Abstract
Pseudocomplementary PNAs containing diaminopurine⋅thiouracil base pairs have been prepared and are shown to bind with high specificity and efficiency to complementary targets in double-stranded DNA by a mechanism termed “double duplex invasion” in which the duplex is unwound and both DNA strands are targeted simultaneously, each by one of the two pseudocomplementary peptide nucleic acids (PNAs). On the basis of our results we predict that (for decameric targets) more than 80% of all sequences can be targeted by straightforward Watson–Crick base pairing by using this approach in its present form. Targeting of pseudocomplementary PNAs to the promoter of the T7 phage RNA polymerase effectively inhibits transcription initiation. These results have important implications in the development of gene therapeutic agents as well as for genetic diagnostic and molecular biology applications.
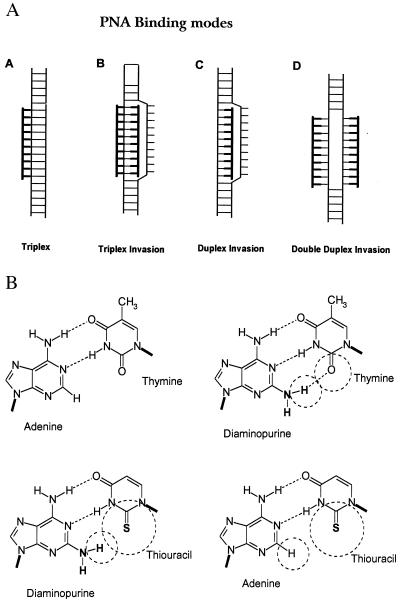
General principles for design of synthetic reagents that sequence-specifically recognize and bind desired targets in double-stranded DNA have been long-sought goals in chemistry and molecular biology (1–3). Three approaches, triplex-forming oligonucleotides (2–4), helix-invading peptide nucleic acids (PNAs) (5, 6), and side-by-side minor groove binders (7, 8), have accomplished impressive progress during the past 10 years, but unresolved limitations on sequence choice and/or target length and specificity still exist for all approaches.
In particular, DNA targeting by triplex-forming oligonucleotides and helix-invading triplex-forming PNAs is still basically limited to homopurine targets because of the involvement of third-strand Hoogsteen recognition (9). However, general sequence recognition by invasion of the DNA double helix exploiting just the Watson–Crick base pairing principle might be accomplished by helix invasion using duplex-forming PNAs (10). Unfortunately, mixed purine-pyrimidine sequence PNAs do not provide sufficient free energy upon hybridization for this mode of binding. However, the required free energy could be gained if both DNA strands were targeted simultaneously (Fig. 1A). Naturally, the two PNAs (or oligonucleotides) would be sequence complementary and if composed of the natural nucleobases A, C, G, and T would “quench” each other by forming a stable duplex. Thus modified nucleobases that are pseudocomplementary to each other are needed—i.e., they should each recognize their natural A⋅T or G⋅C counterpart, but not be able to recognize each other. As also pointed out by Kutyavin et al. (11), a 2,6-diaminopurine⋅2-thiothymine (or thiouracil) base pair could fulfill this requirement. However, in the DNA case efficient DNA invasion with such oligonucleotides could not be demonstrated (11). 2,6-Diaminopurine recognizes thymine more efficiently than does adenine in both DNA and a PNA contexts (12), having an extra hydrogen bond and increased base stacking area (Fig. 1B), and even though sulfur is much larger than oxygen, this should not interfere with the binding of 2-thiouracil to adenine. However, the diaminopurine⋅thiouracil base pair should be significantly destabilized because of steric hindrance (Fig. 1B). To test this recognition principle in a PNA context, we set out to synthesize and study the DNA-binding properties of PNA oligomers in which diaminopurine was substituted for adenine and 2-thiouracil was substituted for thymine.
Figure 1.
(A) Modes by which PNA may recognize double-stranded DNA. (B) Schematic drawing of adenine⋅thymine, diaminopurine⋅thymine, diaminopurine⋅thiouracil, and adenine⋅thiouracil base pairs showing how diaminopurine can form an extra hydrogen bond with thymine, whereas a steric clash occurs between diaminopurine and thiouracil.
