Abstract
Carbohydrate-derivatized self-assembled monolayers (SAMs) are used as a model system to address issues involving cell-surface carbohydrate–protein interactions. Here we examine the influence of carbohydrate surface density on protein-binding avidity. We show that the binding selectivity of Bauhinia purpurea lectin switches from one carbohydrate ligand to another as the surface density of the carbohydrate ligands increases from values of χsugar ≈ 0.1–1.0. Polyvalent binding is possible at all surface densities investigated; hence, the switch in selectivity is not due simply to the achievement of a critical density that permits polyvalent contacts. Instead, secondary interactions at high surface densities promote a switch in carbohydrate-binding selectivity. These findings may have implications for how changes in the composition and the density of cell-surface carbohydrates influence biological recognition processes and regulatory pathways.
The patterns of expression of cell-surface carbohydrates change during development and differentiation (1–3). Changes in the composition of cell-surface carbohydrates also occur during oncogenesis and metastasis (4, 5). Although the biological functions of cell-surface carbohydrates are still incompletely defined, it is believed that they are somehow involved in signaling pathways that determine the temporal and spatial identity of cells. How changing patterns of cell-surface carbohydrates are linked to cell growth and differentiation is not understood.
The early steps in cell-surface carbohydrate-mediated cellular processes are presumed to involve binding events between carbohydrates and receptors in the plasma or on the surface of other cells. Because these binding events are linked to specific cellular responses, one might expect them to be highly specific. However, studies in solution have shown that carbohydrate–protein binding interactions have dissociation constants that are typically in the mM range and that different carbohydrate ligands have similar affinities for the same protein receptor. If carbohydrates bind weakly and with poor selectivity, how can they mediate specific cellular processes?
This paradox has been partially explained by the phenomenon of polyvalency. Carbohydrate–protein binding events usually involve several simultaneous contacts between carbohydrates that are clustered on cell surfaces and protein receptors that contain multiple carbohydrate-binding sites. Numerous studies on model systems have demonstrated that polyvalent displays of carbohydrates can lead to remarkably high binding avidities (6–16). Because avidity and specificity are not necessarily correlated (6, 7), however, polyvalency does not completely resolve the paradox of how marginally selective carbohydrate-binding interactions can produce highly specific responses.
Two mechanisms have been proposed to explain how specificity can be achieved in cell-surface carbohydrate-binding interactions. In one mechanism, polyvalent carbohydrate–protein interactions facilitate the adhesion of cells. The proximity enforced by these carbohydrate–protein interactions permits more specific protein–protein interactions to be established between the cells. The protein–protein interactions actually determine the specific biological response (17). This mechanism circumvents the question of whether the carbohydrate–protein interactions are themselves specific. It has alternatively been suggested that polyvalency can amplify binding specificity as well as binding affinity. According to this hypothesis, protein specificity is achieved because the small energetic differences of individual carbohydrate–protein interactions are greatly magnified in a polyvalent presentation (18).
Because of the complexity of cell surfaces, it has been difficult to evaluate carbohydrate–protein binding events in their native context. Simplified model systems are usually used to study polyvalent carbohydrate–protein interactions (6–16). We recently demonstrated that carbohydrates presented on TentaGel beads exhibit polyvalent binding and may therefore be useful as model systems to investigate the interactions of carbohydrates on surfaces (6, 7). We then synthesized and screened a library of carbohydrate-derivatized TentaGel beads for binding to Bauhinia purpurea (BP) lectin, a carbohydrate-binding protein containing multiple carbohydrate-binding sites. The lectin preferentially bound the unnatural ligand 2 in a library containing 1,300 different di- and trisaccharides, including the natural ligand 1 (Fig. 1). This result showed that polyvalent carbohydrate–protein interactions are remarkably specific. One implication of these model studies is that cell-surface carbohydrates might, in fact, bind selectively enough to proteins to mediate specific biological events.
