Abstract
Soluble glucose dehydrogenase (s-GDH) from the bacterium Acinetobacter calcoaceticus is a classical quinoprotein. It requires the cofactor pyrroloquinoline quinone (PQQ) to catalyze the oxidation of glucose to gluconolactone. The precise catalytic role of PQQ in s-GDH and several other PQQ-dependent enzymes has remained controversial because of the absence of comprehensive structural data. We have determined the crystal structure of a ternary complex of s-GDH with PQQ and methylhydrazine, a competitive inhibitor of the enzyme. This complex, refined at 1.5-Å resolution to an R factor of 16.7%, affords a detailed view of a cofactor-binding site of s-GDH. Moreover, it presents the first direct observation of covalent PQQ adduct in the active-site of a PQQ-dependent enzyme, thereby confirming previous evidence that the C5 carbonyl group of the cofactor is the most reactive moiety of PQQ.
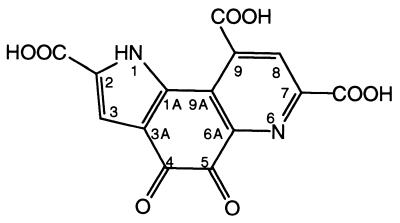
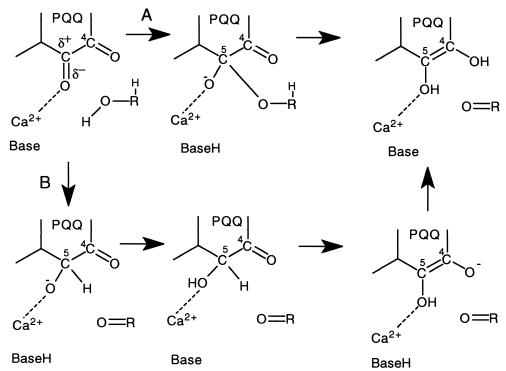
Quinoproteins form a class of dehydrogenases distinct from the NAD(P)+- and flavin-dependent enzymes (1). They use one of four different quinone cofactors for the oxidation of a wide variety of compounds (2–5). The prototypical cofactor pyrroloquinoline quinone (PQQ; Fig. 1) is present in the bacterial quinoproteins methanol dehydrogenase (MDH) and glucose dehydrogenase (GDH) (2, 6, 7). On the basis of a variety of biochemical and kinetic data for MDH and the soluble GDH (s-GDH), two reaction mechanisms have been proposed for PQQ-containing enzymes (Fig. 2). The first mechanism includes general base-catalyzed proton abstraction from the oxidizable hydroxyl group, followed by the formation of a covalent substrate–PQQ complex and product elimination (Fig. 2A) (8–11). The second possible reaction mechanism involves general base-catalyzed proton abstraction in concert with hydride transfer to PQQ and subsequent tautomerization to pyrroloquinoline quinone (PQQH2) (Fig. 2B) (10–12). The substrate or hydride anion is normally considered to form an adduct at the PQQ C5 position (8–11, 13, 14), but Zheng and Bruice (12) have calculated that a mechanism involving hydride transfer from the substrate to the PQQ C4 atom may be energetically favorable. Thus, structural information is required to undisputedly resolve which of the two carbonyl groups of the ortho-quinone group of PQQ is most reactive in an enzymatic environment.
Figure 1.
Structure of PQQ. Atom nomenclature is indicated.
Figure 2.
Possible reaction mechanisms for s-GDH. (A) The addition-elimination mechanism. (B) Mechanism comprising direct hydride transfer and tautomerization to PQQH2 (upper right structure).
s-GDH has long been thought to be exclusively present in the periplasmic space of the bacterium Acinetobacter calcoaceticus, but recently homologous sequences have been identified in the genomes of four other bacteria (15). s-GDH is a basic (pI = 9.5) dimeric enzyme of identical subunits of 50 kDa each (454 residues after cleavage of a 24-residue signal peptide) (16, 17). The enzyme requires calcium for dimerization as well as for binding of PQQ (18). It oxidizes a broad range of aldose sugars to the corresponding lactones, with concomitant reduction of PQQ to PQQH2 (Fig. 2) (19). Several artificial electron acceptors are able to reoxidize the cofactor (16). Stopped-flow kinetics indicated that the oxidation of glucose has a pH optimum of around 7 (11).
