Abstract
In Salmonella typhimurium, the CobU, CobS, CobT, and CobC proteins have been proposed to catalyze the late steps in adenosylcobalamin biosynthesis, which define the nucleotide loop assembly pathway. This paper reports the in vitro assembly of the nucleotide loop of adenosylcobalamin from its precursors adenosylcobinamide, 5,6-dimethylbenzimidazole, nicotinate mononucleotide, and GTP. Incubation of these precursors with the CobU, CobS, and CobT proteins resulted in the synthesis of adenosylcobalamin−5′-phosphate. This cobamide was isolated by HPLC, identified by UV-visible spectroscopy and mass spectrometry, and shown to support growth of a cobalamin auxotroph. Adenosylcobalamin−5′-phosphate was also isolated from reaction mixtures containing adenosylcobinamide-GDP (the product of the CobU reaction) and α-ribazole-5′-phosphate (the product of the CobT reaction) as substrates and CobS. These results allowed us to conclude that CobS is the cobalamin(-5′-phosphate) synthase enzyme in S. typhimurium. The CobC enzyme, previously shown to dephosphorylate α-ribazole-5′-phosphate to α-ribazole, was shown to dephosphorylate adenosylcobalamin−5′-phosphate to adenosylcobalamin. Adenosylcobinamide was converted to adenosylcobalamin in reactions where all four enzymes were present in the reaction mixture. This in vitro system offers a unique opportunity for the rapid synthesis and isolation of cobamides with structurally different lower-ligand bases that can be used to investigate the contributions of the lower-ligand base to cobalamin-dependent reactions.
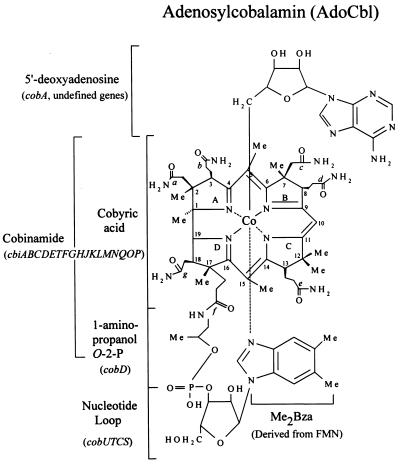
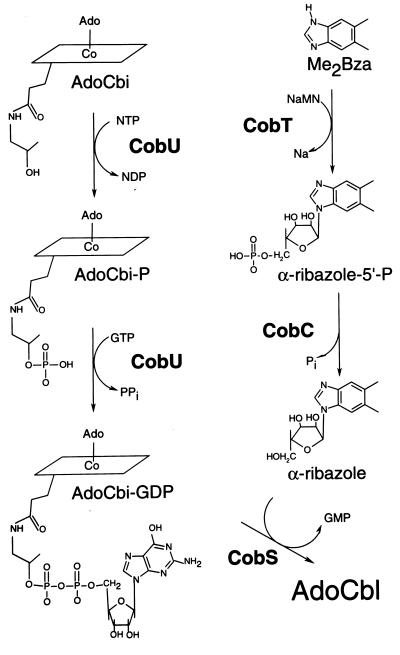
Salmonella typhimurium LT2 is capable of de novo synthesis of adenosylcobalamin (AdoCbl) only under anaerobic growth conditions (1). This bacterium possesses a highly specific transport system that translocates complete and incomplete corrinoids across the outer and inner membranes (2, 3). During aerobic growth, cobinamide (Cbi, Fig. 1) is transported into the cell by this system. Once inside the cell, Cbi is adenosylated before it can serve as substrate for the enzymes that catalyze the assembly of the nucleotide loop (4–6). The nucleotide loop is the structure that joins the lower-ligand base to the 1-amino-2-propanol substituent of the corrin ring (Fig. 1). The conversion of adenosylcobinamide (AdoCbi) to AdoCbl is known as the nucleotide loop assembly (NLA) pathway. Four enzymes have been implicated in this pathway, namely CobU, CobS, CobT, and CobC (Fig. 2). CobU is the AdoCbi kinase/AdoCbi-phosphate guanylyltransferase that activates AdoCbi to AdoCbi-GDP via AdoCbi-phosphate (7, 8). CobT is the nicotinate mononucleotide (NaMN):5,6-dimethylbenzimidazole (DMB) phosphoribosyltransferase that synthesizes N1-(5-phospho-α-d-ribosyl)-DMB (also known as α-ribazole-5′-phosphate; α-ribazole-5′-P; refs. 9 and 10). CobC has α-ribazole-5′-P phosphatase activity (11); and CobS has been proposed to be the putative cobalamin synthase that catalyzes the synthesis of AdoCbl from AdoCbi-GDP and α-ribazole. The assignment of CobS as the cobalamin synthase was based strictly on its homology to the CobV protein of Pseudomonas denitrificans (12–14).