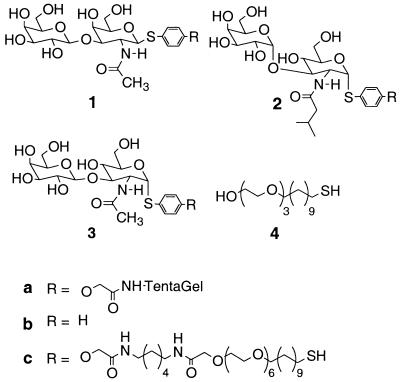
Figure 1.
Structures of natural ligand (1), hit ligand (2), control ligand (3), and tri(ethylene glycol)-terminated alkanethiol (4).
In further studies, we found that the solution affinities of selected carbohydrate ligands for BP lectin did not correlate with their polyvalent avidities. For example, in solution, BP lectin binds slightly better to disaccharide 1 than to disaccharide 2; however, when disaccharides 1 and 2 are presented on TentaGel beads, BP lectin adheres preferentially to beads presenting disaccharide 2. Therefore, the polyvalent presentation of these carbohydrate ligands does not merely amplify the intrinsic binding selectivities observed in solution. Instead, the binding selectivity switches on going from solution to a polyvalent format.
In this paper, we address whether the observed change in selectivity is caused simply by tethering the carbohydrate ligands to the surface (an orientation effect) or by clustering the tethered carbohydrates (a density-dependent polyvalent effect). To distinguish these effects, we monitored protein binding to surfaces presenting the tethered carbohydrate ligands 1c, 2c, and 3c (Fig. 1) at increasing ligand densities. Self-assembled monolayers (SAMs) of alkanethiolates on gold provide a convenient way to display carbohydrates on surfaces with control over the average, in-plane density of the carbohydrate ligand. Protein binding to monolayers can be readily evaluated by using surface plasmon resonance (SPR) (19–24). Remarkably, we found that the same protein binds preferentially to one carbohydrate ligand at low surface densities, but to a different carbohydrate ligand at high surface densities. These results may have implications for how changes in the composition and the density of cell-surface carbohydrates influence biological recognition processes.
Materials and Methods
Materials.
Dichloromethane (CH2Cl2), N,N-dimethylformamide (DMF), tetrahydrofuran (THF), N,N-diisopropylethylamine, triethylamine (TEA), and trifluoroacetic acid (TFA) were purchased from Aldrich. 1-Hydroxybenzotriazole and 2-(1H-benzotriazol-1yl)-1,1,3,3-tetramethyluronium hexafluorophosphate were from Applied Biosystems. Absolute ethanol was purchased from Pharmco Products (Brookfield, CT). Bovine carbonic anhydrase II was obtained from Worthington. PBS, BSA, BP lectin, carbonic anhydrase, and concanavalin A were purchased from Sigma. Electrophoresis-grade SDS (Bio-Rad) was used. PBS (10 mM phosphate/138 mM NaCl/2.7 mM KCl) was prepared in distilled, deionized water. PBST is 0.05% Tween-20 (Fluka) in PBS. Protein solutions were prepared in PBST containing 1% BSA. All solutions were degassed under vacuum and filtered through 0.45-μm filters before use. Titanium (99.99%, Aldrich) and gold (99.999% as machined pellets, Materials Research Corp., Orangeburg, NY) were used as received. Glass cover slips (0.20 mm, no. 2) were from Corning.
Synthesis of Compound 4.
Compound 4 was synthesized with use of a published procedure (25).
Synthesis of Compound 5.
Compound 5 was synthesized in three steps from CH2CHO(CH2CH2O)6H: (i) NaH, ClCH2C(O)NH(CH2)6NHBoc, THF, 0°C to room temperature (RT), 10 h; (ii) thiolacetic acid, 2,2′-azobisisobutyronitrile (AIBN), benzene, 80°C, 48 h; and (iii) 20% TFA/CH2Cl2, RT, 4.5 h.