Recently, we reported the three-dimensional structure of the dimeric apo-form of s-GDH at 1.7 Å (15). The monomers fold into a β-propeller structure consisting of six four-stranded β-sheets. Three calcium ions per monomer are present in the crystal structure, of which two are located close to the dimerization interface, whereas the third calcium ion was predicted to be ligated by PQQ in the active site of the enzyme. Here, we present the three-dimensional structure of s-GDH in complex with PQQ and methylhydrazine (MH), a competitive inhibitor of the protein (hydrazine inhibits the conversion of glucose with a Ki value of 5 ± 0.5 mM; A. Dewanti and J. A. Duine, personal communication). This structure presents the first detailed view of PQQ binding in s-GDH, shows how MH inhibits the enzyme, and identifies the reactive moiety of the functional ortho-quinone group of PQQ.
Methods
Crystallization and Complex Formation.
Colorless crystals of the apo-form of s-GDH were prepared as described previously (15). They belong to space group P21, and the cell dimensions are a = 61.9 Å, b = 94.2 Å, c = 86.5 Å, β = 104.9° at room temperature. A double-soaking procedure was applied in an attempt to obtain crystals of holo-s-GDH in complex with MH. Crystals were first soaked for at least 8 hr in a stabilizing solution containing 1 mM PQQ, 3 mM CaCl2, 120 mM NaCl, and 25% (wt/vol) polyethylene glycol (PEG) 6000 in 50 mM 2-[N-cyclohexylamino]-ethanesulfonic acid (CHES) buffer, pH 9.2, yielding blue crystals of the holoenzyme belonging to space group P21 with similar cell dimensions as described for crystals of the apoenzyme. Subsequently, such a blue crystal was soaked in a solution containing 1 mM MH, 3 mM CaCl2, 120 mM NaCl, and 25% (wt/vol) PEG6000 in 50 mM 2-[N-cyclohexyamino]-ethanesulfonic acid (CHES) buffer, pH 9.2. In the course of the 2.5-hr soaking time, the color of the crystal changed from blue to light green. At this point, the crystal was flash frozen in a cryo-protecting solution consisting of 3 mM CaCl2, 120 mM NaCl, 25% (wt/vol) PEG6000, and 15% (vol/vol) glycerol in 50 mM CHES buffer, pH 9.2.
Data Collection, Processing, and Structure Refinement.
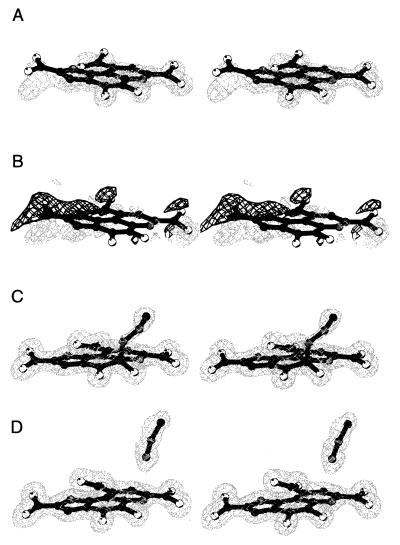
A complete data set was collected to 1.5-Å resolution at beam line ID14–3 at the European Synchrotron Radiation Facility, Grenoble. Data processing and reduction were performed with denzo/scalepack (20). The data set was divided into a work and a test set (21), consisting of 5% of the reflections and including all reflections of the apoenzyme refinement test set. Refinement was initially done with refmac/arp and auxiliary programs from the CCP4 suite (22). The apo-structure, refined at 1.7-Å resolution, was used as an initial model. After a few rounds of model refinement, some manual adjustments were made, and the loop regions of residues A335-A344, B105-B110, and B335-B344, not present in the apo-model because of the absence of convincing electron density, were added to the model by using the program o (23). At this stage, σA-weighted (24) 2Fo − Fc and Fo − Fc electron-density maps unequivocally revealed the presence of PQQ in the crystals. PQQ was manually fitted to the electron density in both monomers and initially refined with planarity restraints on the entire molecule. After initial refinement, σA-weighted (24) 2Fo − Fc and Fo − Fc electron density maps showed that the cofactor is not completely planar in either molecule (Fig. 3 A and B). In the next round of refinement, tight planarity restraints were maintained on each of the three rings of PQQ and the carboxylate groups, but restraints constraining planarity between rings and/or carboxylates were released. The resulting PQQ model clearly fitted the electron density much better (Fig. 3 C and D).
