Abstract
The fidelity of protein synthesis depends on specific tRNA aminoacylation by aminoacyl-tRNA synthetase enzymes, which in turn depends on the recognition of the identity of particular nucleotides and structural features in the substrate tRNA. These features generally reside within the acceptor helix, the anticodon stem-loop, and in some systems the variable pocket of the tRNA. In the alanine system, fidelity is ensured by a G⋅U wobble base pair located at the third position within the acceptor helix of alanine tRNA. We have investigated the activity of mutant alanine tRNAs to explore the mechanism of enzyme recognition. Here we show that the mismatched pair C-C is an excellent substitute for G⋅U in alanine-tRNA-knockout cells. A structural investigation by NMR spectroscopy of the C-C RNA acceptor end reveals that the two cytosines are intercalated into the helix, and that C-C exists in multiple conformations. Structural heterogeneity also is present in the wild-type G⋅U RNA, whereas inactive Watson–Crick helices are structurally rigid. The correlation between functional and structural data suggests that the G⋅U pair provides a distinctive structure and a point of deformability that allow the tRNA acceptor end to fit into the active site of the alanyl-tRNA synthetase. Fidelity is ensured because noncognate and inactive mutant tRNAs are bound in the active site in an incorrect conformation that reduces enzymatic activity.
Aminoacylation of Escherichia coli tRNAAla by its cognate alanyl-tRNA synthetase (AlaRS) enzyme depends on a G⋅U wobble base pair§ in the acceptor helix (Fig. 1); this feature is critical for aminoacylation and can confer partial alanine acceptance on other noncognate tRNAs (1, 2). The features that allow its recognition are likely to include direct interactions with distinctive atomic groups and indirect recognition of the structural and dynamic information encoded in the sequence of the G⋅U pair and its immediate context. The G⋅U wobble base pair provides specific recognition signals in numerous RNAs, ranging from ribosomal RNA and splicing regulatory signals (3) to RNA enzymes (4). A detailed understanding of recognition mechanisms of the G⋅U pair is still lacking. Therefore, tRNAAla provides not only an important system to understand how fidelity in protein synthesis is ensured, but also a paradigm to dissect the features that allow such a widespread role for the G⋅U pair as a distinctive recognition tag.
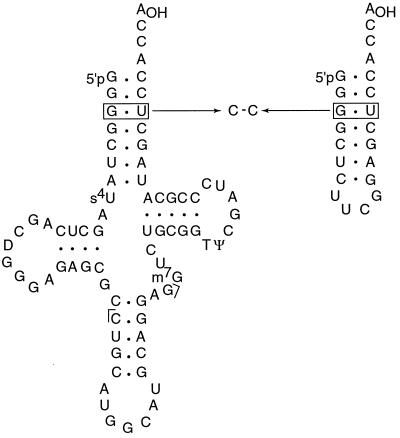
Figure 1.
Diagram of tRNAAla and its tetraloop acceptor minihelix RNA showing the location of the G⋅U wobble base pair and the C-C substitution at the third position. The standard residue numbers of nucleotide at the third position are 3 and 70 (e.g., G3 and U70). Brackets flanking the anticodon indicate endpoints of the Northern blot hybridization probe.
Here we report the remarkable observation that replacing the G⋅U wobble pair with a C-C mispair preserves tRNAAla aminoacylation in vivo. An acceptor minihelix RNA containing the C-C substitution has been analyzed by high-resolution NMR spectroscopy and compared with wild-type G⋅U RNA and inactive G⋅C and A⋅U mutant RNAs. The data demonstrate that the two cytosines are intercalated within the acceptor helix, and that C-C exists in multiple conformations in equilibrium on the μs-ms time scale. The structure at and around G⋅U and C-C is significantly more mobile than that of Watson–Crick base pair substitution mutants. The correlation between structural and functional data suggests that the G⋅U pair provides a distinctive structure and a point of deformability that allow the tRNA acceptor end and AlaRS to adopt a structure that is optimal for catalysis by induced fit.
Methods
Aminoacyl-tRNA Determination.