Synthesis of Compounds 1c and 3c.
Trifluoromethanesulfonic anhydride (Tf2O, 1.5 equivalents) was added dropwise to a solution of sulfoxide (1.5 equivalents) in CH2Cl2 at −78°C. A solution of glycosyl acceptor (one equivalent) and 2,6-di-tert-butyl-4-methylpyridine (DTBMP; three equivalents) in CH2Cl2 was then added and the reaction mixture was warmed to 0°C over 2 h. The disaccharide was deprotected in two steps: (i) 20% TFA/CH2Cl2, RT, 45 min; and (ii) lithium hydroxide, methanol (MeOH), RT, 12 h. After acetylation of the alcohols [catalytic 4-dimethylaminopyridine (DMAP), TEA, acetic anhydride (Ac2O), CH2Cl2, RT, 1 h], the azide was reduced and acetylated (thiolacetic acid, RT, 48 h).
The allyl ether was oxidatively cleaved [tetra(kistriphenylphosphine)palladium, N-methylmorpholine, acetic acid, and chloroform (1:2:5), RT, 12 h] and the resulting phenol was alkylated (cesium carbonate, tert-butyl bromoacetate, DMF, RT, 30 min). The ester was removed (20% TFA/CH2Cl2, RT, 1 h) and the disaccharide was coupled to H2N(CH2)6NHC(O) CH2O(CH2CH2O)6(CH2)11SAc 5 [2-(1H-benzotriazol-lyl)-1,1,3,3-tetramethyluronium hexafluorophosphate; 1-hydroxybenzotriazole; N,N-diisopropylethylamine; DMF; RT, 1 h], and the coupled product was deprotected [sodium methoxide (NaOMe), MeOH, RT, 12 h].
Synthesis of Compound 2c.
Trifluoromethanesulfonic anhydride (1 equivalent) was added dropwise to a solution of sulfoxide and 2,6-di-tert-butyl-4-methylpyridine (2 equivalents) in CH2Cl2 at −78°C. A solution of glycosyl acceptor (1.1 equivalents) in CH2Cl2 was added and then the reaction mixture was warmed to 0°C over 2 h. The disaccharide was deprotected and acetylated in four steps: (i) sodium methoxide, THF, and methanol (2:3), RT, 5 h; (ii) 4-dimethylaminopyridine, TEA, Ac2O, CH2Cl2, RT, 1 h; (iii) 20% TFA/CH2Cl2, RT, 1 h; and (iv) 4-dimethylaminopyridine, TEA, Ac2O, CH2Cl2, RT, 1 h. The azide was reduced (trimethylphosphine, THF, RT, 30 min; drop of water, 45°C, 24 h) and acylated (isovaleryl chloride, TEA, CH2Cl2, RT, 2 h). Allyl ether cleavage and subsequent synthetic steps followed the same procedures as those in the synthesis of carbohydrate ligands 1c and 3c.
Preparation of Gold Substrates.
Gold substrates were prepared by evaporating an adhesion layer of titanium (1.5 nm) and gold (38 nm) onto glass cover slips. Stock solutions (1–2 mM in absolute ethanol) of tri(ethylene glycol)-terminated alkanethiol 4 and disaccharide ligand 1c, 2c, or 3c were combined in different ratios in glass scintillation vials. The gold-coated slides were cut into squares 1 cm2 in size, rinsed with ethanol, and dried with a stream of nitrogen. Two gold substrates were simultaneously immersed in the adsorption solutions for 8–12 h, rinsed with ethanol, and dried with nitrogen. The gold chips were glued into BIAcore cassettes by using 5-Minute Epoxy (Devcon, Danvers, MA).
X-Ray Photoelectron Spectroscopy.