Figure 3.
(A) 2Fo-Fc electron density, contoured at 1.5 σ, for PQQ, refined as a completely planar molecule. (B) Density (3 σ Fo-Fc) (light grey) and −3 σ Fo-Fc density (dark grey) for planar PQQ. (C) Electron density (2Fo-Fc), contoured at 1 σ, for the final model of the carbinolamine type complex in monomer B. (D) Electron density (2Fo-Fc), contoured at 1 σ, for the final model of PQQ and noncovalently bound MH in monomer A. This picture was created with bobscript (30).
At this point, continuous density from PQQ to a water molecule could be recognized in monomer B in the 2Fo − Fc electron density map. A MH molecule, covalently attached to the C5 atom of PQQ with an occupancy of 0.7, fitted this density well (Fig. 3C). Attempts to refine monomer A in an identical fashion did not result in the appearance of continuous electron density. Instead, a MH molecule could be fitted to a large piece of density at a different position in the active site of that monomer (Fig. 3D). Nevertheless, utilization of the bulk solvent correction and anisotropic scaling procedures of x-plor (25) resulted in the appearance of positive Fo − Fc electron density near the unligated PQQ C5 atom in monomer A, suggesting that MH may have partially formed a covalent complex with PQQ in this monomer. Refinement of the occupancies of the two conformations showed that this covalent complex has an occupancy of only 0.26 (occupancy is 0.74 for the noncovalently bound MH conformation). A summary of data collection and structure refinement statistics is given in Table 1.
Table 1.
Statistics of data collection and refinement and quality of the final model
| Space group | P21 |
| Cell dimensions: | |
| a, Å, b, Å, c, Å, β, ° | 60.6, 92.7, 85.7, 105.4 |
| Resolution range, Å | 100–1.50 |
| Total number of reflections | 580,379 |
| No. of unique reflections | 142,076 |
| Rmerge(I)* | 0.033 |
| Rmerge(I)* (1.53–1.50 Å) | 0.340 |
| Completeness of data, % | 97.8 |
| Completeness, % (1.53–1.50 Å) | 91.2 |
| Refinement resolution range, Å | 20–1.50 |
| No. of nonhydrogen atoms in protein model | 7,040 |
| No. of calcium atoms | 6 |
| No. of ligand nonhydrogen atoms | 90 |
| No. of water molecules | 900 |
| Final R factor†/R free‡ | 0.174/0.195 |
| Root mean square deviations from ideality for | |
| Bond lengths, Å | 0.027 |
| C bond angles, ° | 2.9 |
| Dihedral angles, ° | 27.2 |
| Improper dihedral angles, ° | 1.6 |
Rmerge (I) = ∑hkl∑I | Ii(hkl) − 〈I(hkl)〉|/∑hkl∑i/i(hkl)
R factor = ∑hkl||Fobs, work| − k|Fcalc||/∑hkl|Fobs, work|
R free = ∑hkl||Fobs, test| − k|Fcalc||/∑hkl|Fobs, test|
Results and Discussion
Structure Refinement.
The s-GDH-PQQ-MH complex was refined to a crystallographic R factor of 17.4% (with a free R factor of 19.5%). The final model has excellent stereochemistry (Table 1). It comprises two protein monomers, six calcium ions, 900 water molecules, one glycerol, one PQQ, one MH molecule with two conformations, and two carbinolamine type PQQ-MH complexes. Some of these ligand molecules/complexes have partial occupancies. The model contains all protein residues with the exception of residues A105-A110, A451–454, and B453–454.
PQQ Binding.