Materials and Methods
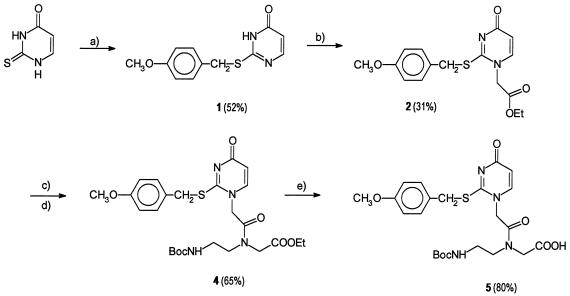
The S-4-methoxybenzyl-protected 2-thiouracil PNA monomer was prepared in five steps as shown in Scheme S1. Standard PNA Boc monomers, HBTU, and methylbenzhydryl amine (MBHA) resin were from PerSeptive Biosystems (Framingham, MA); other reagents were from Aldrich. NMR spectra were recorded in DMSO-d6 on a Varian 400-MHz Unity spectrometer; fast atom bombardment (FAB) mass spectra, on a JEOL HX 110/110 mass spectrometer; and matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectra, on a Kratos MALDI II spectrometer.
Scheme 1.
Synthesis of 2-thiouracil PNA monomer. (a) 4-Methoxybenzyl chloride in alkaline (KOH) water/EtOH (1:1). (b) Ethyl bromoacetate in ethanolic sodium ethoxide followed by precipitation from ethyl acetate. (1H nuclear Overhauser enhancement NMR analysis and comparison of the 13C NMR spectrum with reports in the literature of N-alkylated S-benzylthiouracils (13) confirmed the assignment of the desired product (N1-alkylation.) (c) LiOH in water/MeOH/tetrahydrofuran (THF) (1:3:7). (d) Ethyl N-(2-Boc-aminoethyl)glycinate/O-benzotriazole-1-yl-N,N,N′,N′-tetramethyluronium hexafluorophosphate (HBTU) in dimethylformamide. (e) LiOH in water/MeOH/THF (1:3:7).
Synthesis.
S-(4-Methoxybenzyl)-2-thiouracil, 1.
2-Thiouracil (6.4 g, 50 mmol) was dissolved in water/EtOH (25 ml/25 ml) together with KOH (3.95 g, 60 mmol). The mixture was heated to 45°C, and 4-methoxybenzyl chloride (8.6 g, 55 mmol, 7.5 ml) was added. After 20 min, the mixture is evaporated to dryness, and the crude product was suspended in NaHCO3 (50 ml, 10%), and filtered off. It was washed with water, EtOH, and diethyl ether. Yield: 6.4 g (52%). 1H NMR (δ/ppm): 3.75 (3H, s), 4.35 (2H, s), 6.13 (1H, d), 6.87 (2H, d), 7.32 (2H, d), 7.92 (1H, d), 12.6 (1H, broad s). 13C NMR (δ/ppm): 33.32, 55.14, 113.97, 128.78, 130.93, 158.61. Mass: 249.1 (M + H). Elemental analysis (%): found (calc. for C12H12N2O2S⋅0.1H2O) C 57.59 (57.64), H 4.81 (4.92), N 10.80 (11.20). S 12.83 (12.82).
N-1-(Ethyloxycarboxymethyl)-S-(4-methoxybenzyl)-2-thiouracil,2.
Sodium (620 mg, 27 mmol) was dissolved in refluxing absolute EtOH (50 ml), and S-(4-methoxybenzyl)-2-thiouracil (1) (6.4 g, 26 mmol) was added and dissolved. Ethyl bromoacetate (4.7 g, 27 mmol, 3.1 ml) was added and reflux was continued for 1 h. The mixture was evaporated to dryness and partitioned between water (50 ml) and dichloromethane (150 ml)/MeOH (50 ml). The organic phases was evaporated to dryness, taken up in ethyl acetate/hexane, and again evaporated. The residue was triturated in cold ethyl acetate (10 ml), whereby the product precipitated. It was filtered off, and washed with ethyl acetate. Yield: 2.7 g (31%). 1H NMR (δ/ppm): 1.17 (3H, t), 3.72 (3H, s), 4.14 (2H, q), 4.35 (2H, s), 4.79 (2H, s), 5.94 (1H, d), 6.87 (2H, d), 7.33 (2H, d), 7.67 (1H, d). 13C NMR (δ/ppm): 13.97, 34.45, 52.51, 55.16, 61.85, 108.75, 113.99, 128.00, 130.50, 145.62, 158.78, 162.39, 166.47, 167.06. Mass: 335.2 (M + H). Elemental analysis (%): found (calc. for C16H18N2O4S⋅0.75 H2O) C 55.00 (55.25), H 5.34 (5.65), N 8.01 (8.05), S 9.11 (9.22).