Figure 1.
Structure of AdoCbl.
Figure 2.
NLA pathway.
The CobU and CobT proteins have been isolated and biochemically characterized. The activity for CobC has been documented; however, little progress has been made on the characterization of CobS. In this paper, we address the role of CobS in this pathway and report the development of an in vitro system for the assembly of the nucleotide loop from its precursors, AdoCbi, GTP, NaMN, and DMB. The cobamide generated by this system (AdoCbl-5′-P) was converted to cyanocobalamin (CNCbl)-5′-P, isolated by reverse-phase HPLC (RP-HPLC), identified by UV-visible spectroscopy and mass spectrometry, and shown to support growth of a cobalamin auxotroph. The data obtained allowed us to assign cobalamin−5′-P synthase activity to the CobS protein. We also address the timing of the removal of the 5′-P group by CobC. We show that CobC dephosphorylates AdoCbl-5′-P in vitro. These results suggest that in vivo, the timing of phosphate removal most likely depends on the affinities of CobS and CobC for α-ribazole-5′-P.
The development of the in vitro NLA system, combined with the lack of substrate specificity of CobT, CobC, and CobS, offers a unique opportunity for the rapid synthesis and purification of cobamides with different lower ligands. These cobamides would be valuable tools for the analysis of the contributions of the lower ligand to the function of this essential coenzyme.
Materials and Methods
Bacteria, Culture Media, and Growth Conditions.
S. typhimurium strains JE212 [metE205 ara-9 Δ299(hisG-cob)] and JE2197 [metE205 ara-9 Δ299(hisG-cob) cobC1175∷Tn10Δ16Δ17)] were grown in Luria–Bertani medium supplemented, when appropriate for the maintenance of plasmids, with ampicillin (100 μg/ml) and/or kanamycin (50 μg/ml). For a complete list of strains and plasmids, see Table 1. The production of NLA enzymes from T7 expression vectors in JE2197 was accomplished by including the vector pGP1-2 (15) to provide temperature-sensitive synthesis of T7 RNA polymerase. Cultures were grown in 1-liter batches at 30°C to A600 = 0.6. Cultures were transferred to a 42°C water bath for 45 min to induce synthesis of the T7 polymerase. After 4 h of incubation at 37°C, cells were harvested by centrifugation at 11,700 × g for 10 min in a Sorvall RC-5B refrigerated centrifuge (DuPont). The Escherichia coli strain BL21(λDE3) was used to express His-tagged CobS from pCOBS5 as described (16).
Table 1.
Strain and plasmid list
| Strain | Genotype | Ref. or source |
|---|---|---|
| S. typhimurium | ||
| JE212 | metE205 ara-9 Δ299(hisG-cob) | Lab collection |
| JE2197 | metE205 ara-9 Δ299(hisG-cob)cobC1175∷Tn10Δ16Δ17 | Lab collection |
| JE4793 | JE2197/pGP1-2 | This work |
| Escherichia coli | ||
| DH5α | endA1 hsdR17 (rk− mk+) supE44 thi-1 recA1 gyrA (Nalr) relA1 Δ(lacZYA-argF) U169 deoR (Δ80dlacΔ(lacZ)M15) | 24 |
| JE4064 | DH5α/pNLA1 | This work |
| JE3676 | DH5α/pCOBS4 | This work |
| JE4065 | DH5α/pCOBS5 | This work |
| BL21(λDE3) | F′ ompT hsdSB (rB− mB−) gal dcm (DE3) | Novagen |
| Plasmids | ||
| pNLA1 | cobUST+bla+ (derivative of pT7-7) | This work |
| pCOBS4 | cobS+bla+ (derivative of pT7-7) | This work |
| pCOBS5 | cobS+bla+ (derivative of pT7-6) | This work |
| pJO12 | cobS+cat+ (vector: pSU21) | 13 |
| pJO46 | cobC+bla+ (derivative of pT7-5) | 11 |
| pJO52 | cobU+bla+ (derivative of pT7-7) | 7 |
| pGP1-2 | T7 rpo+ (RNA polymerase) kan+ | 15 |
| pT7-7 | T7 overexpression vector, bla+ | 15 |
| pT7-5 | T7 overexpression vector, bla+ | 15 |
| pT7-6 | T7 overexpression vector, bla+ | 15 |
| pSU19 | cat+, cloning vector | 20 |
| pSU21 | cat+, cloning vector | 20 |
| pET-15b | N-terminal His6 tag vector, bla+ | Novagen |
Bioassays were used to detect product formation during in vitro NLA and cobalamin synthase reactions. Strain JE212, a cobalamin auxotroph, was used as indicator strain in all bioassays. Strain JE212 carries a defective metE gene. The lack of the MetE enzyme requires that methylation of homocysteine to methionine be catalyzed by the cobalamin-dependent MetH methionine synthase enzyme (17). In the absence of cobalamin, strain JE212 is a methionine auxotroph. Cells from an overnight culture in complex medium were washed once with sterile saline. Approximately 108 cells were added to 3 ml of molten 0.7% (wt/vol) agar and overlaid on Vogel–Bonner minimal medium (18) supplemented with 11 mM glucose and 0.1 mM histidine. In vitro reactions were centrifuged to pellet denatured proteins, and 5 μl of each supernatant was spotted onto the overlay. CNCbl (14 pmol) was also applied as a positive control. Growth was assessed after overnight incubation at 37°C.