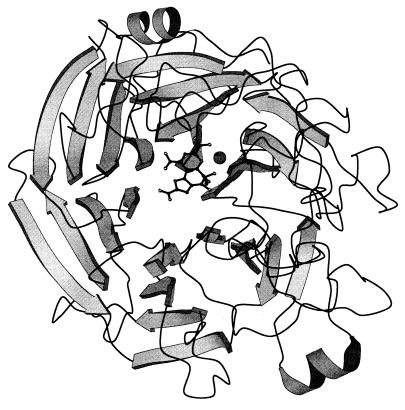
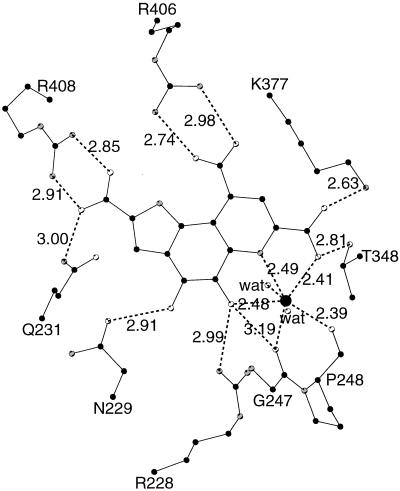
PQQ resides in a deep and broad solvent-accessible cleft in the center of each monomer, near the top of the β-propeller structure (Fig. 4). The binding of the cofactor to s-GDH is predominantly governed by polar interactions (Fig. 5). The C2, C7, and C9 carboxyl groups of PQQ (PQQ nomenclature explained in Fig. 1) form salt bridges with Arg-408, Lys-377, and Arg-406, respectively. The ortho-quinone O4 and O5 atoms are bound by Asn-229 and Arg-228, respectively. The N6, O7A, and O5 atoms of PQQ are ligands for the active-site calcium ion. The other calcium ligands are provided by the two main chain carbonyl oxygen atoms of Gly-247 and Pro-248 and two water molecules.
Figure 4.
Ribbon diagram of one s-GDH monomer. PQQ and MH are shown in ball-and-stick representation. The calcium ion is shown as a large grey sphere. Oxygen, nitrogen, and carbon atoms are shown in white, grey, and black, respectively. This picture was produced with molscript (31).
Figure 5.
Hydrogen bonding interactions in the active site of s-GDH. Distances are given in Ångstroms. Oxygen, nitrogen and carbon atoms are shown in white, grey and black, respectively. This picture was created with molscript (31).
The overall binding of PQQ is similar to that in the active site of MDH (26, 27). In both proteins, equatorial interactions are polar, whereas hydrophobic stacking interactions are formed on both sides of the plane of PQQ. The active-site calcium ion and one arginine side chain are ligated to PQQ in an almost identical fashion.
Conformation of PQQ.
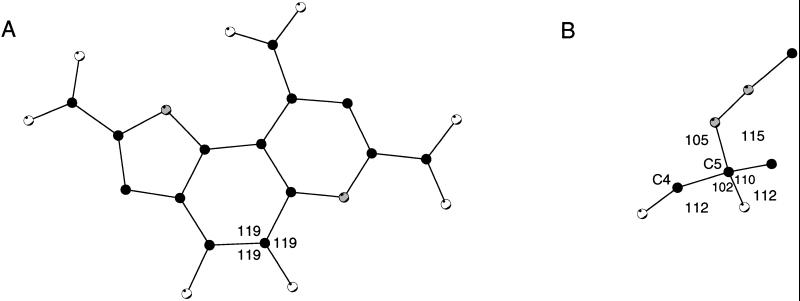
Because the ternary complex is refined at 1.5-Å resolution, it presents a detailed view of the conformation of PQQ in s-GDH (Fig. 6A). A superposition of the two PQQ molecules in the dimer shows that the cofactor binds in an identical fashion in the two active sites (not shown). The conformation of PQQ, mostly unligated in monomer A and complexed with MH in monomer B, is virtually the same in either active site. Only the conformation of the C5 atom varies because of a different hybridization state (sp2 in PQQ vs. sp3 in the covalent adduct; see below). The geometry of the cofactor molecule deviates significantly from planarity in both active sites. The three carboxylate groups are all to some extent twisted of the plane of the monocyclic ring to which they are attached. The orientations of these groups seem optimized for polar interactions with the protein ligands (Fig. 5). The O4 and O5 atoms of the ortho-quinone are also of the plane of the cofactor. The possibility that these oxygen atoms may be positioned of the molecular plane was previously suggested by model studies of Zheng and Bruice (12), who proposed that they may be positioned on opposite sides of the PQQ plane to alleviate repulsive interactions. In the present structure, however, both atoms are positioned on the same side of the plane. This is compensated for by several favorable interactions (Fig. 5). The O4 atom forms a hydrogen bond with the ND2 atom of Asn-229, whereas the O5 atom is within hydrogen bonding distance of the NH1 atom of Arg-228 and the active-site calcium ion. The deformations around the C6A-C9A and C9-C9X bonds orient the carboxylic groups at C9 and C7 optimally for polar interactions with Arg-406 and Lys-377, respectively (Fig. 5). Thus, the nonplanar conformation of the cofactor seems to be stabilized by polar interactions with several protein side chains and the active-site calcium ion of s-GDH.
