Abstract
A three-plasmid system for heterologous expression of 6-deoxyerythronolide B synthase (DEBS) has been developed to facilitate combinatorial biosynthesis of polyketides made by type I modular polyketide synthases (PKSs). The eryA PKS genes encoding the three DEBS subunits were individually cloned into three compatible Streptomyces vectors carrying mutually selectable antibiotic resistance markers. A strain of Streptomyces lividans transformed with all three plasmids produced 6-deoxyerythronolide B at a level similar to that of a strain transformed with a single plasmid containing all three genes. The utility of this system in combinatorial biosynthesis was demonstrated through production of a library of modified polyketide macrolactones by using versions of each plasmid constructed to contain defined mutations. Combinations of these vector sets were introduced into S. lividans, resulting in strains producing a wide range of 6-deoxyerythronolide B analogs. This method can be extended to any modular PKS and has the potential to produce thousands of novel natural products, including ones derived from further modification of the PKS products by tailoring enzymes.
Polyketides are structurally diverse natural products that include important therapeutic agents used as antibacterials (erythromycin), immunosuppressants (FK506), cholesterol-lowering agents (lovastatin), and others (1). Currently, there are about 7,000 identified polyketides, but this represents only a small fraction of what nature is capable of producing. DNA sequencing of genes encoding several of the enzymes that produce type 1 modular polyketide synthases (PKSs) has revealed the remarkably logical organization of these multifunctional enzymes (2–5). This organization has prompted the application of combinatorial techniques to the generation of novel natural products by adding, deleting, or exchanging domains or entire modules. We describe here a practical combinatorial biosynthesis technology that should achieve and perhaps exceed the diversity of modular polyketide structures thus far revealed in nature.
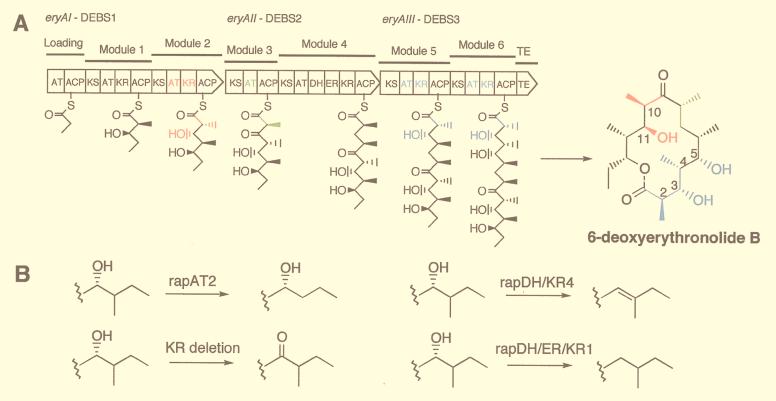
The known modular PKSs have a linear organization of modules, each of which contains the activities needed for one cycle of polyketide chain elongation, as illustrated for 6-deoxyerythronolide B (6dEB) synthase (DEBS) in Fig. 1A. The minimal module contains a ketosynthase (KS), an acyltransferase (AT), and an acyl carrier protein (ACP) that together catalyze a 2-carbon extension of the chain. The specificity of the AT for either malonyl- or an α-alkylmalonyl-CoA determines which 2-carbon extender is used, and thus the nature of the alkyl substituent at the α-carbon of the growing polyketide chain. After each 2-carbon unit condensation, the oxidation state of the β-carbon is either retained as a ketone or modified to a hydroxyl, methenyl, or methylene group by the presence of a ketoreductase (KR), a KR + a dehydratase (DH), or a KR + DH + an enoylreductase (ER), respectively. In effect, the AT specificity and the composition of catalytic domains within a module serve as a “code” for the structure of each 2-carbon unit; the order of the modules in a PKS specifies the sequence of the distinct 2-carbon units, and the number of modules determines the size of the polyketide chain.
Figure 1.