Construction of Expression Plasmids.
The recombinant DNA techniques used to construct the following plasmids have been reported (19).
Plasmid pCOBS4.
This plasmid was constructed by PCR amplification of cobS from plasmid pJO12 (13). The 5′ primer sequence (ggagtcaaaatCaTatgagtaag) created an NdeI site at the putative translation initiation codon for cobS. The 5′ primer and the 3′ (tccggcgcaggaatgtcac) primers were synthesized at the Biotechnology Center at the University of Wisconsin. The PCR product was blunt-end ligated into plasmid pSU19 (20) to yield plasmid pCOBS2. The 850-bp NdeI–HindIII fragment of this plasmid was cloned into vector pT7-7 (15) to produce plasmid pCOBS4. This strategy placed the translation initiation codon of cobS immediately 3′ to the pT7-7 ribosome-binding site.
Plasmid pCOBS5.
This plasmid was constructed by cloning the 810-bp NdeI–BamHI fragment of plasmid pCOBS4 into plasmid pET15b (Novagen) cut with the same enzymes. This generated an N-terminal, His-tagged (His)6CobS fusion protein.
Plasmid pNLA1.
This plasmid was constructed by cloning the 2.2-kilobase EcoRI–XmaI fragment from plasmid pMJ2 (cobUST+) into pSU19 and then moving the EcoRI–HindIII fragment into plasmid pJO52 (cobU+) (7). This strategy placed all three genes under the control of the T7 promoter and ribosome-binding site.
Synthesis of NLA Substrates.
AdoCbi and AdoCbi-GDP.
These corrinoids were synthesized from dicyanocobinamide [(CN)2Cbi] by using homogeneous CobA and CobU enzymes as described (7).
Radiolabeled DMB.
([14C-2]DMB (specific activity = 30.4 Ci/mol) was synthesized as described (10, 21).
α-Ribazole-5′-P.
This precursor was synthesized from DMB and NaMN by using homogeneous phosphoribosyltransferase enzyme CobT (10). α-Ribazole-5′-P was purified by HPLC (Waters) by using a C18 Prodigy column (Phenomenex, Torrance, CA). The column was developed with the following protocol. A 4-min linear gradient from 98% A/2% B to 90% A/10% C was followed by a 6-min linear gradient to 75% A/25%C. Finally, a 30-min linear gradient to 100% C was performed. Solvents A, B, and C were distilled H2O, 1 M NaCl, and 100% (vol/vol) methanol, respectively. α-Ribazole-5′-P eluted 19 min after injection.
Radiolabeled α-ribazole-5′-P.
This precursor was synthesized from [14C-2]DMB and purified by TLC on silica plates (Kodak 13179) by employing two different mobile phases. The first TLC was developed by using a mobile phase of methanol:chloroform (3:2; ref. 9). α-Ribazole-5′-P was extracted from the origin along with unreacted NaMN by using methanol. The second TLC was developed with a mobile phase of 35% (vol/vol) methanol. The relative mobility (Rf) of α-ribazole-5′-P was 0.68; unreacted NaMN had an Rf of 0.81. Radiolabeled α-ribazole-5′-P was extracted from the plate with methanol.
In Vitro NLA Reaction with CobU, CobS, and CobT Enzymes.