Figure 6.
(A) Conformation of unligated PQQ in monomer A. (B) Conformation of the C5 atom of the carbinolamine type complex in monomer B. Angles are given in degrees. Oxygen, nitrogen, and carbon atoms are shown in white, grey, and black, respectively. This picture was created with molscript (31).
Noncovalently Bound MH.
In the majority (74%) of the monomers A in the crystal, MH is bound in a noncovalent fashion, packed against one side of a wide cavity that is present in the surface of the protein directly above PQQ. In this active site, MH is bound in two slightly different conformations, which differ only by a rotation of the methyl group around the N1–N2 bond. This methyl group makes hydrophobic interactions with Tyr-343 and Trp-346, the N2 atom has no interactions with the protein surroundings, and the N1 atom has a water-mediated interaction with the active-site calcium ion. Interestingly, a glycerol solvent molecule is present at a similar position in the crevice above PQQ in the holoenzyme structure (unpublished results). Because this crevice is wide enough to accommodate even larger compounds such as glucose, it may well be the substrate-binding site of s-GDH.
Covalent Carbinolamine Type Complex of PQQ and MH.
In 26% and 70% of the monomers A and B in the crystal, respectively, MH is covalently bound to the C5 atom of PQQ. Compared with noncovalently bound MH, the inhibitor N1 atom has moved over a distance of 2.6 Å in the direction of the PQQ C5 atom. This latter atom has been pushed up above the plane of PQQ and has adopted a tetrahedral configuration (Fig. 6B).
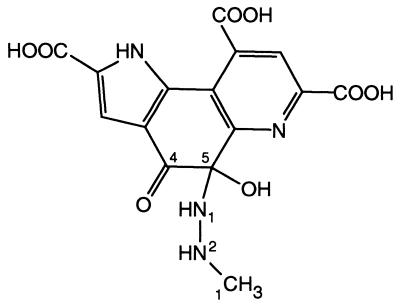
The carbinolamine type PQQ–MH complex (Fig. 7) is the first structure of a covalent adduct of any compound to PQQ in an enzymatic setting. Small molecule structures of PQQ adducts of methanol, acetone, and cyclopropanol have been determined previously (2, 9, 28). Furthermore, spectroscopic evidence indicates that a variety of compounds are able to form adducts at the PQQ C5 position in solution (29). These results show that the reactivity of PQQ toward nucleophiles is highest at the C5 atom, in solution as well as in the active site of s-GDH. Therefore, it seems likely that the reaction mechanism of s-GDH, and possibly other PQQ-dependent proteins, proceeds through nucleophilic attack of a substrate or hydride anion on the PQQ C5 atom and not on the C4 atom, as has been proposed on the basis of ab initio calculations (12).
Figure 7.
Structure of the adduct of PQQ and MH. Atom nomenclature is indicated for the MH atoms and for the carbon atoms of the ortho-quinone group of PQQ.
Two Different Modes of Binding.
MH is bound in two distinct modes in the two active sites of the homodimer. The covalent PQQ–MH complex has been formed in about 26% and 70% of the A and B monomers in the crystal, respectively, whereas noncovalently bound MH is present in the active site of the remaining 74% of the A subunits. This indicates that the reactivity of the two active sites in the dimer is not identical. However, no significant structural differences are present between the two active sites nor in the area between the two cofactors, including the dimerization interface. The most pronounced difference between the active sites is the flexibility of the loops comprising residues 335–344. The B factors of the Cα-atoms of monomer A in this loop are on average 10 Å2 lower than they are in monomer B. The more rigid loop of monomer A makes hydrophobic interactions with the noncovalently bound MH, whereas the more flexible loop of subunit B does not. It thus seems that the binding of MH in a noncovalent form is related to a relatively rigid conformation of the 335–344 loop. Because the loops of the two monomers make different interactions with crystallographically related molecules, the differential reactivity of these monomers toward MH may be a result of differing crystal packing interactions.
Inhibition Mechanism.