Wild-type and mutant forms of the eryA genes and DEBS proteins. (A) The eryAI–eryAIII genes and proteins are shown as broad arrows oriented in the direction of transcription with the domains in modules 1 to 6 of DEBS1-DEBS3, indicated by the symbols explained in the text. The first substrate, propionyl-CoA, is attached to the loading domain ACP and (2S)-2-methylmalonyl-CoA to the module 2 ACP. Then, a decarboxylative condensation between the propionate and methylmalonate takes place followed by reduction of the incipient β-ketone to form the intermediate shown attached to the ACP of module 2. This intermediate is transferred to the ACP of module 3, and the sequence of reactions is repeated at each of the other modules with or without ketone reduction, dehydration, or double bond reduction to form the linear 21-carbon polyketide attached to the ACP of module 6, which then is cyclized and released as 6dEB. (B) Replacement of the one or more of the colored domains in DEBS1, DEBS2, or DEBS3 with one of the three rap PKS domains or cassettes, or deletion of the KR, results in the corresponding functional group changes shown at one or more of the colored positions of 6dEB.
The remarkable structural diversity of polyketides (6) is governed by the combinatorial possibilities of arranging modules containing the various catalytic domains, the sequence and number of modules, and the post-PKS “tailoring enzymes” that accompany the PKS genes. The direct correspondence between the catalytic domains of modules in a PKS and the structure of the resulting biosynthetic product allow rational modification of polyketide structure by genetic engineering.
Over the past several years, each of the elements that code for polyketide structure has been modified (7–11). Recently, a combinatorial library of over 50 novel polyketides was prepared by systematic modification of DEBS, the PKS that produces the macrolide aglycone precursor of erythromycin (12). By using a single plasmid containing the eryAI, -AII, and -AIII genes encoding the three DEBS subunits, ATs and β-carbon processing domains were replaced by counterparts from the rapamycin PKS (4) that encode alternative substrate specificities and β-carbon processing activities. The approach used was to develop single “mutations,” then sequentially combine the single mutations to produce multiple changes in the PKS. It was observed that when two or more single PKS mutants were functional, there was a high likelihood that combinations would also produce the expected polyketide. Although this strategy provided high assurance that the multiple mutants would be productive, the production of each polyketide required a separate engineering. Thus, if X mutants of eryAI, Y mutants of eryAII, and Z mutants at eryAIII were prepared, X + Y + Z separate experiments were required to produce that same number of polyketides. Clearly, the preparation of very large libraries by this approach would be laborious.
Another strategy for preparing large numbers of polyketides is by random digestion–religation leading to “mutagenesis” of the domains or modules of a mixture of PKS genes, including the refinements embodied in the DNA shuffling method (13). The expected low probability of assembling an active PKS by such an approach, however, would demand an extraordinary analytical effort (in the absence of a biological selection) to detect clones that produced polyketides within the much larger number of clones that are nonproducers. Moreover, this method is unlikely to lead to novel polyketides not attainable by the more practical approach presented below.
We describe a strategy for creating a polyketide library that enables the production of large libraries of polyketides while retaining the high probability of obtaining productive clones by combining PKS mutations that are known to be productive. The principle involves cloning mutants of individual ORFs of a PKS on separate compatible plasmids, then coexpressing the separate ORFs in a suitable host to produce the PKS. By using this multiple plasmid approach, with X mutants of ORF 1, Y mutants of ORF 2, and Z mutants of ORF 3, along with the wild-type genes, for instance, a combinatorial library of (X + 1) × (Y + 1) × (Z + 1) mutants plus the wild-type PKS could be created expeditiously.
DEBS was chosen for a test of this multiple-plasmid approach. This PKS consists of three >280-kDa protein subunits, each containing two modules, that are assembled into the complete PKS complex (3). Using two possible AT domains and the four possible β-modifications, i.e., eight at each module, 8 × 8 = 64 permutations for the two modules are possible in each DEBS subunit. Constructing the mutations in each DEBS ORF separately would require that 64 experiments be carried out on each gene, or a total of 192 such experiments. However, by cotransforming a host strain with three plasmids, each bearing the 64 permutations of a different DEBS subunit, one could generate the mutant PKSs necessary to achieve, in theory, a library of 262,144 polyketides (643), as 6dEB analogs (Fig. 1A). In contrast, the same number of mutagenesis experiments performed in the single plasmid system would theoretically yield only 192 polyketides.