The complete reaction mixture contained AdoCbi (1.2 nmol), GTP (40 nmol), DMB (2 nmol), NaMN (20 nmol), Ches buffer [pH 9; 1 μmol of 2-(N-cyclohexylamino)ethanesulfonic acid], MgCl2 (50 nmol), and a source of CobU, CobS, and CobT enzymes. The final volume of the reaction mixture was 20 μl. Cell-free extracts of the expression strain carrying plasmid pNLA1 (overexpresses cobUST+) were made at 4°C in 0.1 M Tris⋅HCl buffer (pH 8.0) containing 5 mM DTT (7); 7.5 μg of total protein were used per reaction. When homogeneous CobU (7) and CobT (10) enzymes were used, 0.17 μg and 0.015 μg were added, respectively. In these reactions, the source of CobS enzyme was either (i) 2.5 μg of cell-free extract of strain JE4793 carrying plasmid pCOBS4, containing cobalamin synthase with a specific activity of 8 (nmol of AdoCbl-5′-P synthesized per min per mg of protein) or (ii) 0.4 μg of protein of a (His)6CobS preparation with a specific activity of 20. Reactions were incubated at 37°C for 20 min. Reactions were stopped by the addition of 20 μl of 20 mM KCN, followed by heating at 80°C for 10 min. This treatment generated the cyano derivatives of all corrinoids. All reactions were performed under dim light or in darkness to prevent photolysis of the C–Co bond between the cobalt ion and 5′-deoxyadenosine. The product of these reactions was detected by using the bioassay described above with strain JE212 as the indicator strain. The in vitro NLA reaction was scaled up to 1 ml for HPLC analysis (described below).
In Vitro NLA Reaction Containing CobU, CobS, CobT, and CobC Enzymes.
These reactions mixtures contained the same components as the reaction described above but scaled up to 1 ml. Protein (75 μg) from a cell-free extract of strain JE4793 carrying plasmid pNLA1 (cobUST+) was used as the source of CobU, CobS, and CobT enzymes. Similarly, 34 μg of cell-free extract protein from strain JE4793 carrying plasmid pJO46 (cobC+; ref. 11) was used as the source of CobC enzyme. The level of CobC enzyme in the extract was unknown. In one experiment, CobC (or a vector-only control extract) was added 1 h after the addition of CobU, CobS, and CobT. Reactions were stopped as described above 1.5 h after the addition of CobC. In a separate experiment, CobC was added simultaneously with CobU, CobS, and CobT. This reaction was incubated for 1 h and was stopped as described above. All reactions were incubated at 37°C. The products of these reactions were analyzed by RP-HPLC (see below).
In Vitro Cobalamin Synthase Assays.
A complete reaction contained AdoCbi-GDP (1.2 nmol), α-ribazole-5′-P (1.2 nmol), Ches buffer (pH 9; 1 μmol), MgCl2 (50 nmol), and a source of CobS enzyme in a final volume of 20 μl. Reactions were stopped by adding 20 μl of 20 mM KCN and incubating at 80°C for 10 min. The product of these reactions was detected by using the bioassay described above. To quantitate cobalamin synthase activity, assays were performed by using a mixture of 0.3 nmol of [14C]α-ribazole-5′-P and 0.9 nmol of unlabeled α-ribazole-5′-P. Reactions were stopped by the addition of 5 μl of 100 mM KCN, followed by heating at 80°C for 10 min. Reactions were applied to polyethyleneimine cellulose TLC plates (CEL 300 PEI, Macherey & Nagel). TLC plates were developed with a mobile phase of 0.1 M potassium phosphate (pH 8.0) containing 10 mM KCN. α-Ribazole-5′-P and CNCbl-5′-P migrated with Rf values of 0.33 and 0.43, respectively. Quantitation was achieved with a Storm 660 PhosphorImager (Molecular Dynamics). A unit of cobalamin synthase activity was defined as the amount of enzyme needed to generate 1 nmol of product per min.
RP-HPLC Analysis of NLA and Cobalamin Synthase Reaction Products.
NLA reaction mixtures were passed over a C18 SepPak column (Waters) previously equilibrated with water. Corrinoids were eluted with methanol, vacuumed to dryness in a SpeedVac Concentrator (Savant), and resuspended in 200 μl of distilled H2O. Corrinoids were separated by using a Waters HPLC system equipped with a Prodigy C18 column (Phenomenex) as described (13). Corrinoids were identified by their spectra by using a Waters photodiode array detector. (CN)2Cbi, (CN)2Cbi-GDP, and CNCbl were used as standards.
Mass Spectrometry.
The HPLC-purified product of the NLA reactions was desalted by adsorbing it onto a C18 SepPak (Waters), washing it with water to remove salt, eluting it with 100% (vol/vol) methanol, and vacuuming it to dryness. The sample was submitted for analysis to the Mass Spectrometry Facility at the University of Wisconsin-Madison Biotechnology Center. The mass spectrum was obtained from a Perkin–Elmer Sciex API365 triple quadrupole instrument with an ion spray source (positive ion mode).
Other Procedures.