E. coli K45slΔArelA knockout cells (5) expressing the indicated tRNAAla gene from plasmid pGFIB were grown in 2× YT medium containing 100 μg/ml ampicillin, 20 μg/ml chloramphenicol, and 25 μg/ml kanamycin at 33°C to mid-log phase (0.6 OD550; ≈2 × 108 cells/ml), RNA was isolated under acidic conditions, and the relative level of aminoacyl-tRNA and deacyl-tRNA was determined by two independent analyses. First, for direct analysis of aminoacyl-tRNAAla, half of an RNA sample was left untreated and the other half was treated with Tris base to strip aminoacyl-tRNAs (0.5 M Tris, pH 9.0, 30 min, 37°C). RNA samples (0.05 OD260) were fractionated on a 6.5% polyacrylamide, 8 M urea gel (pH 5.2) at 4°C, transferred to Nytran Plus membrane, crosslinked, and hybridized at 35°C with 32P-labeled DNA probe complementary to position 28 to 45 of tRNAAla. Second, for direct analysis of deacyl-RNAAla, half of an RNA sample was left untreated and the other half was subjected to periodate oxidation and beta elimination (40 mM NaIO4, 90 min, 0°C then 330 mM rhamnose, 30 min, 0°C followed by ethanol precipitation then resuspension in 1 M lysine, 60 min, 45°C followed by ethanol precipitation) to remove the terminal nucleoside of deacyl-tRNA. RNA samples (0.05 OD260) were fractionated on an 8% polyacrylamide urea DNA sequencing gel (pH 8.3) at ≈80°C (Amersham Pharmacia) and subjected to a Northern blot analysis as above. The gel profiles were quantitated by using a Molecular Dynamics Storm 860 instrument and imagequant 4.1 software (5, 6).
Western Blot Analysis.
Cultures grown to 0.85–1.0 OD595 were harvested, and the cells were broken with lysozyme-EDTA at 4°C. The lysate was clarified by centrifugation, and the supernatant was diluted to 1 mg/ml by Bradford assay with BSA as standard. Serial dilutions were separated by SDS-6% PAGE, transblotted to nitrocellulose membrane, probed with rabbit polyclonal AlaRS antibody and goat anti-rabbit IgG alkaline phosphatase conjugate, and color-developed with an alkaline phosphatase conjugate substrate. The dilution end-point of AlaRS in independent assays of the same mutant tRNA is reproducible to within 2-fold (7).
Knockout Cell Analysis.
E. coli K45slΔArelA knockout cells lack both tRNAAla (GGC anticodon) isoacceptor genes and one of three tRNAAla (UGC anticodon) isoacceptor genes. Knockout cells grow in glucose minimal medium only when active tRNAAla (GGC anticodon) is produced from a plasmid, although they grow in LB rich medium without the tRNA (5). Competent K45slΔArelA cells were prepared after growing in LB broth to 0.4 OD550. The cells were transformed with plasmid pGFIB producing the indicated tRNA and grown in LB broth at 37°C for 1 hr. An aliquot (100 μl) of the transformation mix was spread on glucose minimal medium plus ampicillin plates and incubated at 34°C for 2 days. One transformant colony was picked with a loop into 1 ml of minimal medium, vortexed vigorously, mirocentrifuged, and resuspended in 1 ml of minimal medium. Agar plates containing ampicillin and either glucose minimal or LB medium were spotted with 3 μl of serial dilutions of the cell suspension and then incubated at 34°C for 44 hr.
NMR Spectroscopy.
The 22-mer acceptor end minihelix (Fig. 1) and each of three mutants were synthesized by in vitro transcription using T7 RNA polymerase and purified by using denaturing gel electrophoreses as described (8). The concentrations of samples for NMR spectroscopy were 1–1.5 mM in 10 mM phosphate buffer at pH 6–6.5. NMR spectra were acquired with Bruker AMX-500 or DMX-600 spectrometers and processed by using Felix 2.30 (Micron Separations). Acquisition and processing parameters were essentially as reported in other publications from our laboratory (9). Two-dimensional nuclear Overhauser and exchange spectroscopy (NOESY) spectra in H2O were recorded by using both Watergate (10) and 11 solvent suppression at a mixing time of 150 ms at 4°C to reduce the rate of exchange with solvent of imino and amino resonances. Spectra in D2O were acquired at 27°C; additional spectra at long mixing time (300 ms) also were recorded at different temperatures (17°C and 37°C) to facilitate identification of partially overlapped cross-peaks. A NOESY-build-up series (mixing times 75, 150, and 300 ms) was recorded to allow reliable quantification of cross-relaxation rates for the C-C RNA. Total correlation spectroscopy spectra were recorded with a mixing time of 80 ms. Spectral assignments for all base and anomeric protons and most sugar H2′, H3′, and H4′ resonances were obtained by using standard methods (11).
Structural Modeling.
Experimental NOE-based distance constraints were used to model the conformation of the C-C minihelix. Because spectral assignments were essentially identical between C-C and G⋅U RNAs, with the exception of the third position and the two base pairs flanking it, the list used to calculate the structure of the G⋅U containing acceptor end minihelix sequence (9) was used to prepare an equivalent constraint set for the C-C mutant. The model of the C-C acceptor end minihelix was calculated as in any formal structure determination from our group by using a constraint set that was identical to that used for the G⋅U structure, with the exceptions of the C-C site and the immediately surrounding base pairs. Experimental NOE-based distance constraints for the C-C mispair and its neighboring base pairs were extracted from NOESY data recorded at different mixing times as described (9). NOESY data recorded at different temperatures allowed the resolution of partially overlapped cross-peaks and the identification of most sugar H2′, H3′, and H4′ resonances. Structure calculation methods and constraint statistics were very similar to those already reported for the G⋅U structure (9).