The successful implementation of this multiple-plasmid strategy requires that the DEBS subunits translated from three different mRNAs would faithfully interact to give the active PKS. An analogous independent expression of subunits of the bacterial peptide synthase surfactin has recently been reported (14). Some indication that this would also be the case with DEBS was provided by in vitro experiments that showed that reconstitution of the isolated DEBS1–DEBS2 complex with DEBS3 forms a functional PKS (15). Further, it was recently demonstrated that coexpression of the three subunits of DEBS from two plasmids produced active PKS in vivo (16).
Materials and Methods
Construction of the Expression Plasmids.
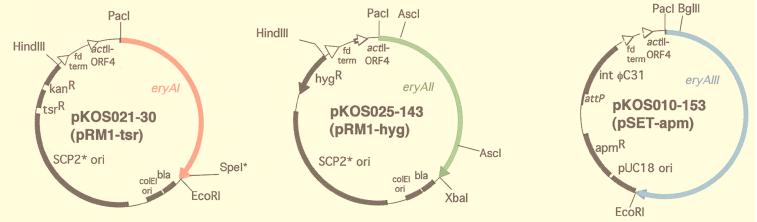
In each vector, the eryA gene is expressed under the control of the upstream actI promoter and actII–ORF4 gene as previously described (16, 17). For testing the high-copy number replicating plasmid possessing the pJV1 origin, the pB45-based Escherichia coli shuttle vector pKOS025-32 was constructed by fusing pB45 with LITMUS 28 (New England Biolabs) at BglII and PstI sites. Into pKOS025-32 the ca. 14-kb HindIII–XbaI fragment containing the actI promoter and actII–ORF4 gene (17) was inserted followed by the eryAII gene to give pKOS025-35. The configuration of the components in the HindIII–XbaI fragment is described in Fig. 2. The same HindIII–XbaI fragment was also cloned into shuttle vector pOSint1/Hygro (18) to yield the eryAII/pSAM2-hyg plasmid, pKOS038-67.
Figure 2.
The three-plasmid expression system for eryA genes. In each vector, the eryA gene is expressed under the control of the upstream actI promoter and actII–ORF4 gene as previously described (16, 17). To facilitate construction of the various eryA mutations, a SpeI site (ACTAGT) was introduced at nt 10366–10371 of the eryAI ORF by making D3455T and A3456S mutations (27) in pKOS025-179 (Q.X., unpublished data). This change enables insertion of the mutated gene segment between the PacI site and the SpeI site of pKOS025-179 (16, 17). After the replacement of the 6-kb fragment between the AscI sites in eryAII (nucleotides 1213 and 7290) with a 6-kb AscI fragment containing a specific AT substitution in an intermediate plasmid, the resulting PacI–XbaI fragment containing the mutant eryAII gene was inserted into pKOS025-143. All eryAIII mutants were constructed by replacing the segment in pKOS010-153 between the unique BglII site at nucleotide 251 and the EcoRI site (nucleotide 9290) that overlaps the stop codon.
Transfer of Mutation Cassettes into the Three-Plasmid System.
The plasmids containing eryAI mutations in module 2 were pKOS025-179 (AT2 → rapAT2), pKOS038-1 (KR → rapDH/KR4), and pKOS038-3 (KR → rapDH/ER/KR1) and were made as follows. The PacI–SpeI fragments containing the corresponding mutations were transferred into the plasmids described in Fig. 2 from plasmids pKOS008-41, pKOS015-56, and pKOS015-57, respectively (R. McDaniel, Kosan Biosciences, personal communication). The plasmids containing the eryAIII mutations in module 5 were pKOS025-1831 (AT → rapAT2), pKOS025-1832 (KR → AT/ACP linker), pKOS025-1833 (KR → rapDH/KR4), and pKOS025-1834 (KR → rapDH/ER/KR1); and in module 6 were pKOS025-1841(KR → rapDH/KR4), pKOS021-106 (KR → AT/ACP linker), and pKOS025-1842 (AT → rapAT2). These were constructed as follows. The BglII–EcoRI fragments containing the corresponding mutations were transferred into the plasmids described in Fig. 2 from plasmids pKOS006-188, pKOS016-12, pKOS006-178, pKOS026-11b, pKOS011-25, pKOS011-13, and pKOS015-53, respectively (R. McDaniel, personal communication). pKOS038-20 contains eryAII with the AT3 → rapAT2 replacement in module 3 and was made by transferring the mutation from a previously prepared subclone pKOS015-28 (R. McDaniel, personal communication).