The two different binding modes of MH in the dimer suggest a mechanism for the inhibition of s-GDH by MH. First, the inhibitor may enter the wide solvent accessible crevice in the protein surface and might pack against the face constituted by residues 343 and 346, making a water-mediated interaction with the active-site calcium, as is observed in monomer A. In the next step, MH could move over a distance of approximately 2.6 Å in the direction of the PQQ C5 atom. The lone pair of the N1 atom of MH could then perform a nucleophilic attack on the C5 atom of PQQ. The reactivity of this C5 atom toward nucleophilic compounds may be enhanced by polarization of the C5–O5 bond (11, 12). Polarization of this bond may be accomplished by the nearby Arg-228 and the active-site calcium ion. Formation of the carbinolamine type PQQ–MH complex could be completed by the net transfer of the proton from the N1 atom of MH to the O5 atom of PQQ. A similar function of increasing the activity of the PQQ C5 atom has been proposed for the presence of positive charge close to the O5 atom in the active site of MDH (27).
This proposed inhibition mechanism is similar to the addition-elimination mechanism (Fig. 2), which is one of the two reaction mechanisms that have been proposed for the oxidation of glucose on the basis of kinetic experiments with s-GDH (11). It is tempting to assume that this PQQ–MH complex provides the lacking structural evidence to substantiate the addition-elimination mechanism. However, glucose and MH are chemically distinct with respect to their oxidizable groups. MH is a much more reactive compound than glucose, because the N1 atom of MH can directly perform a nucleophilic attack with its lone pair, whereas the O1 atom of glucose first has to lose a proton before it can act as a nucleophile. The prerequisite of releasing the proton from the glucose O1 atom before reaction with PQQ may demand a completely different orientation of glucose, which might be unsuitable for the actual addition to occur, and it thus remains to be established whether s-GDH oxidizes its substrates via covalent addition.
Conclusions
Our results suggest that the native state of PQQ in s-GDH is nonplanar and that the substrate binding site is located directly above the cofactor. Moreover, MH is likely to inhibit s-GDH via covalent addition of its N1 atom to the cofactor C5 atom, which adopts a tetrahedral conformation in the resulting carbinolamine type PQQ–MH complex. This structure represents the first covalent adduct in a PQQ-dependent protein and provides conclusive structural evidence that the carbonyl group at the cofactor C5 position is the most reactive group of PQQ in the active site of s-GDH.
Acknowledgments
We are grateful to Dr. W. Burmeister at beam line ID14–3 at the European Synchrotron Radiation Facility (ESRF), Grenoble, for help during data collection, and to the ESRF for support of the work at the ESRF. We thank Hans Duine for providing unpublished kinetic data and for stimulating discussions during the preparation of this manuscript. The investigations were supported by the Netherlands Foundation for Chemical Research (SON) with financial aid from the Netherlands Organization for Scientific Research (NWO).
Abbreviations
- GDH
glucose dehydrogenase
- MDH
methanol dehydrogenase
- MH
methylhydrazine
- PQQ
pyrroloquinoline quinone
- PQQH2
pyrroloquinoline quinone
- s-GDH
soluble GDH
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code ICR4).
References
- 1.Duine J A, Frank Jzn J. Trends Biochem Sci. 1981;2:278–280. [Google Scholar]
- 2.Salisbury S A, Forrest H S, Cruse W B T, Kennard O. Nature (London) 1979;280:843–844. doi: 10.1038/280843a0. [DOI] [PubMed] [Google Scholar]
