Follows the Monomer Pathway to Bind DNA.
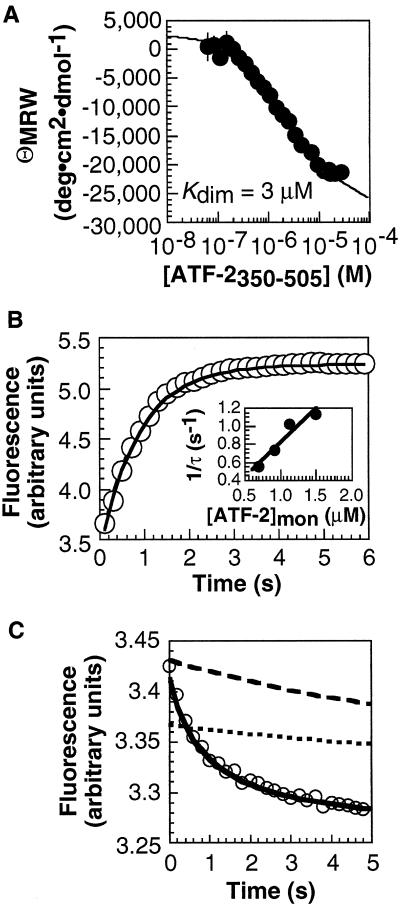
Our previous studies showed that DNA binding by certain bZIP peptides (17) could not proceed through the dimer pathway; additional experiments provided evidence that the monomer pathway was followed (4). These bZIP element peptides formed coiled coils characterized by low thermodynamic stabilities. To evaluate whether a bZIP protein capable of forming a stable coiled coil would also follow the monomer pathway, we studied a 156-residue fragment of ATF-2 (ref. 18), ATF-2350–505. The CD spectrum of ATF-2350–505 was consistent with a fully helical dimerization region at concentrations greater than 10 μM (Fig. 2A). At lower concentrations, the ATF-2350–505 dimer dissociated into unstructured monomers (Kdim = 3 μM) that were detected by an increase in ellipticity at 222 nm. The kinetics of the ATF-2350–505 monomer–dimer transition were determined by using stopped-flow fluorescence spectroscopy. The ATF2350–505 dimer was formed at concentrations above 10 μM and diluted rapidly, and the intrinsic tryptophan fluorescence was monitored to determine the rate of relaxation from dimer to monomer (Fig. 2B). The time-dependent increase in fluorescence was fit to a single exponential to determine the relaxation rate, 1/τ, at each concentration. Association (k3) and dissociation (k−3) rate constants of 1.9 × 105 M−1⋅s−1 and 0.05 s−1 were derived from a plot of 1/τ vs. the concentration of ATF-2350–505 monomer (14) (Fig. 2B Inset). The ratio k−3/k3 (0.3 μM) was comparable to the Kdim value determined in equilibrium experiments (3 μM).
Figure 2.
Analysis of ATF-2350–505 monomer–dimer equilibrium and kinetics and [ATF-2350–505]2⋅CRE24 association kinetics. (A) ATF-2350–505 monomer–dimer equilibrium analyzed by CD. (B) ATF-2350–505 monomer–dimer kinetics analyzed by stopped-flow fluorescence spectroscopy. A solution of [ATF-2350–505]Total = 30 μM was diluted 10-fold, and the fluorescence was monitored as a function of time. (Inset) Relaxation rate as a function of [ATF-2350–505]monomer. (C) [ATF-2350–505]2⋅CRE24 association kinetics analyzed by stopped-flow fluorescence spectroscopy. A 200-nM solution of ATF-2350–505 was mixed rapidly with 100 nM CRE24, and the fluorescence was monitored as a function of time. The relaxation rate (1/τ) was determined from the fit to a single exponential. Simulations of binding of CRE24 by ATF-2350–505 via the monomer (solid line) and dimer pathways are shown. The concentration of [ATF-2350–505]2 at the start of the simulation along the dimer pathway was defined either by the ratio k−3/k3 determined by fluorescence (dotted line) or by Kdim determined by equilibrium CD measurements (dashed line). In neither case can the dimer pathway account for the rate of DNA binding.