Streptomyces Transformation.
Streptomyces lividans K4-114 and K4-155 (19) transformants were prepared according to standard methods by using apramycin (100 μg/ml), thiostrepton (50 μg/ml), and hygromycin (225 μg/ml) in the R5 protoplast regeneration plates (20). In the three-plasmid system for which eryAII/pSAM2-hyg replaced eryAII/SCP2*-hyg, S. lividans was transformed sequentially with pKOS010-153, pKOS038-67, and pKOS021-30.
Diketide Feeding.
The (2S,3R)-2-methyl-3-hydroxyhexanoyl-N-acetylcysteamine (NAC) thioester (propyl diketide) was synthesized by G.A., A. Rico, and I. Chan-Kai (unpublished work). The S. lividans triple transformants containing the KS1 null allele of eryAI (11) were cultured in 5 ml of R5 medium at 30°C for 6 days under appropriate antibiotic selection. On day 4 of the incubation, 300 μl of diketide solution (4.7 mg/ml in 10% DMSO) and 50 μl of pentanoic acid (2.5 mg/ml) were added to the culture.
Production and Analysis of Polyketide Analogs.
The S. lividans triple transformants were cultured in 5 ml of R5 medium (20) at 30°C for 6 days under appropriate antibiotic selection. The cultured solution was extracted twice with 5 ml of ethyl acetate, and the organic layers were combined and concentrated. A 50-μl aliquot of the concentrates was analyzed by HPLC on a reverse-phase C18 column (4.6 mm × 15 cm; Beckman Coulter) using a Perkin–Elmer SCIEX API100 detector based on liquid chromatography/mass spectrometry (LC/MS). Quantitative determination of polyketide yield was quantitated with evaporative light scattering detection (Analtec model 500 ELSD). The polyketides were identified by their mass spectrum and correspondence to the products expected or to known standards. Under the ionization conditions used, 6dEB and its analogs generate signature dehydration patterns. The yield of the 13-ethyl 6dEB analogs varied from 7 to <0.1 mg/liter, and the 13-propyl analogs were produced in a range of 0.2 to 20 mg/liter. The yields of the 12-membered lactones were not determined.
Results and Discussion
Establishment of Three-Plasmid Expression System for DEBS.
The approach required at least three vectors that could be separately introduced into a Streptomyces host strain and concomitantly express functional PKS subunits. At the outset, we had tested only two such vectors: the autonomously replicating SCP2*-based plasmid pRM1 (17) and the integrating bacteriophage ΦC31-based plasmid pSET152 (21). We therefore examined several additional plasmids, each of which contained identical configurations of an eryA gene downstream of the Streptomyces coelicolor actI promoter and actII-ORF4 transcriptional activator as described (16). pB45, a high-copy replicating plasmid possessing the pJV1 origin (22), carrying eryAII was introduced into S. lividans harboring eryAI on pRM1 and eryAIII in the pSET152 integration site; less than 0.1 mg/liter of 6dEB was produced in this system compared with 50 mg/liter for the single-plasmid system (17). Subsequently, we found that two SCP2*-type plasmids carrying different antibiotic markers can coexist and express PKS subunits in a Streptomyces sp., as had been reported, but using high-copy number plasmids (23). Cotransformation of S. lividans with eryAIII/pSET-apm, eryAI/pRM1-tsr, and eryAII/pRM1-hyg (Fig. 2) yielded a strain that also produced 50 mg/liter 6dEB (17). Further, we showed that after over 24 generations under double antibiotic selection, both replicating plasmids could be rescued by standard procedures with unchanged restriction maps (20). We also cloned the eryAII gene into a pSAM2 site-specific integrating plasmid (24). Sequential transformation of S. lividans with eryAIII/pSET-apm, eryAII/pSAM-hyg, and eryAI/pRM1-tsr provided a strain that produced 40–50 mg/liter 6dEB. A potential advantage of this system over the two-replicating vector system is that it requires one fewer antibiotic and avoids potential problems in maintaining two plasmids containing the same ori in a Streptomyces host (25). Nevertheless, for the present study, we chose to use the two SCP2*-based plasmids + the pSET-derived vector.