Results
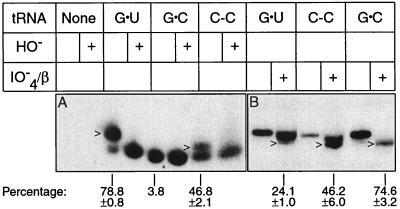
We have investigated how fidelity in protein synthesis is ensured in the alanine system by studying the activity of mutant tRNAs in vivo. Extensive analyses revealed that C-A and G-A mispairs could partially substitute for G⋅U in amber suppressor tRNA and knockout tRNA systems (5, 12, 13). In the present work, we first assessed the aminoacylation efficiency of other mutant tRNAs. The steady-state concentration of cellular aminoacyl-tRNA and deacyl-tRNA reflects the balance between rates of tRNA aminoacylation and turnover during protein synthesis. We assay the steady-state level of alanyl-tRNAAla in tRNAAla knockout cells by a Northern blot analysis of a polyacrylamide gel. The aminoacyl bond is relatively stable in the acidic conditions of analysis and protonation of the amino acid causes aminoacyl-tRNA to migrate more slowly than deacyl-tRNA during gel electrophoresis. The Northern hybridization probe covers the GGC anticodon of tRNAAla and thus hybridizes this tRNAAla isoacceptor in large preference to other cellular tRNAs. Fig. 2A shows that the steady-state level of alanyl-C-C tRNAAla was 46.8 ± 2.1% in comparison with 78.8 ± 0.8% for alanyl-G⋅U tRNAAla. The identity of alanyl-tRNAAla was confirmed by its sensitivity to mild alkaline hydrolysis (Fig. 2). The reduced separation between aminoacyl and deacyl forms of C-C tRNAAla relative to G⋅U tRNAAla is a property of the mismatch and also is observed in the amber suppressor tRNA (13). The use of periodate oxidation to remove the 3′ terminal adenosine distinctive of deacyl tRNAAla provided an independent assessment of the fractions of chains present as aminoacyl-tRNAAla (Fig. 2B).
Figure 2.
Steady-state level of alanyl-tRNAAla. Total low molecular weight RNA was isolated from knockout cells producing the indicated form of tRNAAla under acidic conditions to preserve aminoayl-tRNA. (A) Direct identification of aminoacyl-tRNA and deacyl-tRNA by fractionation on an acidic polyacrylamide gel and detected by Northern blot analysis. HO− indicates deacylation by Tris base. The arrowhead indicates alanyl-tRNAAla. The percent aminoacyl-tRNAAla (100 × aminoacyl-tRNAAla/{aminoacyl-tRNAAla + deacyl-tRNAAla}) shown is the average and standard error of three determinations, except the G⋅C sample is based on one measurement. (B) Direct identification of deacyl-tRNAAla by 3′ nucleoside resection (giving N-1 length chains) and resolution from full-length (N-length chains) tRNAAla on a DNA sequencing gel. IO4−/β indicates treatment with periodate and β elimination. The arrowhead indicates N-1 tRNAAla. The percent deacyl-tRNAAla [100 × (N-1/{N-1 + N})] shown is the average and standard error of four determinations (two for G⋅C).
Aminoacylation measurements in the knockout system may be more reliable than analogous measurements in the amber suppressor system where aminoacyl-tRNA turnover is lower because of a reduced abundance of UAG codons in messenger RNAs. However, the levels of alanyl-tRNAAla observed in the amber suppressor system for C-C tRNAAla (59.1 ± 6.1%) and G⋅U tRNAAla (82.1 ± 4.8%) (5, 13) are not statistically different at the 95% confidence interval from the values reported here for the knockout system. Attempts to determine alanyl-tRNAAla levels for C-A and G-A tRNAs in the knockout system have been hampered by severe tRNA aggregation during gel electrophoresis, but aggregation is not observed for these tRNAs in the amber suppressor system. Previous analysis showed that amber suppressor C-C tRNAAla accepts glutamine and alanine in vivo (12). The partial glutamine acceptance results from the U in the middle anticodon position (CUA) of amber suppressor tRNA and is curtailed by the GGC anticodon of tRNAAla. Cell viability considerations make it very likely that G⋅U tRNAAla and C-C tRNAAla molecules are specifically acylated with alanine, although this assumption is not established by the Northern blot analysis (see below).