To evaluate whether the ATF-2350–505 dimer could function as an intermediate during formation of a specific DNA complex, we monitored association of ATF-2350–505 with the oligonucleotide duplex CRE24 by stopped-flow fluorescence spectroscopy. Equilibrium measurements showed that the intrinsic fluorescence of ATF-2350–505 diminished upon binding to CRE24 (data not shown). Kinetic experiments showed that binding of 200 nM ATF-2350–505 and 100 nM CRE24 occurred rapidly; the time-dependent decrease in fluorescence was best described by a single exponential with a relaxation rate of 1 s−1 (Fig. 2C).
Binding of ATF-2350–505 to CRE24 via the monomer and dimer pathways was simulated by using numerical integration (15). To simulate the dimer pathway, we employed experimentally determined values of k3 (1.9 × 105 M−1⋅s−1) and k−3 (0.05 s−1). The rate of DNA binding must be fast to account for the overall kinetics of the association reaction, so k4 was set at 1010 M−1⋅s−1, a diffusion-limited second-order rate constant (19) that takes into account the potential influence of rate-enhancing parameters such as electrostatic guidance. On the basis of the overall equilibrium dissociation constant of the [ATF-2350–505]2⋅CRE24 complex (2.9 × 10−18 M2) and the relationship Kd = (k−3/k3)⋅(k−4/k4), k−4 was calculated to be 0.11 s−1. The concentration of ATF-2350–505 dimer at the start of the simulation was defined by k−3/k3 or by Kdim. In either case, the dimer pathway could account for the observed rate of DNA binding only if the rate constant for dimerization of ATF-2350–505 (k3) were more than 30 times greater than the value inferred from experiment. By contrast, simulation of binding along the monomer pathway, using rate constants consistent with the overall equilibrium dissociation constant and the estimated DNA affinity of a bZIP monomer (20), reproduced the experimental results (Fig. 2C). The values of k1 = 7.5 × 106 M−1⋅s−1, k−1 = 200 s−1, k2 = 1 × 1010 M−1⋅s−1, and k−2 = 0.0011 s−1 do not represent a unique solution: a range of reasonable rates allowed the experimental data to be simulated under the monomer pathway. Like simpler bZIP peptide models, we propose that ATF-2350–505 follows the monomer pathway to bind specific DNA.
The bHLHZip Protein Max Follows the Monomer Pathway to Bind DNA.
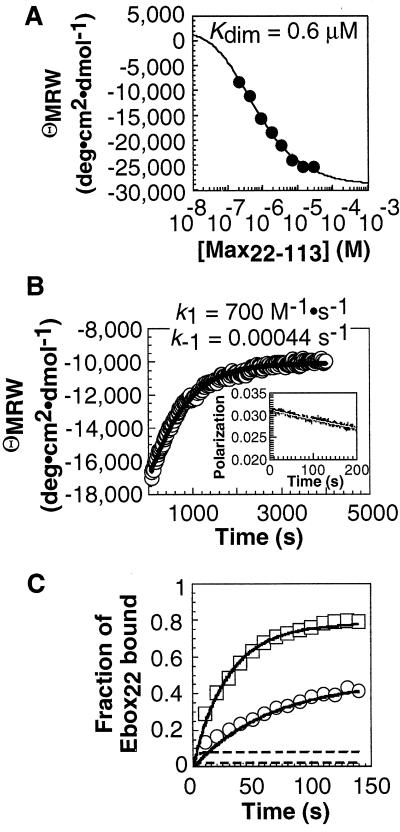
We performed an analogous set of experiments with the bHLHZip protein Max (21) to examine whether a protein possessing a more globular dimerization domain also preferred the monomer pathway. The CD spectrum of Max22–113 was consistent with a fully helical dimerization domain at concentrations greater than 10 μM. At lower concentrations, the Max22–113 dimer dissociated into unstructured monomers (Kdim = 0.6 μM) (22, 23) that were detected by an increase in ellipticity at 222 nm (Fig. 3A).
Figure 3.