Construction of Plasmids Comprising Mutant eryA Genes.
A demonstration library composed of three single mutations in eryAI (module 2), one in eryAII (module 3), and seven in eryAIII (modules 5 or 6) as well as wild-type ORFs was created by using this three-plasmid system. To facilitate cloning, vectors were prepared that contained restriction sites that allowed transfer of DNA cassettes from previously prepared mutant eryA genes (12) (Fig. 2). Fourteen expression vectors, comprising the three wild-type and eleven mutant ORFs (Table 1) were constructed by cassette transfers from plasmids previously prepared in the single-plasmid system (12). In eryAI, module 2 was modified by replacing the AT by rapAT2, and the KR by rapDH/KR4 or rapDH/ER/KR1. In eryAII, the AT of module 3 was replaced by rapAT2; in eryAIII, module 5 was modified by replacing the AT by rapAT2, and the KR by rapDH/KR4 or rapDH/ER/KR1 or the AT/ACP linker that eliminates the ery module 5 KR activity. Also, in module 6, the AT was replaced with rapAT2, and the KR by rapDH/KR4 or the AT/ACP linker. The consequence of introducing each of these rapamycin PKS gene cassettes into DEBS is shown in Fig. 1B; at a given α-position in the growing polyketide carbon chain, a methyl group can be removed, or a β-hydroxyl can be changed to a ketone or removed by dehydration to produce a double bond, or the double bond resulting from dehydration can then be reduced to a methylene group.
Table 1.
Geneotype of the plasmids containing DEBS genes
| Vector: | pKOS021 | pKOS025 | pKOS010 |
|---|---|---|---|
| Gene/protein: | eryAI(DEBS1) | eryAII(DEBS2) | eryAIII(DEBS3) |
| Module: | 1 2 | 3 | 5 6 |
| 1. Wild type | |||
| 2. AT → rapAT2 | |||
| 3. KR → rapDH/KR4 | |||
| 4. KR → rapDH/ER/KR1 | |||
| 5. KS1°* | |||
| 6. Wild type | |||
| 7. AT3 → rapAT2 | |||
| 8. Wild type | |||
| 9. AT → rapAT2 | |||
| 10. KR → AT/ACP linker | |||
| 11. KR → rapDH/KR4 | |||
| 12. KR → rapDH/ER/KR1 | |||
| 13. Module 5 + TE† | |||
| 14. AT → rapAT2 | |||
| 15. KR → AT/ACP linker | |||
| 16. KR → rapDH/KR4 | |||
The plasmids with mutant DEBS genes were constructed by cloning the specified DNA segments into one of the three vectors shown in Fig. 2. Arrows signify that the wild-type domain was replaced with the one indicated.
*The Cys-729 → Ala null allele was created by site-specific mutagenesis.
†Module 6 was deleted to fuse its TE to the ACP of module 5.
Combinatorial Approach to Generating “Unnatural” Natural Polyketide Libraries.
The approach was to first individually introduce the eight eryAIII variants into the pSET integration site of S. lividans, then to individually cotransform the resulting strains with each of four eryAI variants on SCP2*-tsr vectors and two eryAII variants on SCP2*-hyg vectors. Thus, from the 14 vectors prepared, we obtained 64 triple transformants. Recombinant cells were grown under appropriate antibiotic selection, and extracts were analyzed for polyketide production by LC/MS. Of the 64 triple transformants, 46 (72%) produced detectable levels of one or more polyketides under the test conditions employed (Fig. 3), and 43 different polyketides were produced. These included 6dEB (1) and products arising from 11 single (2–12), 26 double (13–38), and 5 (39–43) triple mutants of DEBS. Twenty-eight (1–28) of the 43 polyketides produced have been previously prepared by using the single-plasmid system (12). Fifteen (29–43) were novel polyketides readily identifiable by mass spectra and correspondence to the products expected based on the cassettes used in the mutagenesis.
Figure 3.
