Abstract
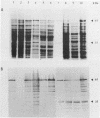
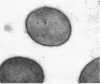
The chromosomal pepN gene encoding lysyl-aminopeptidase activity in Lactococcus lactis has been identified in a lambda EMBL3 library in Escherichia coli by using an immunological screening with antiserum against a purified aminopeptidase fraction. The pepN gene was localized and subcloned in E. coli on the basis of its expression and hybridization to a mixed-oligonucleotide probe for the previously determine N-terminal amino acid sequence of lysyl-aminopeptidase (P. S. T. Tan and W. N. Konings, Appl. Environ. Microbiol. 56:526-532, 1990). The L. lactis pepN gene appeared to complement an E. coli strain carrying a mutation in its pepN gene. High-level expression of the pepN gene in E. coli was obtained by using the T7 system. The overproduction of the 95-kDa aminopeptidase N could be visualized on sodium dodecyl sulfate-polyacrylamide gels and immunoblots. Cloning of the pepN gene on a multicopy plasmid in L. lactis resulted in a 20-fold increase in lysyl-aminopeptidase activity that corresponded to several percent of total protein. Nucleotide sequence analysis of the 5' region of the pepN gene allowed a comparison between the deduced and determined amino-terminal primary sequences of aminopeptidase N. The results show that the amino terminus of PepN is not processed and does not possess the characteristics of consensus signal sequences, indicating that aminopeptidase N is probably an intracellular protein. The intracellular location of aminopeptidase N in L. lactis was confirmed by immunogold labeling of lactococcal cells.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Casadaban M. J., Cohen S. N. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980 Apr;138(2):179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- Chang A. C., Cohen S. N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978 Jun;134(3):1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exterkate F. A. Location of Peptidases Outside and Inside the Membrane of Streptococcus cremoris. Appl Environ Microbiol. 1984 Jan;47(1):177–183. doi: 10.1128/aem.47.1.177-183.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasson M. J. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J Bacteriol. 1983 Apr;154(1):1–9. doi: 10.1128/jb.154.1.1-9.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karn J., Brenner S., Barnett L., Cesareni G. Novel bacteriophage lambda cloning vector. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5172–5176. doi: 10.1073/pnas.77.9.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kok J. Genetics of the proteolytic system of lactic acid bacteria. FEMS Microbiol Rev. 1990 Sep;7(1-2):15–42. doi: 10.1111/j.1574-6968.1990.tb04877.x. [DOI] [PubMed] [Google Scholar]
- Konings W. N., Poolman B., Driessen A. J. Bioenergetics and solute transport in lactococci. Crit Rev Microbiol. 1989;16(6):419–476. doi: 10.3109/10408418909104474. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Latil M., Murgier M., Lazdunski A., Lazdunski C. Isolation and genetic mapping of Escherichia coli aminopeptidase mutants. Mol Gen Genet. 1976 Oct 18;148(1):43–47. doi: 10.1007/BF00268544. [DOI] [PubMed] [Google Scholar]
- Mayo B., Kok J., Venema K., Bockelmann W., Teuber M., Reinke H., Venema G. Molecular cloning and sequence analysis of the X-prolyl dipeptidyl aminopeptidase gene from Lactococcus lactis subsp. cremoris. Appl Environ Microbiol. 1991 Jan;57(1):38–44. doi: 10.1128/aem.57.1.38-44.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardi M., Chopin M. C., Chopin A., Cals M. M., Gripon J. C. Cloning and DNA sequence analysis of an X-prolyl dipeptidyl aminopeptidase gene from Lactococcus lactis subsp. lactis NCDO 763. Appl Environ Microbiol. 1991 Jan;57(1):45–50. doi: 10.1128/aem.57.1.45-50.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remaut E., Stanssens P., Fiers W. Plasmid vectors for high-efficiency expression controlled by the PL promoter of coliphage lambda. Gene. 1981 Oct;15(1):81–93. doi: 10.1016/0378-1119(81)90106-2. [DOI] [PubMed] [Google Scholar]
- Rosenberg S. M. Improved in vitro packaging of lambda DNA. Methods Enzymol. 1987;153:95–103. doi: 10.1016/0076-6879(87)53050-6. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon D., Chopin A. Construction of a vector plasmid family and its use for molecular cloning in Streptococcus lactis. Biochimie. 1988 Apr;70(4):559–566. doi: 10.1016/0300-9084(88)90093-4. [DOI] [PubMed] [Google Scholar]
- Studier F. W., Rosenberg A. H., Dunn J. J., Dubendorff J. W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan P. S., Konings W. N. Purification and Characterization of an Aminopeptidase from Lactococcus lactis subsp. cremoris Wg2. Appl Environ Microbiol. 1990 Feb;56(2):526–532. doi: 10.1128/aem.56.2.526-532.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos P., Boerrigter I. J., Buist G., Haandrikman A. J., Nijhuis M., de Reuver M. B., Siezen R. J., Venema G., de Vos W. M., Kok J. Engineering of the Lactococcus lactis serine proteinase by construction of hybrid enzymes. Protein Eng. 1991 Apr;4(4):479–484. doi: 10.1093/protein/4.4.479. [DOI] [PubMed] [Google Scholar]
- Vos P., van Asseldonk M., van Jeveren F., Siezen R., Simons G., de Vos W. M. A maturation protein is essential for production of active forms of Lactococcus lactis SK11 serine proteinase located in or secreted from the cell envelope. J Bacteriol. 1989 May;171(5):2795–2802. doi: 10.1128/jb.171.5.2795-2802.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood W. B. Host specificity of DNA produced by Escherichia coli: bacterial mutations affecting the restriction and modification of DNA. J Mol Biol. 1966 Mar;16(1):118–133. doi: 10.1016/s0022-2836(66)80267-x. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- de Vos W. M., Boerrigter I., van Rooyen R. J., Reiche B., Hengstenberg W. Characterization of the lactose-specific enzymes of the phosphotransferase system in Lactococcus lactis. J Biol Chem. 1990 Dec 25;265(36):22554–22560. [PubMed] [Google Scholar]
- de Vos W. M., Vos P., de Haard H., Boerrigter I. Cloning and expression of the Lactococcus lactis subsp. cremoris SK11 gene encoding an extracellular serine proteinase. Gene. 1989 Dec 21;85(1):169–176. doi: 10.1016/0378-1119(89)90477-0. [DOI] [PubMed] [Google Scholar]
- van Asseldonk M., Rutten G., Oteman M., Siezen R. J., de Vos W. M., Simons G. Cloning of usp45, a gene encoding a secreted protein from Lactococcus lactis subsp. lactis MG1363. Gene. 1990 Oct 30;95(1):155–160. doi: 10.1016/0378-1119(90)90428-t. [DOI] [PubMed] [Google Scholar]
- von Heijne G. Transcending the impenetrable: how proteins come to terms with membranes. Biochim Biophys Acta. 1988 Jun 9;947(2):307–333. doi: 10.1016/0304-4157(88)90013-5. [DOI] [PubMed] [Google Scholar]