Analysis of Max22–113 monomer–dimer equilibrium and kinetics and [Max22–113]2⋅Ebox22 association kinetics. (A) Max22–113 monomer–dimer equilibrium analyzed by CD. (B) Max22–113 monomer–dimer kinetics analyzed by CD and stopped-flow fluorescence. A 50-μM solution of Max22–113 was diluted 50-fold into PBS buffer at 25°C, and the negative ellipticity at 222 nm was monitored as a function of time. (Inset) Max22–105SFlu monomer–dimer relaxation kinetics monitored by stopped-flow fluorescence polarization within the first 200 sec of the reaction. A solution of 5 μM Max22–105SFlu was diluted 10-fold into PBS buffer at 25°C, and the fluorescence anisotropy was monitored as a function of time. (C) Association of 1 nM (○) and 2 nM (□) Max22–113 with 50 pM Ebox22 at 25°C. Each point represented the average of at least three independent experiments. Error bars shown represent the SD. Similar association kinetics were observed with a 135-bp DNA fragment containing a single Ebox site (CACGTG). Simulations of binding of Ebox22 by Max22–113 via the monomer (solid lines) and dimer (dashed lines) pathways are shown.
The kinetics of the Max22–113 monomer–dimer transition were monitored using CD and fluorescence polarization spectroscopy. The Max22–113 dimer was formed at concentrations above 5 μM, diluted rapidly, and the ellipticity at 222 nm or the extent of fluorescence polarization was monitored. Initial fluorescence polarization experiments (24) using a fluorescein-labeled Max derivative, Max22–105SFlu, indicated that relaxation was slow (1/τ < 0.002 s−1; Fig. 3B Inset). A precise relaxation rate of 1/τ = 0.0013 ± 0.0001 s−1 was determined by CD (Fig. 3B). This value, in combination with the independently measured equilibrium dissociation constant of the Max22–113 dimer (0.6 μM), indicated that the Max22–113 dimer formed slowly (k3 = 700 M−1 s−1) and was kinetically stable (k−3 = 0.00044 s−1).
To evaluate whether the Max22–113 dimer could function as an intermediate during formation of the specific DNA complex, we monitored association of Max22–113 with 32P end-labeled Ebox22 ([32P]Ebox22) by gel electrophoresis (Fig. 3C). Max22–113 bound [32P]Ebox22 rapidly at all concentrations tested, with half-lives between 25 (2 nM Max22–113) and 60 (0.75 nM Max22–113) sec.
Formation of the [Max22–113]2⋅Ebox22 complex along the monomer and dimer pathways was simulated by using numerical integration (15) (Fig. 3C). For simulation of the dimer pathway, we employed experimentally determined values for k3 (700 M−1⋅s−1) and k−3 (0.00044 s−1); k4 was set at 1010 M−1⋅s−1, the largest allowed second-order rate constant (19) and k−4 was defined to be 0.016 s−1 so that the overall equilibrium dissociation constant calculated from the rates matched the experimentally determined equilibrium dissociation constant of the complex (10−18 M2). This simulation indicated that, along the dimer pathway, 7,000 sec were required for association of 2 nM Max22–113 with one-half of the available 50 pM Ebox22 (Fig. 3C). This length of time was at least 150 times longer than the experimentally determined half-life at this concentration (25 sec) (Fig. 3C). By contrast, simulation of Ebox22 binding along the monomer pathway using rate constants (k1 = 8 × 107 M−1⋅s−1, k−1 = 80 s−1, k2 = 8 × 109 M−1⋅s−1, and k−2 = 0.008 s−1) that were physically reasonable and consistent with the overall equilibrium dissociation constant reproduced the experimental results. These simulations indicate that the monomer pathway provides rapid access to the [Max22–113]2⋅Ebox22 complex whereas the dimer pathway does not (Fig. 3C).
Why Is the Monomer Pathway Preferred?
The results described above indicate that the monomer pathway is favored by bZIP and bHLHZip proteins in vitro. We proposed a physical explanation for the dominance of the monomer pathway based on electrostatic guidance (25–27) in an early stage of the reaction (4). Both the first and the second steps in the monomer pathway are promoted by strong electrostatic interactions, with slow formation of the dimer occurring as an isomerization on the DNA. But what benefit might the monomer pathway provide in vivo? Together, the monomer and dimer pathways constitute a thermodynamic cycle, and no gain in equilibrium specificity can result from partitioning along the two pathways (4). However, the two pathways proceed through different intermediates, which could interact at different rates with nonspecific proteins and DNA. Therefore, the monomer and dimer pathways could generate unequal levels of specificity prior to equilibrium. The rate at which the protein searches for and locates its specific site in the more elaborate cellular milieu will depend on the relative rates of interaction with specific and nonspecific competitors.