X-ray photoelectron spectroscopy (XPS) spectra were obtained on an SSX-100 spectrometer (Surface Science Instruments, Mountain View, CA) by using an AL Kα source, a quartz monochromator, a concentric hemispherical analyzer in transmission mode, and a multichannel detector. The spectra were accumulated at a takeoff angle of 35° relative to the surface and a pressure of (2–8) × 10−9 Torr. Spectra were fitted by using software from Surface Science Instruments. N(1s) peaks were modeled as a Gaussian function. The mole fractions of 1c, 2c, or 3c in each mixed SAM were calculated by comparing the area under the N(1s) photoelectron peak of the mixed SAM with the area under the N(1s) peak of the pure carbohydrate SAM.
SPR Measurement of Protein Binding.
A BIAcore 1000 (Pharmacia) instrument was used to measure protein binding to the carbohydrate-derivatized surfaces. The BIAcore instrument reports θm in resonance units (RU, 1° = 10,000 RU). The resolution of the instrument is ≈0.0001°.
Protein solutions of carbonic anhydrase (33 mM), concanavalin A (513 nM), and BP lectin (513 nM) were prepared in PBST containing 1% BSA. In the BIAcore instrument, the carbohydrate-derivatized surface was cleaned with a solution of detergent (SDS, 10 mg/ml in PBST containing 1% BSA) at a constant flow rate of 5 μl/min for 2 min. A solution of PBST buffer containing 1% BSA was allowed to flow through the cell for 3 min, followed by a flow of protein in the same buffer for 3 min, and then by a flow of buffer for another 3 min.
Determination of IC25 Values.
BP lectin (5 mg) was dissolved in 5 ml of PBST buffer containing 1% BSA. Inhibitor solutions of seven different concentrations were made from a stock solution of Galβ1,3GalNAcβ-thiophenyl glycoside 1b (1 mg/ml in PBST buffer containing 1% BSA). A solution of lectin (100 μl) was added to each inhibitor solution to afford a final lectin concentration of 100 μg/ml (513 nM). The seven lectin-inhibitor solutions were separately injected over individual flow cells. Protein adsorption to carbohydrate-derivatized SAMs was measured by allowing a solution of PBST buffer containing 1% BSA to flow through the cell at a constant flow rate of 5 μl/min for 3 min followed by a flow of lectin-inhibitor solution for 10 min, and then reintroducing the buffer to initiate dissociation. Duplicate experiments were performed on a second set of SAMs prepared from the original adsorbent solution. From the SPR response curves, the response values (RU) at t = 633.5 s were determined for each concentration of the inhibitor 1b and normalized as RU/RUmax, where RUmax is the RU value in the absence of 1b. RU/RUmax values in duplicate were plotted against the concentration of 1b for each ligand density. IC25 values were determined as the concentration of 1b required to decrease the RU/RUmax value by 25% from its maximum value. IC25 values were obtained for the three carbohydrate ligands with values of χsugar ranging from 0.1–1.0. Protein avidity is reported in terms of IC25 values rather than IC50 values because the change in protein binding is not a linear function of soluble ligand concentration, and the concentrations can be determined more accurately at lower inhibition levels.
Determination of Kinetic Constants.
The SPR response curves were analyzed by using biacore evaluations software version 2.1. The region of the curve corresponding to the dissociation phase was fitted to a nonlinear exponential decay function to determine the apparent dissociation constant (kdis). The validity of the binding model was evaluated through examination of the residual plots. Residual plots show the differences between the experimental data and the calculated fit for each point in the fit. If the model is appropriate, the residual values will be of low magnitude and randomly scattered. Comparison of different binding models provided dissociation rate constants of similar magnitude.
Results
Preparation of Mixed SAMs Containing Disaccharides 1c, 2c, and 3c.
We first synthesized the specific disaccharides 1c and 2c as well as the nonspecific control disaccharide 3c with an appropriate alkanethiol linker at the anomeric position (Fig. 2). Sets of mixed SAMs presenting different mole fractions of disaccharide 1c, 2c, or 3c (χsugar) were then prepared by immersing gold substrates in solutions containing mixtures of disaccharide and tri(ethylene glycol)-terminated alkanethiol 4. The component 4 was used in the mixed SAMs because it resists the nonspecific adsorption of protein (25, 26). The mole fractions of disaccharide in each surface were determined by using x-ray photoelectron spectroscopy as described in Materials and Methods.