- 3.McIntire W S, Wemmer D E, Chistoserdov A, Lidstrom M E. Science. 1991;252:817–824. doi: 10.1126/science.2028257. [DOI] [PubMed] [Google Scholar]
- 4.Janes S M, Mu D, Wemmer D, Smith A J, Kaur S, Maltby D, Burlingame A L, Klinman J P. Science. 1990;248:981–987. doi: 10.1126/science.2111581. [DOI] [PubMed] [Google Scholar]
- 5.Wang S X, Mure M, Medzihradsky K F, Burlingame A L, Brown D E, Dooley D M, Smith A J, Kagan H M, Klinman J P. Science. 1996;273:1078–1084. doi: 10.1126/science.273.5278.1078. [DOI] [PubMed] [Google Scholar]
- 6.Duine J A, Frank J, van Zeeland J K. FEBS Lett. 1979;108:443–446. doi: 10.1016/0014-5793(79)80584-0. [DOI] [PubMed] [Google Scholar]
- 7.Duine J A, Frank J, Verwiel P E. Eur J Biochem. 1980;108:187–192. doi: 10.1111/j.1432-1033.1980.tb04711.x. [DOI] [PubMed] [Google Scholar]
- 8.Frank Jzn J, Dijkstra M, Duine J A, Balny C. Eur J Biochem. 1988;174:331–338. doi: 10.1111/j.1432-1033.1988.tb14102.x. [DOI] [PubMed] [Google Scholar]
- 9.Frank J, Jr, van Krimpen S H, Verwiel P E, Jongejan J A, Mulder A C, Duine J A. Eur J Biochem. 1989;184:187–195. doi: 10.1111/j.1432-1033.1989.tb15006.x. [DOI] [PubMed] [Google Scholar]
- 10.Anthony C. Biochem J. 1996;320:697–711. doi: 10.1042/bj3200697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Olsthoorn A J J, Duine J A. Biochemistry. 1998;37:13854–13861. doi: 10.1021/bi9808868. [DOI] [PubMed] [Google Scholar]
- 12.Zheng Y-J, Bruice T C. Proc Natl Acad Sci USA. 1997;94:11881–11886. doi: 10.1073/pnas.94.22.11881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Itoh S, Kawakami H, Fukuzumi S. J Am Chem Soc. 1997;119:439–440. [Google Scholar]
- 14.Itoh S, Fukuzumi S. Biochemistry. 1998;37:6562–6571. doi: 10.1021/bi9800092. [DOI] [PubMed] [Google Scholar]
- 15.Oubrie A, Rozeboom H J, Kalk K H, Duine J A, Dijkstra B W. J Mol Biol. 1999;289:319–333. doi: 10.1006/jmbi.1999.2766. [DOI] [PubMed] [Google Scholar]
- 16.Dokter P, Frank Jzn J, Duine J A. Biochem J. 1986;239:163–167. doi: 10.1042/bj2390163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cleton-Jansen A M, Goosen N, Vink K, van de Putte P. Mol Gen Genet. 1989;217:430–436. doi: 10.1007/BF02464914. [DOI] [PubMed] [Google Scholar]
- 18.Olsthoorn A J J, Otsuki T, Duine J A. Eur J Biochem. 1997;247:659–665. doi: 10.1111/j.1432-1033.1997.00659.x. [DOI] [PubMed] [Google Scholar]
- 19.Matsushita K, Shinagawa E, Adachi O, Ameyama M. Biochemistry. 1989;28:6276–6280. doi: 10.1021/bi00441a020. [DOI] [PubMed] [Google Scholar]
- 20.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 21.Brünger A T. Nature (London) 1992;355:472–475. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
- 22.Collaborative Computational Project Number 4. Acta Crystallogr D. 1994;50:760–763. [Google Scholar]
- 23.Jones T A, Zou J-Y, Cowan S W, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 24.Read R J. Acta Crystallogr A. 1986;42:140–149. [Google Scholar]
- 25.Brünger A T, Kuriyan J, Karplus M. Science. 1987;235:458–460. doi: 10.1126/science.235.4787.458. [DOI] [PubMed] [Google Scholar]
- 26.Ghosh M, Anthony C, Harlos K, Goodwin M G, Blake C. Structure (London) 1995;3:177–187. doi: 10.1016/s0969-2126(01)00148-4. [DOI] [PubMed] [Google Scholar]
- 27.Xia Z-X, Dai W-W, Zhang Y-E, White S A, Boyd G D, Mathews F S. J Mol Biol. 1996;259:480–501. doi: 10.1006/jmbi.1996.0334. [DOI] [PubMed] [Google Scholar]
- 28.Itoh S, Ogino M, Fukui Y, Murao H, Komatsu M, Ohshiro Y, Inoue T, Kai Y, Kasai N. J Am Chem Soc. 1993;115:9960–9967. [Google Scholar]
- 29.Dekker R H, Duine J A, Frank Jzn J, Verwiel P E, Westerling J. Eur J Biochem. 1982;125:69–73. doi: 10.1111/j.1432-1033.1982.tb06652.x. [DOI] [PubMed] [Google Scholar]
- 30.Esnouf R M. J Mol Graph. 1997;15:132–134. doi: 10.1016/S1093-3263(97)00021-1. [DOI] [PubMed] [Google Scholar]
- 31.Kraulis P. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]