The purification of (His)6CobS was performed by nickel affinity chromatography according to the manufacturers’ instructions (Novagen). The concentration of proteins in samples was determined as described (22). SDS/PAGE was performed as reported (23).
Results
In Vitro Cobamide Synthesis from AdoCbi and DMB.
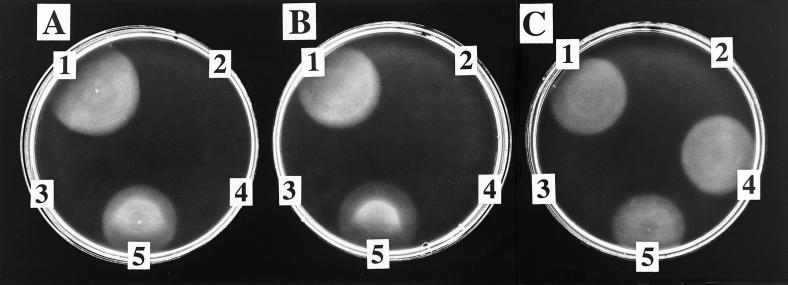
Incubation of AdoCbi, GTP, DMB, and NaMN with cell-free extract of a strain carrying plasmid pNLA1 (cobUST+) yielded a cobamide that supported cobalamin-dependent growth of strain JE212 (Fig. 3A). No growth of strain JE212 was observed when the same substrates were incubated with cell-free extract of a strain carrying plasmid pT7-7 (vector-only control). If any one of the substrates was omitted, growth was not observed. Cell-free extract of the strain carrying plasmid pNLA1 was replaced by homogeneous CobU and CobT proteins and either a cell-free extract of a strain carrying plasmid pCOBS4 (cobS+) or purified (His)6CobS protein (Fig. 3B). Cobalamin biosynthetic activity was observed over a pH range from 7 to 10 (data not shown). Although the qualitative assessment of activity was not ambiguous, quantitation of the amount of product synthesized in vitro was difficult because of variability in the bioassay (thickness of agar plate and cells per ml in the overlay).
Figure 3.
Bioassay for the detection of cobamides synthesized in vitro. (A and B) Detection of cobamides synthesized from AdoCbi and DMB (i.e., NLA reactions). (C) Detection of cobamides synthesized from AdoCbi-GDP and α-ribazole-5′-P (i.e., cobalamin synthase reactions). A 5-μl sample of each reaction mixture was applied to an overlay of strain JE212 on minimal glucose medium supplemented with 0.1 mM histidine. (A) Results from reaction mixtures containing cell-free extract (from a strain carrying pNLA1) as the source of CobU, CobS, and CobT. Shown are the complete reaction mixture (1); the vector-only control reaction (2); a reaction mixture lacking DMB (3); 70 pmol (CN)2Cbi standard (4); and 14 pmol CNCbl standard (5). (B) Results from reactions performed by using homogeneous CobU and CobT and partially purified (His)6CobS. Shown are the complete reaction (1); reactions in which CobU (2), CobS (3), and CobT (4) were individually omitted; and 14 pmol CNCbl standard (5). (C) Shown are the complete reaction with a cell-free extract of a strain carrying pCOBS4 as a source of CobS (1); the vector-only control reaction (2); a reaction mixture lacking α-ribazole-5′-P (3); the complete reaction with (His)6CobS (4); and 14 pmol CNCbl standard (5).
In Vitro Cobamide Synthesis from AdoCbi-GDP and α-Ribazole-5′-P.
Incubation of AdoCbi-GDP (the product of the CobU reaction) and α-ribazole-5′-P (the product of the CobT reaction) with cell-free extract of a strain carrying pCOBS4 (cobS+) yielded a cobamide that supported cobalamin-dependent growth of strain JE212 (Fig. 3C). Growth of JE212 was not observed when the same substrates were incubated with cell-free extract of a control strain carrying plasmid pT7-7, the cloning vector used to construct pCOBS4. A purified (His)6CobS preparation (described further below) was substituted for the pCOBS4 cell-free extract in these reactions, and the same results were obtained. To quantitate the level of cobalamin synthase activity present in the cell-free extracts, radiolabeled cobalamin synthase reactions were performed. Extracts from cells carrying plasmids pNLA1 (cobUST+) or pCOBS4 (cobS+) contained cobalamin synthase with specific activities of 22 or 8 nmol of product per min per mg of protein, respectively. These results allowed us to conclude that CobS had cobalamin synthase activity.
Purification of His-Tagged Cobalamin Synthase.