Structures of macrolactones produced by S. lividans strains containing assorted combinations of three plasmids (Fig. 2 and Table 1). The positions in 6dEB (1) that are altered are color coded to correspond to the genetic characteristics of modules 2, 3, 5, and 6 of DEBS that are illustrated in Fig. 1A and Fig. 2, and listed in Table 1. Structures 1–40 and 49–56 are 14-membered lactones, and structures 44–48 are 12-membered lactones. Compounds 49–59 with the C13 propyl group were produced by mutational biosynthesis with the eryAI KS1o null allele.
In 43 of the 46 transformants that produced polyketides, the isolated polyketides had structures expected of the mutation(s). However, as observed in corresponding single mutations of the eryA gene in the single-plasmid system (12), additional products were observed with certain mutations. In most cases, the rapAT2 domain in module 3 recognized and processed both malonyl- and methylmalonyl-CoA and gave the expected 8-desmethyl analogs plus lesser amounts of the 8-methyl analogs. In a few cases, only the 8-methyl analogs were formed. The relaxed specificity of rapAT2 appears to be module-dependent because only the expected products were observed with the same rapAT2 sequence at modules 2 or 5 (12, 26). Likewise, when the KR of module 5 was replaced by either the rapDH/KR4 or rapDH/ER/KR1 domains to give the expected 4,5-anhydro and 5-deoxy analogs, respectively, 5-keto analogs were formed in addition to the expected products. This possibly results from transfer of the β-ketothioester intermediate from KR5 to ACP5 at a rate competitive with its reduction by the foreign rapKR4 domain. Because aberrant products were not observed with rapDH/KR4 at modules 2 or 6, the nonspecificity appears to be module-dependent. Finally, when the KR of module 2 was replaced with the rapDH/ER/KR1 domain to provide the 11-deoxy analogs, the 10,11-dehydro analogs were often observed as minor but significant products. Here, the intermediate is processed by the foreign rapKR and DH domains in module 2, but transfer of the 10,11-dehydro intermediate to KS3 must be competitive with the ER-catalyzed reduction. Interestingly, with rapDH/ER/KR1 in module 2 and rapAT2 in module 3, we did not detect 10,11-dehydro by-products. Either the levels produced were too low for detection with the methods employed, or the aberrant dehydro intermediate of module 2 was not processed by module 3 containing the rapAT2 substitution. Of the 18 transformants that did not produce polyketides at levels detectable in these experiments, 2 were double mutants and 16 were triple mutants; only 1 of these mutants was previously prepared in the single-plasmid system, where it also failed to produce a detectable polyketide (R. McDaniel, personal communication). We believe that, as previously reported with the single-plasmid system (12), an increased number of mutations resulted in a decrease in yield to a level undetectable by the analytical method used.
A major advantage of the multiple plasmid system is that once a number of plasmids encoding functional mutants of PKS subunits are available, they can be rapidly combined with one or more additional mutants to expand the library of polyketides. In one example, we prepared a single Cys-729 → Ala mutation at the KS1 domain of DEBS1 module 1 (11). The inactive KS1 prevents propagation of the starter unit and permits introduction of exogenous synthetic diketide thiol esters into positions 12 and 13 of the 14-membered macrolide product. The plasmid encoding the KS1 null allele of eryAI (Table 1) was introduced by cotransformation into S. lividans with the 1 eryAII mutant and 7 eryAIII mutants (Table 1) to provide 16 transformants. Treatment of each of these with a propyl-diketide N-acetylcysteine thioester, following the work of Jacobsen et al. (11), provided 11 novel 13-propyl polyketide analogs (Fig. 3, compounds 49–59). To prepare these same PKSs by the single-plasmid system would have required preparation of 16 individual mutants rather than the single KS1 null mutant used here. In another example, we leveraged our already prepared variants of eryAI and eryAII to prepare a small library of 12-membered macrolactones. It was previously shown in the eryA single-plasmid system that omission of module 6, along with fusion of module 5 to the thioesterase domain of DEBS3 (Fig. 1A) results in formation of a 12-membered macrolactone. We introduced a truncated eryAIII gene containing module 5-TE (27) (Table 1) into S. lividans and transformed the strain with permutations of the 4 eryAI and 2 eryAII variants described above. Of the 8 12-membered lactones that could have been produced, 5 were observed (compounds 44–48). Compound 44 results from combining the wild-type eryAI and eryAII genes with the module 5-TE construct, and has previously been prepared in the single-plasmid system; compounds 45–48 are novel products that more than double the number of known 12-membered macrolides.