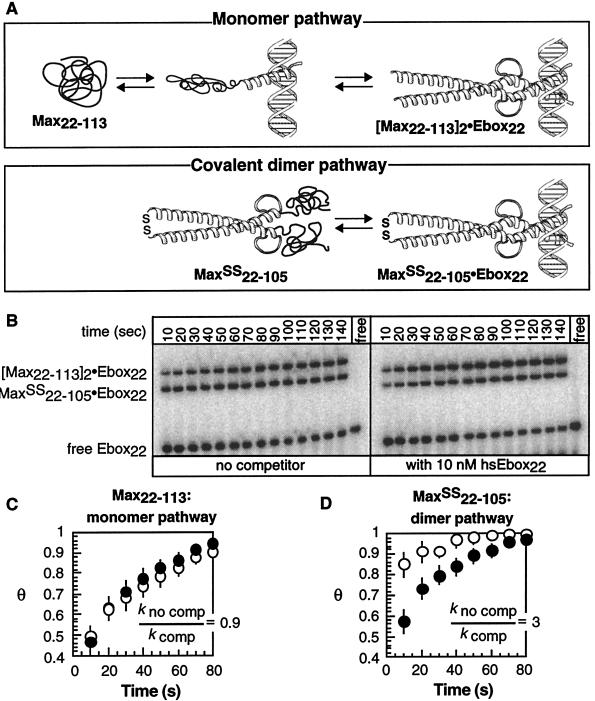
To compare the rates at which a protein locates its specific site via the monomer and dimer pathways, we prepared a variant of Max (Max22–105SS) that allowed us to observe DNA binding along the dimer pathway that we propose is not normally utilized (Fig. 4A). By virtue of a disulfide bond covalently linking the coiled coil, Max22–105SS bypassed the slow, initial step along the dimer pathway (Fig. 4A). We then compared the rates with which Max22–105SS and Max22–113 bound [32P]Ebox22 in the presence and absence of competitor DNA containing two mutations within the 6-bp Ebox site (hsEbox22). The fraction of [32P]Ebox22 bound (Θ) as a function of time by either Max22–113 (Fig. 4C) or Max22–105SS (Fig. 4D) in the absence or presence of 10 nM competitor hsEbox22 was normalized to a value of Θ = 1 at equilibrium and fit to single exponentials to give kno comp and kcomp, respectively. In the absence of hsEbox22 competitor, binding of [32P]Ebox22 by Max22–113 (Fig. 3C) through the two-step monomer pathway competed well with one-step binding by the constitutive dimer Max22–105SS (Fig. 3D). The presence of hsEbox22 caused the preformed dimer, Max22–105SS, to bind [32P]Ebox22 more slowly (Fig. 4D), whereas no effect was observed on the rate of binding by the Max22–113 monomer (Fig. 4C). Similar results were obtained for competition with calf thymus DNA or with HeLa cell nuclear extracts (data not shown). Even low concentrations of nonspecific proteins and DNA selectively decreased the rate of specific complex formation by a transcription factor that followed the dimer pathway. No decrease was observed for a protein that followed the monomer pathway. The dimer pathway was slowed, presumably by formation of long-lived but nonproductive intermediates—those between the preformed dimer and nonspecific protein and DNA partners.
Figure 4.
Comparison of the rate of DNA binding through the monomer and dimer pathways in the presence and absence of excess nonspecific competitor DNA. (A) Scheme illustrating the monomer-binding pathway utilized by Max22–113 and the covalent dimer pathway utilized by Max22–105SS. (B) Phosphorimage illustrating the relative rate of [32P]Ebox22 binding by Max22–113 and Max22–105SS in the presence or absence of competitor hsEbox22. The fraction of DNA bound (Θ) by either Max22–113 (C) or Max22–105SS (D) at each time point in the absence (○) or presence (●) of hsEbox22 DNA was normalized to a value of Θ = 1 at equilibrium and fit to a single exponential to give values of kno comp and kcomp, respectively. Each point represented the average of at least four independent experiments. Error bars represent SE.
Implications for Transcription.