CobS was not expressed from plasmid pCOBS4 at levels that were visible in Coomassie blue-stained SDS/PAGE gels. The same system failed to detect (His)6CobS protein encoded by plasmid pCOBS5. Use of standard nickel affinity chromatography resulted in 2.4% of the total cobalamin synthase activity binding to the column. Therefore, the increase in specific activity in the eluate represented a 10-fold purification (specific activity = 20.4 vs. 1.9 in the cell-free extract loaded onto the column), despite the fact that only three contaminating proteins were visible in the SDS/PAGE gel. A vector-only control purification (with pET-15b) showed these same three contaminants, indicating that (His)6CobS was still not visible after purification but that its activity was readily detectable. Cell-free extracts of the strain carrying the cloning vector without cobS contained no cobalamin synthase activity.
Isolation of Products from the NLA and Cobalamin Synthase Reactions.
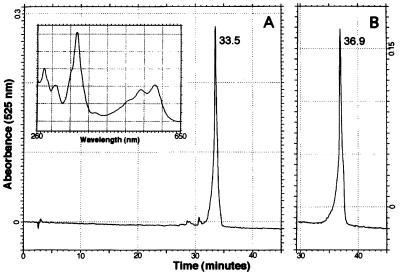
Corrinoids were isolated from a NLA assay mixture by RP-HPLC after derivatization of all corrinoids with KCN. Fig. 4A shows that the reaction product eluted 33.5 min after injection. Authentic CNCbl eluted at 36.9 min (Fig. 4B). Standards of the substrate (CN)2Cbi and the NLA intermediate (CN)2Cbi-GDP eluted at 32.3 and 29 min, respectively (data not shown). Although the UV-visible spectrum of the reaction product was identical to that of authentic CNCbl (Fig. 4A Inset), we concluded that this corrinoid was a modified form of CNCbl, because it eluted off the column 3.4 min faster than the CNCbl standard. The same approach was used to identify the product of the CobS reaction when AdoCbi-GDP and α-ribazole-5′-P were used as substrates. HPLC analysis revealed similar results, with corrinoids eluting at 29 min [(CN)2Cbi-GDP] and 33.6 min (reaction product). The retention time and spectrum of the product of the cobalamin synthase reaction indicated that this molecule was identical to the product of the NLA reaction.
Figure 4.
RP-HPLC analysis of the product of the NLA reaction. Shown are chromatograms of the elution of the product of the NLA reaction (A) and authentic (CN)2Cbl (15 nmol) (B). A cell-free extract of a strain carrying pNLA1 (cobUST+) was used as the source of enzyme. The corrinoid shown in A eluted 33.5 min after injection. (B) Authentic CNCbl eluted at 36.9 min. (Inset) The spectrum of the reaction product (A) was identical to that of authentic CNCbl (data not shown).
Identification of Reaction Product as CNCbl-5′-P.
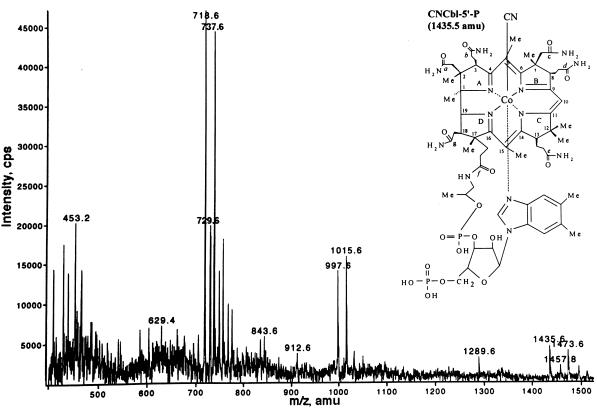
The corrinoid that eluted at 33.5 min was analyzed by electrospray ionization mass spectrometry, and the results are presented in Fig. 5. Corrinoids were imparted with either a +1 or +2 charge during ionization, giving rise to two peaks per species. Signals with m/z ratios of 718.6 and 1,435.6; 729.6 and 1457.8; and 737.6 and 1,473.6 were consistent with molecular masses for CNCbl-5′-P, NaCNCbl-5′-P, and KCNCbl-5′-P, respectively. The signal with an m/z ratio of 1,289.6 agreed with the molecular mass of CNCbl-5′-P in which the DMB has been removed. The signals at 997.6 and 1,015.6 were not assigned. No signals were observed that were consistent with the molecular masses of CNCbl or its various salts.
Figure 5.
Mass spectrum of the product of the NLA reaction. Shown is the positive-ion electrospray ionization mass spectrum of the HPLC-purified reaction product. Signals with m/z values of 1,435.6, 1,457.8, and 1,473.6 were consistent with the molecular masses of CNCbl-5′-P, NaCNCbl-5′-P, and KCNCbl-5′-P, respectively, where z = +1. Signals with m/z values of 718.6, 729.6, and 737.6 were consistent with the masses of the same three compounds, where z = +2. (Inset) The structure and predicted molecular mass of CNCbl-5′-P.