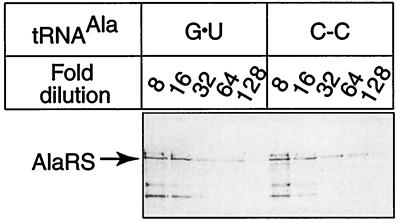
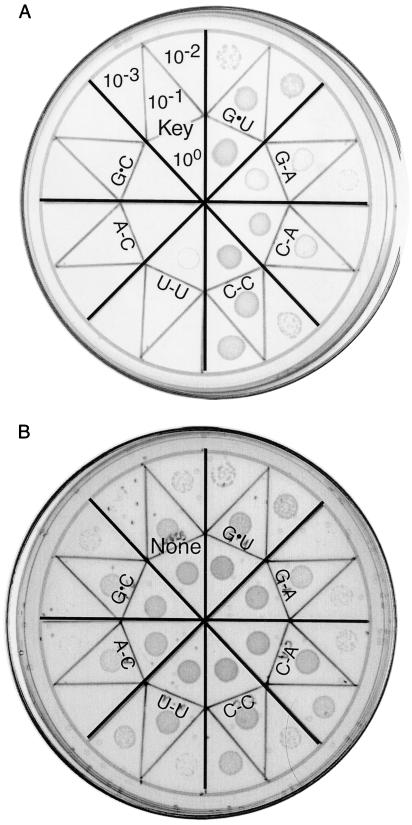
We next considered the possibility that AlaRS is up-regulated in a tRNA-sequence-dependent manner such that cells expressing C-C tRNAAla contain increased amounts of enzyme. If this occurred, aminoacylation of C-C tRNAAla could result from changes in AlaRS protein levels rather than from the intrinsic substrate quality of the mutant tRNA. To address this possibility, we used a Western blot analysis of AlaRS to determine cellular levels of the protein. Fig. 3 shows that AlaRS protein was present in similar levels in separate cell cultures expressing G⋅U tRNAAla and C-C tRNAAla. This result confirms that the observed level of alanyl-C-C tRNAAla reflects the molecule’s substantial substrate quality rather than enzyme overproduction compensating for impaired tRNA acceptor capacity. The final functional test of tRNAAla relied on the availability of E. coli tRNAAla knockout cells (5). Fig. 4 shows that C-C tRNAAla supported growth of knockout cells nearly as well as G⋅U tRNAAla. In view of the cell’s demand for proteins with accurate primary sequences, the result also confirms the alanine acceptor specificity of C-C tRNAAla. Fig. 4 also shows that C-C tRNAAla performed better in this assay than did tRNAAla with C-A or G-A substitutions, the other known G⋅U mimics. The negative controls where cells did not grow are tRNAs with U-U, A-C, and G⋅C substitutions and the plasmid without a tRNA gene insert.
Figure 3.
Western blot analysis of AlaRS in cells expressing the indicated tRNAAla. Each crude extract (1.0 mg/ml) shows a dilution end-point of 1/64. The arrow marks the position of AlaRS determined in other blots.
Figure 4.
Growth properties of tRNAAla knockout cells expressing tRNAAla with G⋅U or the indicated substitution. Knockout cells do not grow on minimal medium unless active tRNAAla is produced from a plasmid. Agar plates containing (A) glucose minimal medium or (B) LB rich medium were spotted with serial dilutions of cultures expressing the indicated tRNAAla. As the growth patches in A developed, cells expressing G⋅U tRNA grew better than those with C-C tRNA, but this difference is more subtle after 44 hr (shown above) when growth also becomes apparent for cells expressing G-A and C-A tRNAs. The sector in A corresponding to the plasmid without a tRNA gene insert shows the dilution key. The speckles in B are occluded air bubbles.