Figure 2.
Synthesis of the carbohydrate ligands. Conditions for ligands 1c and 3c were as follows: (A) trifluoromethanesulfonic anhydride, 2,6-di-tert-butyl-4-methylpyridine, CH2Cl2, −78°C to −30°C; 20% TFA/CH2Cl2, RT, 30 min; LiOH, MeOH, RT, 8 h; Ac2O, TEA, DMAP, CH2Cl2, RT, 1 h; thiolacetic acid, RT, 48 h; (B) Pd(Ph3)4, N-methylmorpholine, AcOH, CHCl3, RT, 8 h; Cs2CO3, t-butyl bromoacetate, DMF, RT, 1 h; 30% TFA/CH2Cl2, RT, 30 min; 1-hydroxybenzotriazole/2-(1H-benzotriazol-lyl)-1,1,3,3-tetramethyluronium hexafluorophosphate; N,N-diisopropylethylamine, DMF, RT, 3 h; and sodium methoxide, MeOH, RT, 8 h. Conditions for ligand 2c were as follows: (C) trifluoroethanesulfonic anhydride, 2,6-di-tert-butyl-4-methylpyridine, CH2Cl2, −78°C to −30°C; sodium methoxide, THF, MeOH, RT, 30 min; 20% TFA/CH2Cl2, RT, 30 min; Ac2O, TEA, 4-dimethylaminopyridine, CH2Cl2, RT, 1 h; Me3P, THF, H2O, RT, 48 h; isovaleryl chloride, TEA, CH2Cl2, 0°C to RT, 1 h; and B.
Mixed SAMs as Model Systems for Assessing Protein Binding to Surface-Bound Carbohydrates.
Initial experiments addressed whether the mixed SAMs were an appropriate model system to investigate specific interactions between surface-bound carbohydrates and proteins. Previous studies on TentaGel beads had shown that BP lectin recognizes ligands 1a and 2a in the presence of other structurally similar ligands, suggesting a high degree of specificity in the polyvalent interactions (6, 7). Mixed SAMs presenting 1c (Fig. 3) or 2c (results not shown) were found to adsorb BP lectin (27, 28), with protein binding initially increasing as a function of surface density and then decreasing again. Decreased protein binding at high ligand densities is characteristic of interactions involving SAMs. This decrease has been attributed to steric effects that alter access of individual carbohydrate ligands to the protein binding pockets, or to changes in the character and packing of the surface of the SAM as a result of lateral interactions between ligands (29). In contrast to the results with BP lectin, the mixed SAMs did not adsorb the enzyme carbonic anhydrase or the mannose-binding protein concanavalin A at low or high surface densities (Fig. 3); hence, the mixed SAMs displayed protein-specific binding and had the required characteristics to serve as models for investigating the relationship between carbohydrate surface density and binding of BP lectin.
Figure 3.
SPR curves for protein binding to mixed SAMs containing 1c. Solid line, BP lectin; dashed lines, concanavalin A and carbonic anhydrase, respectively.
Measurement of Protein Binding at Increasing Ligand Densities.
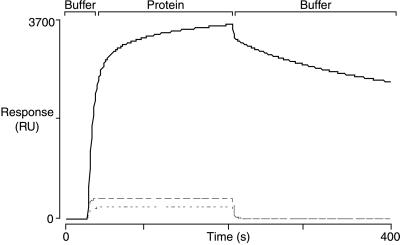
We determined the relative avidities of BP lectin for mixed SAMs containing carbohydrates 1c, 2c, or 3c at different surface densities by estimating the concentration of soluble competitor ligand 1b required to reduce the maximum SPR response by 25% (the IC25). BP lectin was mixed with increasing concentrations of 1b and the solutions were allowed to flow over mixed SAMs with values of χsugar ranging from 0.1 to 1.0. The flow was continued for 10 min and then the protein-inhibitor solution was replaced with buffer. Fig. 4 shows a representative set of SPR response curves obtained for a mixed SAM presenting ligand 2c with a value of χsugar ≈ 0.1.
