Abstract
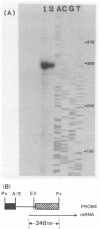
An allosteric L-(+)-lactate dehydrogenase gene of Lactobacillus casei ATCC 393 was cloned in Escherichia coli, and the nucleotide sequence of the gene was determined. The gene was composed of an open reading frame of 981 bp, starting with a GTG codon and ending with a TAA codon. The sequences for the promoter and ribosome binding site were identified, and a sequence for a structure resembling a rho-independent transcription terminator was also found.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barstow D. A., Clarke A. R., Chia W. N., Wigley D., Sharman A. F., Holbrook J. J., Atkinson T., Minton N. P. Cloning, expression and complete nucleotide sequence of the Bacillus stearothermophilus L-lactate dehydrogenase gene. Gene. 1986;46(1):47–55. doi: 10.1016/0378-1119(86)90165-4. [DOI] [PubMed] [Google Scholar]
- Chassy B. M., Gibson E., Giuffrida A. Evidence for extrachromosomal elements in Lactobacillus. J Bacteriol. 1976 Sep;127(3):1576–1578. doi: 10.1128/jb.127.3.1576-1578.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke A. R., Atkinson T., Holbrook J. J. From analysis to synthesis: new ligand binding sites on the lactate dehydrogenase framework. Part I. Trends Biochem Sci. 1989 Mar;14(3):101–105. doi: 10.1016/0968-0004(89)90131-x. [DOI] [PubMed] [Google Scholar]
- Clarke A. R., Atkinson T., Holbrook J. J. From analysis to synthesis: new ligand binding sites on the lactate dehydrogenase framework. Part II. Trends Biochem Sci. 1989 Apr;14(4):145–148. doi: 10.1016/0968-0004(89)90147-3. [DOI] [PubMed] [Google Scholar]
- Clarke A. R., Smith C. J., Hart K. W., Wilks H. M., Chia W. N., Lee T. V., Birktoft J. J., Banaszak L. J., Barstow D. A., Atkinson T. Rational construction of a 2-hydroxyacid dehydrogenase with new substrate specificity. Biochem Biophys Res Commun. 1987 Oct 14;148(1):15–23. doi: 10.1016/0006-291x(87)91070-9. [DOI] [PubMed] [Google Scholar]
- Eventoff W., Rossmann M. G., Taylor S. S., Torff H. J., Meyer H., Keil W., Kiltz H. H. Structural adaptations of lactate dehydrogenase isozymes. Proc Natl Acad Sci U S A. 1977 Jul;74(7):2677–2681. doi: 10.1073/pnas.74.7.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley D. K., McClure W. R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983 Apr 25;11(8):2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensel R., Mayr U., Stetter K. O., Kandler O. Comparative studies of lactic acid dehydrogenases in lactic acid bacteria. I. Purification and kinetics of the allosteric L-lactic acid dehydrogenase from Lactobacillus casei ssp. casei and Lactobacillus curvatus. Arch Microbiol. 1977 Feb 4;112(1):81–93. doi: 10.1007/BF00446658. [DOI] [PubMed] [Google Scholar]
- Hensel R., Mayr U., Woenckhaus C. Affinity labelling of the allosteric site of the L-lactate dehydrogenase of Lactobacillus casei. Eur J Biochem. 1983 Sep 15;135(2):359–365. doi: 10.1111/j.1432-1033.1983.tb07662.x. [DOI] [PubMed] [Google Scholar]
- Hensel R., Mayr U., Yang C. Y. The complete primary structure of the allosteric L-lactate dehydrogenase from Lactobacillus casei. Eur J Biochem. 1983 Aug 15;134(3):503–511. doi: 10.1111/j.1432-1033.1983.tb07595.x. [DOI] [PubMed] [Google Scholar]
- Kunai K., Machida M., Matsuzawa H., Ohta T. Nucleotide sequence and characteristics of the gene for L-lactate dehydrogenase of Thermus caldophilus GK24 and the deduced amino-acid sequence of the enzyme. Eur J Biochem. 1986 Oct 15;160(2):433–440. doi: 10.1111/j.1432-1033.1986.tb09991.x. [DOI] [PubMed] [Google Scholar]