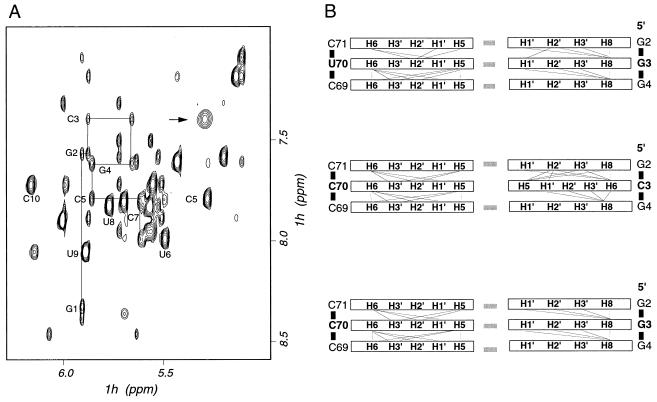
The results presented above emphasize the ability of C-C to mimic G⋅U. To investigate how the C-C pair can functionally substitute for the G⋅U pair, we used NMR spectroscopy to compare the structure of acceptor end minihelices containing either G⋅U or C-C mispairs. Acceptor stem helices containing G⋅C or A⋅U base pairs also were studied to provide comparison with inactive variants. An exhaustive investigation of the structure of the wild-type (G⋅U) acceptor end minihelix already has been reported (9). The NMR data were used in that study to determine the structure for the wild-type acceptor end RNA oligonucleotide to high precision. Assignments of the base, anomeric, and sugar (H2′, H3′, and H4′) regions of the NMR spectra of the active (C-C) and inactive (G⋅C and A⋅U) mutant RNAs were obtained by analyzing NOESY and total correlation spectroscopy spectra by using standard procedures (11). With the exception of position three and the base pairs that immediately surround that site, NMR spectral assignments were essentially identical for each of the four RNAs examined. This result suggests very strongly that the structure of the acceptor end minihelix is unperturbed by the substitutions outside of the substitution site itself and its immediate surroundings. Even at position three, we observed NOE connectivities very similar to those expected for a fully base-paired helix not only for the inactive G⋅C and A⋅U variants (as expected) and for G⋅U RNA (as previously reported) (9), but also for the C-C RNA (Fig. 5). This result demonstrates that the Watson–Crick base pairs on either side of the C-C substitution are in the same conformation as in the wild-type G⋅U minihelix and the inactive Watson–Crick paired RNAs, and that both C3 and C70 are intercalated within the double helix. Unfortunately, we were unable to obtain direct experimental evidence supporting the formation of a non-Watson–Crick base pair between C3 and C70 under the conditions of the NMR experiments (pH 6–6.5). When the pH was lowered to stabilize possible protonated C-C paired structures, we detected large changes in the NMR spectra suggestive of a global change in RNA conformation. A similar observation was reported in the study of a DNA duplex containing the C-C mismatch (14).
Figure 5.
(A) Region of the NOESY spectrum of the C-C minihelix containing signals from base and anomeric RNA resonances. Data were collected at 27°C in 10 mM phosphate buffer (pH 6–6.5) and 300-ms mixing time. Resonances belonging to nucleotides at the 5′ end of the minihelix are connected by a thin line to explicitly show sequential NOE connectives that were used to obtain spectral assignments. The broad base H5-H6 resonance of cytosine C3 is indicated by an arrow. The resonance of C70 overlaps other resonances at this temperature. However, the broad H5-H6 resonances of C70 was observed at a lower temperature. (B) Schematic diagram of NOE connectivities patterns observed for G⋅U, C-C, and G⋅C minihelices. Hydrogen bonding between base pairs is indicated by gray rectangles.
The similarities in the patterns of NOE interactions between C-C and G⋅U minihelices as well as inactive G⋅C and A⋅U variants (Fig. 5B) reveal that the two structures are very similar to each other and to A-form RNA. To obtain further insight into the structure at and around the C-C mispair, we modeled the structure of the C-C minihelix. With the exception of positions two, three, and four, we used the same set of NOE distance constraints used in the calculation of the G⋅U structure (9). The constraints involving base pairs two, three, and four were obtained experimentally as described in Methods, and the computational procedure was not different from a formal structure determination. No assumption was introduced that would bias in any way the result of the calculation. Despite the absence of base-pairing constraints for the C-C RNA, the global structure of the acceptor end minihelix was not perturbed to any significant extent by the substitution. However, significant local structural differences were present between the G⋅U and C-C structures and also between each of the mutant structures and A-form RNA.
A striking result was the observation of conformational exchange at and near the C-C mispair. Conformational exchange in the μs-ms time scale results in very substantial broadening of the resonances corresponding to the base protons of both C residues (Fig. 5A). Broadening is not likely to result from protonation exchange involving C3 and/or C70 in the physiological pH range. The base resonances of C3 remained significantly broader than other pyrimidine resonances between pH 6–6.5 (Fig. 5A) and pH 8 (not shown). Similar observations also were reported in the study of a C-C DNA duplex (14) and provide clear indication that the two bases are experiencing conformational exchange between distinct conformers. In contrast, we found no evidence of conformational exchange for the Watson–Crick-paired, inactive mutants (data not shown). The analysis of multidimensional data on isotopically labeled RNA recorded on the wild-type G⋅U minihelix had allowed us to conclude that the conformation of the phosphodiester backbone of G3 was in exchange between distinctive conformations (9). The functional implications of these observations are described below.