Other Applications.
Other potential uses of the multiple plasmid system are as follows. (i) The expression and genetic engineering of very large PKS genes, such as those involved in the biosynthesis of rapamycin (14 modules) (4) or rifamycin (10 modules) (5), would be predicted. The ORFs for each of the subunits of these and other PKSs could be cloned and expressed in the manner used here to greatly simplify genetic manipulations and the structure/function analysis. (ii) Similarly, it would be desirable to mix PKS genes from entirely different pathways to facilitate production of heterologous and hybrid PKSs, as precedented by the work of Tang et al. involving hybrid erythromycin and picromycin PKS genes (L. Tang, Q.X., H. Fu, and R. McDaniel, unpublished work). Such work could include the thus far untested possibility of extending the carbon chain by the addition of modules. (iii) The PKS libraries generated could be leveraged and expanded by introducing genes for tailoring enzymes that oxidize, methylate, acylate, or glycosylate the product of the PKS. (iv) Finally, the method should be useful with any system consisting of multimodular proteins, such as the large family of nonribosomal peptide antibiotics. These important antibiotics are made by multifunctional synthetases, consisting of complexes of proteins containing between 1 and 11 modules, comparable to the modular PKSs (28).
The multiplasmid technology enables the realization of the full potential of modular PKSs, and thus libraries containing a complete repertoire of polyketides. The achievement of this objective requires only the construction of a limited number of highly expressing, productive single mutants that will assure adequate polyketide production when the mutations are combined. Because all the elements for producing an extraordinarily large polyketide library are contained within this facile system, there is neither need nor benefit to embark on developing more complicated, less reliable systems to reach the same objective.
Acknowledgments
We thank Luis Servin-Gonzalez for pB45; Mark Burlingame and Sally Ou for assistance with LC/MS and fermentations; and Robert McDaniel, Lu Liu, Mary Betlach, and David Hopwood for helpful discussions.
Abbreviations
- PKS
polyketide synthase
- AT
acyltransferase
- KR
ketoreductase
- DH
dehydratase, ER, enoylreductase
- ACP
acyl carrier protein
- DEBS
6-deoxyerythronolide B synthase
- 6dEB
6-deoxyerythronolide B
- KS
ketosynthase
References
- 1.Katz L, Donadio S. Annu Rev Microbiol. 1993;47:875–912. doi: 10.1146/annurev.mi.47.100193.004303. [DOI] [PubMed] [Google Scholar]
- 2.Cortes J, Haydock S F, Roberts G A, Bevitt D J, Leadlay P F. Nature (London) 1990;348:176–178. doi: 10.1038/348176a0. [DOI] [PubMed] [Google Scholar]
- 3.Donadio S, Staver M J, McAlpine J B, Swanson S J, Katz L. Science. 1991;252:675–679. doi: 10.1126/science.2024119. [DOI] [PubMed] [Google Scholar]
- 4.Schwecke T, Aparicio J F, Molnar I, Konig A, Khaw L E, Haydock S F, Oliynyk M, Caffrey P, Cortes J, Lester J B, et al. Proc Natl Acad Sci USA. 1995;92:7839–7843. doi: 10.1073/pnas.92.17.7839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.August P R, Tang L, Yoon Y J, Ning S, Muller R, Yu T W, Taylor M, Hoffmann D, Kim C G, Zhang X, et al. Chem Biol. 1998;5:69–79. doi: 10.1016/s1074-5521(98)90141-7. [DOI] [PubMed] [Google Scholar]