Figure 4.
A typical set of SPR response curves for the binding of BP lectin to SAMs presenting carbohydrate ligands. The amount of bound lectin decreases in the presence of the soluble competitor ligand 1b.
Plots of protein binding as a function of inhibitor concentration for 1c, 2c, or 3c at low and high surface densities (χsugar ≈ 0.1 and 0.6, respectively) are shown in Fig. 5. At low surface densities, the protein binds better to mixed SAMs containing disaccharide 1c than to mixed SAMs containing 2c or 3c, as indicated by the higher concentrations of soluble inhibitor required to block binding. As the surface density increases, the avidity of BP lectin for monolayers containing disaccharide 1c decreases. At the same time, the avidity for monolayers containing 2c increases. At surface densities above χsugar ≈ 0.4, 2c becomes the preferred ligand, with the maximum differential in binding avidity occurring at χsugar ≈ 0.6; hence, the relative avidity of BP lectin for ligands 1c and 2c switches as the surface density of the carbohydrate ligands increases above a critical value.
Figure 5.
Relative binding avidities of BP lectin for mixed SAMs derivatized with carbohydrate ligands 1c, 2c, or 3c at low and high densities (values of χsugar ≈ 0.1 and 0.6) as judged by IC25 values.
Evidence for Polyvalent Binding at All Surface Densities.
The SPR results verified our previous findings that the selectivity of BP lectin for ligands 1 and 2 depends on the context of the carbohydrate ligands (6, 7). The SPR experiments further suggested that the observed change in binding selectivity on going from solution to a TentaGel bead was not caused by a simple orientation effect. If tethering the carbohydrate ligand to a surface had caused the switch in selectivity, 2c should have shown higher binding avidity for BP lectin at all surface densities. Instead, the better ligand in solution, 1, remained the preferred ligand until the surface density reached a critical value. At that critical value, the binding selectivity switched.
One plausible explanation for the switch in selectivity is that the critical density reflects the point at which polyvalent interactions begin to dominate the protein binding events. Calculations suggest, however, that even at χligand ≈ 0.01, the ligands in mixed SAMs should be close enough on average to permit polyvalent interactions (20, 22). The calculations are based on a model in which a monolayer of protein is adsorbed onto the SAM. From the size of the protein, it is possible to determine the surface density of carbohydrate ligands that would provide for one binding interaction per protein (i.e., monovalent binding). The lowest carbohydrate surface densities used in these experiments are approximately ten times higher than the surface density that would support monovalent binding; therefore, assuming a uniform distribution of carbohydrate ligands, polyvalent binding is possible for virtually all protein binding events.
Kinetic measurements support the idea that polyvalent binding between BP lectin and the mixed SAMs occurs at all surface densities. Slow dissociation rates are a hallmark of multivalent interactions. All the carbohydrate-derivatized surfaces, even at the lowest densities, exhibited slow off-rates for the binding of BP lectin. For example, the off-rate for BP lectin binding to mixed SAMs of 2c with χsugar ≈ 0.1 was found to be 2 ×10−3 s−1. The off-rate did not change when a soluble competitor ligand was included in the buffer during the dissociation phase. The similarity in the off-rates in the absence and presence of soluble inhibitor indicates that the binding is interaction controlled rather than mass transport controlled. We have concluded, therefore, that the slow calculated off-rates support the hypothesis that polyvalent interactions predominate even at the lowest surface densities.