Discussion
The G⋅U base pair within the acceptor helix marks tRNAAla for aminoacylation by its cognate AlaRS enzyme. The G⋅U pair presents a distinctive array of hydrogen bond donors and acceptors in the major and minor grooves that could be specifically recognized by complementary functionalities in AlaRS (direct recognition). Alternatively, or in addition, unusual structural features of the G⋅U pair may influence AlaRS binding to the acceptor helix and/or position the 3′ terminal nucleotide of tRNAAla in the enzyme active site for catalysis (indirect recognition). It will remain impossible to fully understand recognition until the structures of the tRNAAla-AlaRS complex and of several functional mutants become available. Nevertheless, the structural and functional data presented here point toward the importance of indirect recognition.
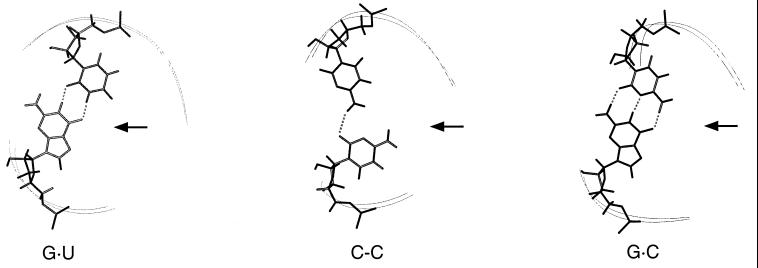
Here we show that tRNAAla with a C-C mispair is functionally very similar to G⋅U tRNAAla. Previous data had shown that tRNAAla with C-A and G-A substitutions are substantially functional, whereas G⋅C, A⋅U, and U⋅G substitutions are inactive (5, 12, 13). The new results show that C-C is the best mimic of G⋅U (Fig. 4). The NMR data demonstrate that each of two cytosines at the third position is intercalated in the double helix and that base-stacking interactions within each strand typical of double-stranded nucleic acids are preserved throughout the mutant acceptor stem. An important observation concerns the glycosidic bond angle, which was found in the anticonformation for both nucleotides. Taken together, these results demonstrate unambiguously that the exocyclic amino group of each cytosine is in the major groove of the acceptor helix (Fig. 6). The exocyclic amino group of the guanosine within the G⋅U pair of tRNAAla was shown to be critical for aminoacylation; the requirement for this amino group was proposed to result from just its direct recognition by AlaRS (15, 16). However, our data show that the G⋅U wobble pair and its C-C mimic do not project common atomic groups into the major or minor groove of double-helical RNA that could guide direct recognition by AlaRS (Fig. 6). This is also likely to be true for the other functional substitutions, C-A and G-A, but remains to be proven rigorously. Therefore, the present data provide strong support for the notion that indirect recognition of the structure of the acceptor end is an important element in how AlaRS aminoacylates the correct tRNA and discriminates against noncognate substrates.
Figure 6.
View of the C-C mispair in a model structure calculated by using the experimental NMR constraints and comparison with corresponding structures of the active wild-type minihelix (G⋅U) and inactive (G⋅C) mutant. Arrows point to the major groove side of each structure. Shown are the nucleotide pairings at position three and the backbones of positions 2–5. Although we found no direct evidence for hydrogen bonding of the C-C mispair, the hydrogen bond indicated in the figure was observed in several structures calculated by using the experimental constraints. Other hydrogen bonding schemes are consistent with the experimental data and were observed in other calculated structures, but all structures had the amino group of both C residues in the major groove.
Indirect recognition would require a mechanism to transmit the structural information encoded within the G⋅U pair to the enzyme active site. Discrimination in the alanine system occurs both at the level of Km and kcat (17), suggesting that the conformation of the enzyme active site is affected by the identity of the bound tRNA. Recognition of the distinctive structural features of the active tRNAs could be important to accommodate the 3′ terminal nucleotide of the tRNA in the enzyme’s active site. Supporting indirect recognition is the observation of structural differences between active G⋅U or C-C containing RNAs and inactive Watson–Crick containing RNAs at and around the third position of the acceptor helix. Although small, these structural differences could have significant functional consequences because they are amplified at a distance. For example, the substitution of U⋅G for G⋅U reverses the helical twist with respect to the flanking base pairs and causes a 2-Å displacement in an RNA double helix just 5 bp from the wobble pair (18). Therefore, one role of the G⋅U pair that U⋅G and other inactive substitutions cannot mimic is to provide a tRNA conformation that is most favorable for binding and positioning the tRNA acceptor end in the enzyme catalytic site.