- Mayr U., Hensel R., Deparade M., Pauly H. E., Pfleiderer G., Trommer W. E. Structure-function relationship in the allosteric L-lactate dehydrogenases from Lactobacillus casei and Lactobacillus curvatus. Eur J Biochem. 1982 Sep 1;126(3):549–558. doi: 10.1111/j.1432-1033.1982.tb06816.x. [DOI] [PubMed] [Google Scholar]
- Melançon P., Leclerc D., Destroismaisons N., Brakier-Gingras L. The anti-Shine-Dalgarno region in Escherichia coli 16S ribosomal RNA is not essential for the correct selection of translational starts. Biochemistry. 1990 Apr 3;29(13):3402–3407. doi: 10.1021/bi00465a037. [DOI] [PubMed] [Google Scholar]
- Minowa T., Iwata S., Sakai H., Masaki H., Ohta T. Sequence and characteristics of the Bifidobacterium longum gene encoding L-lactate dehydrogenase and the primary structure of the enzyme: a new feature of the allosteric site. Gene. 1989 Dec 21;85(1):161–168. doi: 10.1016/0378-1119(89)90476-9. [DOI] [PubMed] [Google Scholar]
- Moran C. P., Jr, Lang N., LeGrice S. F., Lee G., Stephens M., Sonenshein A. L., Pero J., Losick R. Nucleotide sequences that signal the initiation of transcription and translation in Bacillus subtilis. Mol Gen Genet. 1982;186(3):339–346. doi: 10.1007/BF00729452. [DOI] [PubMed] [Google Scholar]
- Ono M., Matsuzawa H., Ohta T. Nucleotide sequence and characteristics of the gene for L-lactate dehydrogenase of Thermus aquaticus YT-1 and the deduced amino acid sequence of the enzyme. J Biochem. 1990 Jan;107(1):21–26. doi: 10.1093/oxfordjournals.jbchem.a123004. [DOI] [PubMed] [Google Scholar]
- Platt T. Transcription termination and the regulation of gene expression. Annu Rev Biochem. 1986;55:339–372. doi: 10.1146/annurev.bi.55.070186.002011. [DOI] [PubMed] [Google Scholar]
- Rhee S. K., Pack M. Y. Effect of environmental pH on fermentation balance of Lactobacillus bulgaricus. J Bacteriol. 1980 Oct;144(1):217–221. doi: 10.1128/jb.144.1.217-221.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprengart M. L., Fatscher H. P., Fuchs E. The initiation of translation in E. coli: apparent base pairing between the 16srRNA and downstream sequences of the mRNA. Nucleic Acids Res. 1990 Apr 11;18(7):1719–1723. doi: 10.1093/nar/18.7.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas T. D., Ellwood D. C., Longyear V. M. Change from homo- to heterolactic fermentation by Streptococcus lactis resulting from glucose limitation in anaerobic chemostat cultures. J Bacteriol. 1979 Apr;138(1):109–117. doi: 10.1128/jb.138.1.109-117.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas T. D., Turner K. W., Crow V. L. Galactose fermentation by Streptococcus lactis and Streptococcus cremoris: pathways, products, and regulation. J Bacteriol. 1980 Nov;144(2):672–682. doi: 10.1128/jb.144.2.672-682.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Wallace R. B., Miyada C. G. Oligonucleotide probes for the screening of recombinant DNA libraries. Methods Enzymol. 1987;152:432–442. doi: 10.1016/0076-6879(87)52050-x. [DOI] [PubMed] [Google Scholar]
- Wilks H. M., Hart K. W., Feeney R., Dunn C. R., Muirhead H., Chia W. N., Barstow D. A., Atkinson T., Clarke A. R., Holbrook J. J. A specific, highly active malate dehydrogenase by redesign of a lactate dehydrogenase framework. Science. 1988 Dec 16;242(4885):1541–1544. doi: 10.1126/science.3201242. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]