The CobC Protein Can Dephosphorylate AdoCbl-5′-P in Vitro.
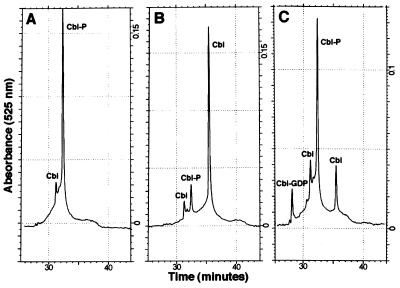
A cell-free extract of a strain expressing CobC from plasmid pJO46 was prepared and included in the NLA reaction. AdoCbl-5′-P was generated in situ after a 1-h preincubation of the NLA substrates with a cell-free extract providing CobU, CobS, and CobT. The CobC extract (or a vector-only control extract) was then added, and the reactions were incubated for an additional 1.5 h. Fig. 6 shows the HPLC analysis of the reaction products after derivatization with KCN. In the presence of CobC, most of the AdoCbl-5′-P was dephosphorylated to AdoCbl. No dephosphorylation was observed in the vector-only control. When CobC was added simultaneously with CobU, CobS, and CobT for an incubation of 1 h, both CNCbl-5′-P and CNCbl were observed in the HPLC chromatogram.
Figure 6.
RP-HPLC analysis of NLA reactions mixtures containing CobC. (A and B) Chromatograms indicating the fate of AdoCbl-5′-P after a 1.5-h incubation with cell-free extract from a strain carrying plasmid pT7-5 (vector-only control; A) or plasmid pJO46 (cobC+; B). The chromatogram in C shows the elution profile obtained when cell-free extracts of strains carrying pNLA1 (cobUST+) and pJO46 (cobC+) were incubated simultaneously with NLA substrates for 1 h. (CN)2Cbi-GDP, (CN)2Cbi, CNCbl-5′-P, and CNCbl eluted at 28.2, 31.2, 32.4, and 35.5 min, respectively. Cbl, cobalamin.
Discussion
The Nucleotide Loop of Cobalamin Can Be Assembled by CobU, CobT, and CobS in the Absence of CobC.
This paper presents in vitro evidence for the assembly of the nucleotide loop of cobalamin by either crude cell-free extracts or purified enzymes. The in vitro assembly of the entire nucleotide loop of cobalamin from AdoCbi, GTP, DMB, and NaMN resulted in the formation of AdoCbl-5′-P, which corrected the nutritional requirement of cobalamin auxotrophs. This result is of interest for two reasons: (i) it shows that the CobS cobalamin synthase enzyme can use α-ribazole-5′-P as substrate, and (ii) it brings into focus the timing of the reaction catalyzed by CobC. The ability of CobS to synthesize AdoCbl-5′-P from AdoCbi-GDP and α-ribazole-5′-P provides biochemical evidence that supports the assignment of cobalamin synthase function to this protein.
Timing of the CobC Reaction.
The experimental conditions of our in vitro NLA system allowed cobalamin synthase (CobS) to synthesize AdoCbl-5′-P, suggesting that this enzyme can use the nucleotide (α-ribazole-5′-P) in the absence of the CobC phosphatase enzyme. The kinetic analysis of the CobS reaction with α-ribazole or α-ribazole-5′-P and CobC with α-ribazole-5′-P or AdoCbl-5′-P as substrates is needed to define the timing of phosphate removal in vivo more accurately. In the absence of this information and of a quantitative assessment of the level of CobC enzyme activity, the preponderance of AdoCbl-5′-P over AdoCbl does not necessarily mean that AdoCbl-5′-P is the penultimate intermediate in the pathway.
Potential Use of the in Vitro NLA System for the Synthesis of Modified Cobamides.
Tzrebiatowski and Escalante-Semerena (10) demonstrated the synthesis of cobamides with alternative lower ligands by using guided biosynthesis. These in vivo experiments showed that CobT and CobS can use a variety of lower-ligand bases as substrates yielding physiologically active cobamides. Now that the entire nucleotide loop can be assembled in vitro, the incorporation of these alternative lower ligands into cobamides and the subsequent activity of these cobamides with cobalamin-dependent enzymes can be assessed. By exploiting this lack of specificity of CobT and CobS through the synthesis of cobamides with structurally different lower-ligand bases, we may come to a better understanding of the contributions of the lower-ligand base to cobalamin-dependent reactions.
Acknowledgments
This work was supported by National Institutes of Health Grant GM40313 (to J.C.E.-S.). L.M.-H. was supported by a National Science Foundation predoctoral fellowship. We thank Z. Kong for obtaining the electrospray ionization mass spectrum at the University of Wisconsin-Madison Biotechnology Center.
