Abstract
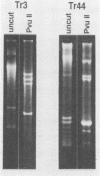
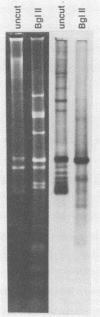
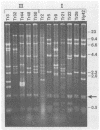
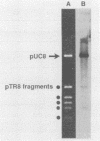
Mitochondrial DNA (mtDNA) was purified from 12 isolates of the Trichoderma viride aggregate and found to be, on the average, 32.7 kb in size. Plasmids were present in the mtDNA preparations from 8 of 12 strains of T. viride examined. Plasmids in four of the strains produced ladderlike banding patterns on gels, and these plasmids were studied in detail. The ladderlike patterns were produced by single molecules that were supercoiled to various degrees. Plasmids from two of the strains do not have homology with the mtDNA but do have a limited amount of homology with each other. No phenotype could be associated with the presence of a plasmid. Restriction endonuclease digestion of the mtDNAs produced patterns in which the presence or absence of certain fragments correlated with the classification of the strains into T. viride group I or II. Phenetic cluster analysis and parsimony analysis of the fragment patterns produced groups that corresponded to T. viride groups I and II. The fragment patterns were very diverse, with nearly all strains having a unique pattern. However, two strains of T. viride group I from widely different geographical locations did have identical restriction patterns for all the enzymes used in this study. This result indicates that it may not be possible to use mtDNA restriction patterns alone to identify Trichoderma strains.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bruns T. D., Palmer J. D., Shumard D. S., Grossman L. I., Hudspeth M. E. Mitochondrial DNAs of Suillus: three fold size change in molecules that share a common gene order. Curr Genet. 1988;13(1):49–56. doi: 10.1007/BF00365756. [DOI] [PubMed] [Google Scholar]
- Collins R. A., Saville B. J. Independent transfer of mitochondrial chromosomes and plasmids during unstable vegetative fusion in Neurospora. Nature. 1990 May 10;345(6271):177–179. doi: 10.1038/345177a0. [DOI] [PubMed] [Google Scholar]
- May G., Taylor J. W. Independent transfer of mitochondrial plasmids in Neurospora crassa. Nature. 1989 May 25;339(6222):320–322. doi: 10.1038/339320a0. [DOI] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Taylor J. W., Smolich B. D., May G. An evolutionary comparison of homologous mitochondrial plasmid DNAs from three Neurospora species. Mol Gen Genet. 1985;201(2):161–167. doi: 10.1007/BF00425654. [DOI] [PubMed] [Google Scholar]
- Taylor J. W., Smolich B. D. Molecular cloning and physical mapping of the Neurospora crassa 74-OR23-1A mitochondrial genome. Curr Genet. 1985;9(7):597–603. doi: 10.1007/BF00381173. [DOI] [PubMed] [Google Scholar]