Another mechanism by which indirect recognition could operate is through induced fit. The observation that the structure of the active C-C RNA is more flexible than those of inactive G⋅C and A⋅U RNAs suggests that helix deformability may be required to allow for induced fit between AlaRS and the tRNA acceptor end. We detected conformational exchange on a time scale of μs-ms for the C-C mispair. Similar conclusions had been reached in NMR and molecular dynamics studies of a DNA duplex containing a C-C mispair, where conformational flexibility was attributed to the ability of C-C to form several base-pairing interactions of comparable energy (14). Here, conformational flexibility of the C-C mispair indicates that this site could be easily deformed by AlaRS in a process of mutual conformational adaptation. Consistent with this suggestion, the conformations of the backbone surrounding G3 and A73 of the wild-type G⋅U RNA also were found to be flexible (9). Different backbone conformations and base stacking arrangements also have been observed in structures of G⋅U pairs from different sequence contexts, as well in different crystal forms of yeast tRNAPhe (19–24). Taken together, these observations suggest very strongly that the structure at and around a G⋅U pair is “soft” and easily adaptable to its chemical and structural environment. Discrimination against rigid, inactive mutants could result from steric or electrostatic clashes that could not be relieved by induced fit and would lead to a conformation of the enzyme active site that is suboptimal for catalysis.
A role for RNA conformational flexibility in acceptor-end recognition has been proposed for tRNASer based on biochemical data (25) and for tRNAAla from the comparison of NMR properties of G⋅U and C-A acceptor-end RNAs (26). In addition, changes in the structure of the acceptor helix are known to affect the catalytic efficiency of several class II aminoacyl-tRNA synthetases (27) to which the alanine system belongs, those of aspartic acid (28, 29) (D. Moras, personal communication) and serine (25, 30). Further, the observation of a conformational change in the tRNA acceptor helix in the glutamine system (31), a class I aminoacyl-tRNA synthetase (27), suggests that conformational adaptation between the acceptor end and the enzyme catalytic domain may be a common feature of the tRNA-enzyme interaction.
The indirect recognition mechanisms described here do not exclude a role for direct recognition of specific atomic groups in either the major or the minor groove (32) by AlaRS. The G⋅U wobble pair in tRNAAla is widely conserved in evolution (2) and no known tRNAAla sequence contains the C-C pair (33). This feature may reflect coevolution of the tRNA and aminoacyl-tRNA synthetase and/or the superiority of G⋅U as a recognition tag. Because none of the G⋅U mimics are functionally identical to the wild-type sequence, the enzyme also may directly recognize one or more functional groups on the G⋅U pair.
Acknowledgments
We thank A. Ramos for help and suggestions in the structural analysis of the minihelices, K. Gabriel and J. Schneider for help in the functional analysis, and R. Sankaranarayanan and D. Moras for providing information before publication. This work was supported by U. S. Public Health Service Grant GM42123.
Abbreviations
- AlaRS
alanyl-tRNA synthetase
- NOE
nuclear Overhauser effect
- NOESY
two-dimensional NOE and exchange spectroscopy
Footnotes
Throughout this paper ⋅ represents the hydrogen bonding of Watson–Crick and G⋅U wobble base pairs, and - between base symbols represents a mismatch pair.
References
- 1.Hou Y-M, Schimmel P. Nature (London) 1988;333:140–145. doi: 10.1038/333140a0. [DOI] [PubMed] [Google Scholar]
- 2.McClain W H, Foss K. Science. 1988;240:793–796. doi: 10.1126/science.2452483. [DOI] [PubMed] [Google Scholar]
- 3.Li B, Vilardell J, Warner J R. Proc Natl Acad Sci USA. 1996;93:1596–1600. doi: 10.1073/pnas.93.4.1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doudna J A, Cormack B P, Szostak J W. Proc Natl Acad Sci USA. 1989;86:7402–7406. doi: 10.1073/pnas.86.19.7402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gabriel K, Schneider J, McClain W H. Science. 1996;271:195–197. doi: 10.1126/science.271.5246.195. [DOI] [PubMed] [Google Scholar]