In vivo, the mass action of excess nonspecific DNA in the nucleus favors formation of nonspecific protein–DNA complexes (2). The relatively long lifetime of a nonspecific complex containing a transcription factor dimer decreases the efficiency with which the preformed dimer locates its specific site and increases the possibility of an inappropriate transcriptional response. Binding through the monomer pathway allows a dimeric transcription factor to respond rapidly to stimuli and to locate its target site quickly without becoming entrapped kinetically at a nonspecific site (28). This method of kinetic discrimination is reminiscent of the immunological phenomenon of T cell activation (29). In the immune response, an antigen may be converted from an agonist to an antagonist by increasing the rate of dissociation from a T cell receptor. It has been suggested that coreceptor proteins slow the ligand dissociation rate and thereby stimulate T cell activation (29, 30). In the realm of transcription, a large number of viral accessory proteins, including HTLV-I Tax (31–33) and hepatitis B virus pX (34), interact with bZIP proteins and deregulate cellular transcription. The mechanism of action of these viral accessory proteins may involve decreasing the dissociation rate of bZIP proteins from nonspecific DNA as well as stabilizing bZIP dimers, which possess a less efficient search mechanism than bZIP monomers.
Acknowledgments
J.J.K. and S.J.M. thank the National Science Foundation and the Organic Chemistry division of the ACS, respectively, for predoctoral fellowships. We are grateful to Stephen Burley (Rockefeller University) for clones encoding Max22–113 and Max22–105SH, to James Hoeffler (University of Colorado, Health Science Center) for the clone encoding ATF-2350–505, and to Hays S. Rye (Department of Genetics, Yale University School of Medicine) for assistance acquiring fluorescence polarization data. This work was supported by the National Institutes of Health.
Abbreviations
- bZip
basic region leucine zipper
- bHLHZip
basic region helix–loop–helix zipper
- ATF-2
activating transcription factor 2
Note Added in Proof
Rentzeperis et al. (35) reported that the Arc repressor also binds DNA by following the monomer pathway.
References
- 1.Tjian R, Maniatis T. Cell. 1994;77:5–8. doi: 10.1016/0092-8674(94)90227-5. [DOI] [PubMed] [Google Scholar]
- 2.Bray D, Lay S. Proc Natl Acad Sci USA. 1997;94:13493–13498. doi: 10.1073/pnas.94.25.13493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim B, Little J W. Science. 1992;255:203–206. doi: 10.1126/science.1553548. [DOI] [PubMed] [Google Scholar]
- 4.Metallo S J, Schepartz A. Nat Struct Biol. 1997;4:115–117. doi: 10.1038/nsb0297-115. [DOI] [PubMed] [Google Scholar]
- 5.Berger C, Piubelli L, Haditsch U, Bosshard H R. FEBS Lett. 1998;425:14–18. doi: 10.1016/s0014-5793(98)00156-2. [DOI] [PubMed] [Google Scholar]
- 6.Wu X, Spiro C, Owen W G, McMurray C. J Biol Chem. 1998;273:20820–20827. doi: 10.1074/jbc.273.33.20820. [DOI] [PubMed] [Google Scholar]
- 7.Wendt H, Thomas R M, Ellenberger T. J Biol Chem. 1998;273:5735–5743. doi: 10.1074/jbc.273.10.5735. [DOI] [PubMed] [Google Scholar]
- 8.Rastinejad F, Perlmann T, Evans R M, Sigler P B. Nature (London) 1995;375:203–211. doi: 10.1038/375203a0. [DOI] [PubMed] [Google Scholar]
- 9.Holmbeck S M A, Dyson H J, Wright P E. J Mol Biol. 1998;284:533–539. doi: 10.1006/jmbi.1998.2207. [DOI] [PubMed] [Google Scholar]