Discussion
Model systems presenting polyvalent carbohydrate ligands have been used to address questions related to binding events involving cell-surface carbohydrates and proteins (6–8, 11–13, 15, 16). Most of these studies focus on the role of polyvalency in determining protein-binding avidity. It has been amply demonstrated that polyvalency increases protein-binding avidities dramatically; however, the influence of carbohydrate surface density on protein-binding selectivity has not been examined. The investigations reported above grew out of a previous study showing that the selectivity of BP lectin for two different carbohydrate ligands switched on going from solution to a polyvalent format. We have now found that the binding selectivity of BP lectin for the carbohydrate ligands depends on the surface density of the ligands in the mixed SAMs, even though binding is polyvalent at all densities investigated. The fact that the carbohydrate-derivatized SAMs exhibit nonstatistical binding suggests that secondary interactions contribute significantly to protein avidity. As the density of the carbohydrate ligands increases, interactions between the carbohydrates may affect the individual binding interactions with the protein. Alternatively, protein–protein interactions may be established at high carbohydrate surface densities. These protein–protein interactions may well differ for different carbohydrate ligands. Regardless of their precise nature, secondary interactions could have a significant effect on protein binding, with the result that the binding selectivity switches at high surface densities.
Our findings raise the possibility that cell-surface carbohydrates may be involved in the regulation of biological pathways in a more complex manner than has been considered previously. Because avid protein binding requires polyvalent interactions, it has been speculated that nature uses carbohydrate surface density as an “on-off” switch to regulate biological events. At low ligand density, carbohydrate ligands are not close enough to permit polyvalent protein binding so a biological pathway is in the off state. As the surface density of a particular carbohydrate ligand increases, protein binding occurs and the pathway is switched on. Our results suggest that the carbohydrate expression levels can modulate far more complicated response patterns. For example, the observation that the binding selectivity of a protein depends on both the carbohydrate ligand and its surface density suggests that the same protein could trigger at least two different types of responses by binding differentially to two different carbohydrate ligands. A protein that is bound to one carbohydrate ligand could be recruited to a different ligand on the same cell surface as expression levels of the second ligand increase, with the concomitant activation of a different biological pathway. It is even conceivable that different proteins may bind to the same carbohydrate ligand, albeit at different surface densities. We propose that changes in the expression levels of cell-surface carbohydrates may permit switching not just from an off state to an on state, but from one on state to another on state.
Acknowledgments
This work was supported by the Office of Naval Research and the National Institutes of Health.
Abbreviations
- SAMs
self-assembled monolayers
- SPR
surface plasmon resonance
- BP lectin
Bauhinia purpurea lectin
- RU
resonance units, DMF, N,N-dimethylformamide
- TEA
triethylamine
- TFA
trifluoroacetic acid
- RT
room temperature
References