- 6.Varshney U, Lee C-P, RajBhandary U L. J Biol Chem. 1991;266:24712–24718. [PubMed] [Google Scholar]
- 7.McClain W H, Schneider J, Bhattacharya S, Gabriel K. Proc Natl Acad Sci USA. 1998;95:460–465. doi: 10.1073/pnas.95.2.460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Price S R, Oubridge C, Varani G. In: RNA-Protein Interactions: A Practical Approach. Smith C W J, editor. New York: Oxford Univ. Press; 1998. pp. 37–74. [Google Scholar]
- 9.Ramos A, Varani G. Nucleic Acids Res. 1997;25:2083–2090. doi: 10.1093/nar/25.11.2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu M, Mao X-A, Ye C, Huang H, Nicholson J K, Lindon J C. J Magn Reson. 1998;132:125–129. doi: 10.1006/jmre.1997.1246. [DOI] [PubMed] [Google Scholar]
- 11.Varani G, Tinoco I., Jr Q Rev Biophys. 1991;24:479–532. doi: 10.1017/s0033583500003875. [DOI] [PubMed] [Google Scholar]
- 12.McClain W H, Chen Y-M, Foss K, Schneider J. Science. 1988;242:1681–1684. doi: 10.1126/science.2462282. [DOI] [PubMed] [Google Scholar]
- 13.McClain W H, Jou Y-Y, Bhattacharya S, Gabriel K, Schneider J. J Mol Biol. 1999;290:391–409. doi: 10.1006/jmbi.1999.2884. [DOI] [PubMed] [Google Scholar]
- 14.Boulard Y, Cognet J A H, Fazakerley G V. J Mol Biol. 1997;268:331–347. doi: 10.1006/jmbi.1997.0975. [DOI] [PubMed] [Google Scholar]
- 15.Musier-Forsyth K, Usman N, Scaringe S, Doudna J, Green R, Schimmel P. Science. 1991;253:784–786. doi: 10.1126/science.1876835. [DOI] [PubMed] [Google Scholar]
- 16.Schimmel P, Musier-Forsyth K. Nature (London) 1996;384:422. doi: 10.1038/384422a0. [DOI] [PubMed] [Google Scholar]
- 17.Park S J, Hou Y-M, Schimmel P. Biochemistry. 1989;28:2740–2746. doi: 10.1021/bi00432a056. [DOI] [PubMed] [Google Scholar]
- 18.Masquida, B., Sauter, C. & Westhof, E. (1999) RNA5, in press. [DOI] [PMC free article] [PubMed]
- 19.Westhof E, Dumas P, Moras D. J Mol Biol. 1985;184:119–145. doi: 10.1016/0022-2836(85)90048-8. [DOI] [PubMed] [Google Scholar]
- 20.Moras D, Dumas P, Westhof E. In: Structure and Dynamics of RNA. vanKippenberg P H, Hilbers C W, editors. New York: Plenum; 1986. pp. 113–124. [Google Scholar]
- 21.Allain F H-T, Varani G. Nucleic Acids Res. 1995;23:341–350. doi: 10.1093/nar/23.3.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Biswas R, Wahl M C, Ban C, Sundaralingam M. J Mol Biol. 1997;267:1149–1156. doi: 10.1006/jmbi.1997.0936. [DOI] [PubMed] [Google Scholar]
- 23.Trikha J, Filman D J, Hogle J H. Nucleic Acids Res. 1999;27:1728–1739. doi: 10.1093/nar/27.7.1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mueller U, Schübel H, Sprinzl M, Heinemann U. RNA. 1999;5:670–677. doi: 10.1017/s1355838299982304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saks M E, Sampson J R. EMBO J. 1996;15:2843–2849. [PMC free article] [PubMed] [Google Scholar]
- 26.Vogtherr M, Schübel H, Limmer S. FEBS Lett. 1998;429:21–26. doi: 10.1016/s0014-5793(98)00542-0. [DOI] [PubMed] [Google Scholar]
- 27.Eriani G, Delarue M, Poch O, Gangloff J, Moras D. Nature (London) 1990;347:203–206. doi: 10.1038/347203a0. [DOI] [PubMed] [Google Scholar]
- 28.Pütz J, Puglisi J D, Florentz C, Giegé R. Science. 1991;252:1696–1699. doi: 10.1126/science.2047878. [DOI] [PubMed] [Google Scholar]
- 29.Cavarelli J, Eriani G, Rees B, Ruff M, Boeglin M, Mitschler A, Martin F, Gangloff J, Thierry J-C, Moras D. EMBO J. 1994;13:327–337. doi: 10.1002/j.1460-2075.1994.tb06265.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cusack S, Yaremchuk A, Tukalo M. EMBO J. 1996;15:2834–2842. [PMC free article] [PubMed] [Google Scholar]
- 31.Rould M A, Perona J J, Söll D, Steitz T A. Science. 1989;246:1135–1142. doi: 10.1126/science.2479982. [DOI] [PubMed] [Google Scholar]
- 32.Sankaranarayanan R, Dock-Bregeon A-C, Romby P, Caillet J, Springer M, Rees B, Ehresmann C, Ehresmann B, Moras D. Cell. 1999;97:371–381. doi: 10.1016/s0092-8674(00)80746-1. [DOI] [PubMed] [Google Scholar]
- 33.Sprinzl M, Steegborn C, Hübel F, Steinberg S. Nucleic Acids Res. 1996;24:68–72. doi: 10.1093/nar/24.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]