Abbreviations
- Cbi
cobinamide
- NLA
nucleotide loop assembly
- CNCbl
cyanocobalamin
- RP-HPLC
reverse-phase HPLC
- NaMN
nicotinate mononucleotide
- DMB
5,6-dimethylbenzimidazole
- α-ribazole-5′-P
N1-(5-phospho-α-ribosyl)-5,6-dimethylbenzimidazole
- (CN)2Cbi
dicyanocobinamide
- AdoCbi
adenosylcobinamide
- AdoCbl
adenosylcobalamin
- -5′-P
-5′-phosphate
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Jeter R M, Olivera B M, Roth J R. J Bacteriol. 1984;159:206–213. doi: 10.1128/jb.159.1.206-213.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.DeVeaux L C, Clevenson D S, Bradbeer C, Kadner R J. J Bacteriol. 1986;167:920–927. doi: 10.1128/jb.167.3.920-927.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heller K, Kadner R J. J Bacteriol. 1985;161:904–908. doi: 10.1128/jb.161.3.904-908.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Escalante-Semerena J C, Suh S-J, Roth J R. J Bacteriol. 1990;172:273–280. doi: 10.1128/jb.172.1.273-280.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Suh S-J, Escalante-Semerena J C. Gene. 1993;129:93–97. doi: 10.1016/0378-1119(93)90701-4. [DOI] [PubMed] [Google Scholar]
- 6.Suh S-J, Escalante-Semerena J C. J Bacteriol. 1995;177:921–925. doi: 10.1128/jb.177.4.921-925.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O’Toole G A, Escalante-Semerena J C. J Biol Chem. 1995;270:23560–23569. doi: 10.1074/jbc.270.40.23560. [DOI] [PubMed] [Google Scholar]
- 8.Thompson T B, Thomas M G, Escalante-Semerena J C, Rayment I. Biochemistry. 1998;37:7686–7695. doi: 10.1021/bi973178f. [DOI] [PubMed] [Google Scholar]
- 9.Trzebiatowski J R, O’Toole G A, Escalante-Semerena J C. J Bacteriol. 1994;176:3568–3575. doi: 10.1128/jb.176.12.3568-3575.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Trzebiatowski J R, Escalante-Semerena J C. J Biol Chem. 1997;272:17662–17667. doi: 10.1074/jbc.272.28.17662. [DOI] [PubMed] [Google Scholar]
- 11.O’Toole G A, Trzebiatowski J R, Escalante-Semerena J C. J Biol Chem. 1994;269:26503–26511. [PubMed] [Google Scholar]
- 12.Roth J R, Lawrence J G, Rubenfield M, Kieffer-Higgins S, Church G M. J Bacteriol. 1993;175:3303–3316. doi: 10.1128/jb.175.11.3303-3316.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O’Toole G A, Rondon M R, Escalante-Semerena J C. J Bacteriol. 1993;175:3317–3326. doi: 10.1128/jb.175.11.3317-3326.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blanche F, Debussche L, Famechon A, Thibaut D, Cameron B, Crouzet J. J Bacteriol. 1991;173:6066–6073. doi: 10.1128/jb.173.19.6066-6073.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tabor S. In: Current Protocols in Molecular Biology. Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Vol. 2. New York: Wiley; 1990. pp. 16.2.1–16.2.11. [Google Scholar]
- 16.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 17.Taylor R R. In: B12. Dolphin D, editor. Vol. 2. New York: Wiley; 1982. pp. 307–356. [Google Scholar]
- 18.Vogel H J, Bonner D M. J Biol Chem. 1956;218:97–106. [PubMed] [Google Scholar]
- 19.Brushaber K R, O’Toole G A, Escalante-Semerena J C. J Biol Chem. 1998;273:2684–2691. doi: 10.1074/jbc.273.5.2684. [DOI] [PubMed] [Google Scholar]
- 20.Martínez E, Bartolomé B, de la Cruz F. Gene. 1988;68:159–162. doi: 10.1016/0378-1119(88)90608-7. [DOI] [PubMed] [Google Scholar]
- 21.Stupperich E, Kräutler B. Arch Microbiol. 1988;149:213–217. [Google Scholar]
- 22.Kunitz M J. J Gen Physiol. 1952;35:423–450. doi: 10.1085/jgp.35.3.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 24.Raleigh E A, Lech K, Brent R. In: Current Protocols in Molecular Biology. Ausubel F A, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Vol. 1. New York: Wiley; 1989. pp. 1.4.1–1.4.14. [Google Scholar]