- 10.Meinke G, Sigler P B. Nat Struct Biol. 1999;6:471–477. doi: 10.1038/8276. [DOI] [PubMed] [Google Scholar]
- 11.Hoeffler J P, Meyer T E, Yun Y, Jameson J L, Habener J F. Science. 1988;242:1430–1433. doi: 10.1126/science.2974179. [DOI] [PubMed] [Google Scholar]
- 12.Ferre-D’Amare A R, Prendergast G C, Ziff E B, Burley S K. Nature (London) 1993;363:38–45. doi: 10.1038/363038a0. [DOI] [PubMed] [Google Scholar]
- 13.Zitzewitz J A, Bilsel O, Luo J, Jones B E, Matthews C R. Biochemistry. 1995;34:12812–12819. doi: 10.1021/bi00039a042. [DOI] [PubMed] [Google Scholar]
- 14.Bernasconi C F. Relaxation Kinetics. New York: Academic; 1976. [Google Scholar]
- 15.Barshop B A, Wrenn R F, Frieden C. Anal Biochem. 1983;130:134–145. doi: 10.1016/0003-2697(83)90660-7. [DOI] [PubMed] [Google Scholar]
- 16.Gill G, Tjian R. Cell. 1991;65:333–340. doi: 10.1016/0092-8674(91)90166-v. [DOI] [PubMed] [Google Scholar]
- 17.Harrison S C. Nature (London) 1991;353:715–719. doi: 10.1038/353715a0. [DOI] [PubMed] [Google Scholar]
- 18.Hai T W, Liu F, Allegretto E A, Karin M, Green M R. Genes Dev. 1988;2:1216–1226. doi: 10.1101/gad.2.10.1216. [DOI] [PubMed] [Google Scholar]
- 19.von Hippel P H, Berg O G. J Biol Chem. 1989;264:675–678. [PubMed] [Google Scholar]
- 20.Zondlo N J, Schepartz A. J Am Chem Soc. 1999;121:6938–6939. [Google Scholar]
- 21.Blackwood E M, Eisenman R N. Science. 1991;251:1211–1217. doi: 10.1126/science.2006410. [DOI] [PubMed] [Google Scholar]
- 22.Fairman R, Bernan-Steed R K, Anthony-Cahill S J, Lear J D, Stafford W F, DeGrado W F, Benfield P A, Brenner S L. Proc Natl Acad Sci USA. 1993;90:10429–10433. doi: 10.1073/pnas.90.22.10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horiuchi M, Kurihara Y, Katahira M, Maeda T, Saito T, Uesugi S. J Biochem (Tokyo) 1997;122:711–716. doi: 10.1093/oxfordjournals.jbchem.a021813. [DOI] [PubMed] [Google Scholar]
- 24.James B R, Williams R J P. J. Chem. Soc. 1961. 2007–2019. [Google Scholar]
- 25.Schreiber G, Fersht A R. Nat Struct Biol. 1996;3:427–431. doi: 10.1038/nsb0596-427. [DOI] [PubMed] [Google Scholar]
- 26.Getzoff E D, Tainer J A, Weiner P K, Kollman P A, Richardson J S, Richardson D C. Nature (London) 1983;306:287–290. doi: 10.1038/306287a0. [DOI] [PubMed] [Google Scholar]
- 27.Getzoff E D, Cabelli D E, Fisher C L, Parge H E, Viezzoli M S, Banci L, Hallewell R A. Nature (London) 1992;358:347–351. doi: 10.1038/358347a0. [DOI] [PubMed] [Google Scholar]
- 28.Pomerantz J L, Wolfe S A, Pabo C O. Biochemistry. 1998;37:965–970. doi: 10.1021/bi972464o. [DOI] [PubMed] [Google Scholar]
- 29.Rabinowitz J D, Beeson C, Lyons D S, Davis M M, McConnell H M. Proc Natl Acad Sci USA. 1996;93:1401–1405. doi: 10.1073/pnas.93.4.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vignali D A A, Strominger J L. J Exp Med. 1994;179:1945–1956. doi: 10.1084/jem.179.6.1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perini G, Wagner S, Green M R. Nature (London) 1995;376:602–605. doi: 10.1038/376602a0. [DOI] [PubMed] [Google Scholar]
- 32.Baranger A M, Palmer C R, Hamm M K, Giebler H A, Brauweiler A, Nyborg J K, Schepartz A. Nature (London) 1995;376:606–608. doi: 10.1038/376606a0. [DOI] [PubMed] [Google Scholar]
- 33.Wagner S, Green M R. Science. 1993;262:395–399. doi: 10.1126/science.8211160. [DOI] [PubMed] [Google Scholar]
- 34.Palmer C R, Gegnas L D, Schepartz A. Biochemistry. 1997;36:15349–15355. doi: 10.1021/bi972076m. [DOI] [PubMed] [Google Scholar]
- 35.Rentzeperis D, Jonsson T, Sauer R T. Nat Struct Biol. 1999;6:569–573. doi: 10.1038/9353. [DOI] [PubMed] [Google Scholar]