- 1.Varki A. Glycobiology. 1993;3:97–130. doi: 10.1093/glycob/3.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hakomori S. Annu Rev Biochem. 1981;50:733–764. doi: 10.1146/annurev.bi.50.070181.003505. [DOI] [PubMed] [Google Scholar]
- 3.Fukuda M, Hindsgaul O, editors. Molecular Glycobiology. New York: Oxford Univ. Press; 1994. [Google Scholar]
- 4.Hakomori S, Zhang Y. Chem Biol. 1997;4:97–104. doi: 10.1016/s1074-5521(97)90253-2. [DOI] [PubMed] [Google Scholar]
- 5.Fukuda M. Cancer Res. 1996;56:2237–2244. [PubMed] [Google Scholar]
- 6.Liang R, Yan L, Loebach J, Ge M, Uozumi Y, Sekanina L, Horan N, Gildersleeve J, Thompson C, Smith A, et al. Science. 1996;274:1520–1522. doi: 10.1126/science.274.5292.1520. [DOI] [PubMed] [Google Scholar]
- 7.Liang R, Loebach J, Horan N, Ge M, Thompson C, Yan L, Kahne D. Proc Natl Acad Sci USA. 1997;94:10554–10559. doi: 10.1073/pnas.94.20.10554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kingery-Wood J E, Williams K W, Sigal G B, Whitesides G W. J Am Chem Soc. 1992;114:7303–7305. [Google Scholar]
- 9.Mahal L K, Yarema K J, Bertozzi C R. Science. 1997;276:1125–1128. doi: 10.1126/science.276.5315.1125. [DOI] [PubMed] [Google Scholar]
- 10.Kanai M, Mortell K H, Kiessling L L. J Am Chem Soc. 1997;119:9931–9932. [Google Scholar]
- 11.DeFrees S A, Phillips L, Guo L, Zalipsky S. J Am Chem Soc. 1996;118:6101–6104. [Google Scholar]
- 12.Spevak W, Foxall C, Charych D H, Dasgupta F, Nagy J O. J Med Chem. 1996;39:1018–1020. doi: 10.1021/jm950914+. [DOI] [PubMed] [Google Scholar]
- 13.Zanini D, Roy R. J Org Chem. 1998;63:3486–3491. [Google Scholar]
- 14.Glick G D, Toogood P L, Wiley D C, Skehel J J, Knowles J R. J Biol Chem. 1991;266:23660–23669. [PubMed] [Google Scholar]
- 15.Matrosovich M N, Mochalova L V, Marinina V P, Byramova N E, Bovin N V. FEBS Lett. 1990;272:209–212. doi: 10.1016/0014-5793(90)80486-3. [DOI] [PubMed] [Google Scholar]
- 16.Choi S-K, Mammen M, Whitesides G M. Chem Biol. 1996;3:97–104. doi: 10.1016/s1074-5521(96)90285-9. [DOI] [PubMed] [Google Scholar]
- 17.Lawrence M B, Springer T A. Cell. 1991;65:859–873. doi: 10.1016/0092-8674(91)90393-d. [DOI] [PubMed] [Google Scholar]
- 18.Mortell K H, Weatherman R V, Kiessling L L. J Am Chem Soc. 1996;118:2297–2298. [Google Scholar]
- 19.Roberts C, Chen C S, Mrksich M, Martichonok V, Ingber D E, Whitesides G M. J Am Chem Soc. 1998;120:6548–6555. [Google Scholar]
- 20.Mrksich M, Grunwell J R, Whitesides G M. J Am Chem Soc. 1995;117:12009–12010. [Google Scholar]
- 21.Schlereth D D, Kooyman R P H. J Electroanal Chem. 1998;444:231–240. [Google Scholar]
- 22.Bamdad C. Biophys J. 1998;75:1989–1996. doi: 10.1016/S0006-3495(98)77640-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mann D A, Kanai M, Maly D J, Kiessling L L. J Am Chem Soc. 1998;120:10575–10582. [Google Scholar]
- 24.Houseman B T, Mrksich M. Angew Chem Int Ed Engl. 1999;38:782–785. doi: 10.1002/(SICI)1521-3773(19990315)38:6<782::AID-ANIE782>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 25.Pale-Grosdemange C, Simon E S, Prime K L, Whitesides G M. J Am Chem Soc. 1991;113:13–20. [Google Scholar]
- 26.Prime K L, Whitesides G M. J Am Chem Soc. 1993;115:10714–10721. [Google Scholar]
- 27.Osawa T, Irimura T, Kawaguchi T. Methods Enzymol. 1978;50:367–372. doi: 10.1016/0076-6879(78)50044-x. [DOI] [PubMed] [Google Scholar]
- 28.Wu A M, Kabat E A, Gruezo F G, Allen H J. Arch Biochem Biophys. 1980;204:622–639. doi: 10.1016/0003-9861(80)90074-0. [DOI] [PubMed] [Google Scholar]
- 29.Sigal G B, Bamdad C, Barberis A, Strominger J, Whitesides G M. Anal Chem. 1996;68:490–497. doi: 10.1021/ac9504023. [DOI] [PubMed] [Google Scholar]