- 6.O’Hagan D. The Polyketide Metabolites. Chichester, U.K.: Ellis Horwood; 1991. [Google Scholar]
- 7.Kao C M, Pieper R, Cane D E, Khosla C. Biochemistry. 1996;35:12363–12368. doi: 10.1021/bi9616312. [DOI] [PubMed] [Google Scholar]
- 8.Liu L, Thamchaipenet A, Fu H, Betlach M, Ashley G. J Am Chem Soc. 1997;119:10553–10554. [Google Scholar]
- 9.McDaniel R, Kao C M, Fu H, Hevezi P, Gustafsson C, Betlach M, Ashley G, Cane D E, Khosla C. J Am Chem Soc. 1997;119:4309–4310. [Google Scholar]
- 10.Marsden A F, Wilkinson B, Cortés J, Dunster N J, Staunton J, Leadlay P F. Science. 1998;279:199–202. doi: 10.1126/science.279.5348.199. [DOI] [PubMed] [Google Scholar]
- 11.Jacobsen J R, Hutchinson C R, Cane D E, Khosla C. Science. 1997;277:367–369. doi: 10.1126/science.277.5324.367. [DOI] [PubMed] [Google Scholar]
- 12.McDaniel R, Thamchaipenet A, Gustafsson C, Fu H, Betlach M, Betlach M, Ashley G. Proc Natl Acad Sci USA. 1999;96:1846–1851. doi: 10.1073/pnas.96.5.1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Patten P A, Howard R J, Stemmer W P C. Curr Opin Biotechnol. 1997;8:724–733. doi: 10.1016/s0958-1669(97)80127-9. [DOI] [PubMed] [Google Scholar]
- 14.Guenzi E, Galli G, Grgurina I, Pace E, Ferranti P, Grandi G. J Biol Chem. 1998;273:14403–14410. doi: 10.1074/jbc.273.23.14403. [DOI] [PubMed] [Google Scholar]
- 15.Pieper R, Luo G, Cane D E, Khosla C. Nature (London) 1995;378:263–266. doi: 10.1038/378263a0. [DOI] [PubMed] [Google Scholar]
- 16.Ziermann, R. & Betlach, M. (1999) J. Ind. Microbiol. Biotechnol., in press.
- 17.Kao C M, Katz L, Khosla C. Science. 1994;265:509–512. doi: 10.1126/science.8036492. [DOI] [PubMed] [Google Scholar]
- 18.Raynal A, Tuphile K, Gerbaud C, Luther T, Guerineau M, Pernodet J. Mol Microbiol. 1998;28:333–342. doi: 10.1046/j.1365-2958.1998.00799.x. [DOI] [PubMed] [Google Scholar]
- 19.Ziermann R, Betlach M. BioTechniques. 1999;26:106–110. doi: 10.2144/99261st05. [DOI] [PubMed] [Google Scholar]
- 20.Hopwood D A, Bibb M J, Chater K F, Kieser T, Bruton C J, Kieser H M, Lydiate D J, Smith C P, Ward J M, Schrempf H. Genetic Manipulation of Streptomyces: A Laboratory Manual. Norwich, U.K.: John Innes Foundation; 1985. [Google Scholar]
- 21.Bierman M, Logan R, O’Brien K, Seno E T, Rao R N, Schoner B E. Gene. 1992;116:43–49. doi: 10.1016/0378-1119(92)90627-2. [DOI] [PubMed] [Google Scholar]
- 22.Servin-Gonzalez L, Sampieri A I, Cabello J, Galvan L, Jarez V, Castro C. Microbiology. 1995;141:2499–2510. doi: 10.1099/13500872-141-10-2499. [DOI] [PubMed] [Google Scholar]
- 23.Rajgarhia V B, Strohl W R. J Bacteriol. 1997;179:2690–2696. doi: 10.1128/jb.179.8.2690-2696.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smokvina T, Mazodier P, Boccard F, Thompsom C J, Guerineau M. Gene. 1990;94:53–59. doi: 10.1016/0378-1119(90)90467-6. [DOI] [PubMed] [Google Scholar]
- 25.Baltz R. Biotechnology of Antibiotics. 2nd. Ed. New York: Marcel Dekker; 1997. pp. 49–62. [Google Scholar]
- 26.Ruan X, Pereda A, Stassi D L, Zeidner D, Summers R G, Jackson M, Shivakumar A, Kakavas S, Staver M J, Donadio S, Katz L. J Bacteriol. 1997;179:6416–6425. doi: 10.1128/jb.179.20.6416-6425.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kao C M, Luo G, Katz L, Cane D E, Khosla C. J Am Chem Soc. 1995;117:9105–9106. [Google Scholar]
- 28.Konz D, Marahiel M A. Chem Biol. 1999;6:39–48. doi: 10.1016/S1074-5521(99)80002-7. [DOI] [PubMed] [Google Scholar]