N-1-(Carboxymethyl)-S-(4-methoxybenzyl)-2-thiouracil, 3.
N-1-(Ethyloxycarboxymethyl)-S-(4-methoxybenzyl)-2-thiouracil (2) (2.7 g, 8.1 mmol) was dissolved in MeOH/tetrahydrofuran (100 ml, 1:2). LiOH (10.0 ml, 2.0 M) was added, and after 300 s (5 min), HCl (10.0 ml, 2.0 M) was added. The volume was reduced to 20 ml, whereby the product precipitated. Yield: 1.85 g (75%). 1H NMR (δ/ppm): 3.72 (3H, s), 4.34 (2H, s), 4.70 (2H, s), 6.11 (1H, d), 6.87 (2H, d), 7.33 (2H, d), 7.89 (1H, d), 12.7 (1H, broad s). 13C NMR (δ/ppm): 34.39, 52.60, 55.16, 108.61, 114.00, 128.01, 130.51, 145.73, 158.76, 162.40, 166.58, 168.41. Mass: 304.9 (M − H). Elemental analysis (%): found (calc. for C14H14N2O4S) C 54.71 (54.86), H 4.59 (4.61), N 9.09 (9.15), S 10.49 (10.47).
S-(4-Methoxybenzyl)-2-thiouracil-PNA monomer ethyl ester, 4.
N-1-(Carboxymethyl)-S-(4-methoxybenzyl)-2-thiouracil (3) (1.7 g, 5.5 mmol) was dissolved in dimethylformamide (15 ml) and triethylamine (1 ml). Ethyl N-(Boc-aminoethyl)glycinate (1.45 g, 6 mmol) (14) and HBTU (2.28 g, 6.0 mmol) were added and the mixture was gently heated to 45°C for 30 min. Ethyl acetate (100 ml) was added and the mixture was extracted with sodium citrate (50 ml, 10%, pH 4.5) and NaHCO3 (50 ml, 10%). The organic phase was evaporated to dryness and purified on a silica column with MeOH (15–20%) in dichloromethane. Yield: 2.45 g (86%). 1H NMR (δ/ppm): 1.14 (3H, t), 1.32 and 1.35 (9H, two s), 3.02 and 3.18 (2H, two q), 3.37 (2H, t), 3.72 (3H, s), 4.02–4.34 (4H, overlapping signals), 4.76 and 4.96 (2H, two s), 5.91–5.93 (1H, two d), 6.86 (1H, broad t), 6.87 (2H, d), 7.33 (2H, two d), 7.46 (1H, two d). 13C NMR (δ/ppm): 14.10, 28.23, 34.59, 35.87, 55.17, 60.66, 113.96, 130.49 Mass: 557.2 (M + Na).
S-(4-Methoxybenzyl)-2-thiouracil-PNA monomer, 5.
S-(4-Methoxybenzyl)-2-thiouracil-PNA monomer ethyl ester (4) (2.38 g, 4.45 mmol) was dissolved in tetrahydrofuran (42 ml) and water (16 ml) and cooled to 0°C. LiOH (2.45 ml, 2.00 M, 1.1 eq) was added, and stirring was continued for 15 min on ice. HCl (2.45 ml, 2.0 M) was added and the mixture was freeze dried. This produced a white powder, which was suspended in ice water (15 ml), stirred for 10 min, and then filtered off and dried. Yield 1.8 g (80%). 1H NMR (δ/ppm): 1.29 and 1.32 (9H, two s), 3.02 and 3.14 (2H, two q), 3.28 and 3.37 (2H, two t), 4.12 and 4.32 (2H, 2 s), 4.73 and 4.92 (2H, two s), 5.91–5.93 (1H, two d), 6.67 (1H, broad t), 6.85 (2H, d), 7.33 (2H, d), 7.46 (1H, two d). 13C NMR (δ/ppm): 28.51, 35.03, 55.51, 108.81, 114.31, 130.88, 146.48, 159.04 ≈163.44. Mass: 505.15 (M − H).
PNA oligomer synthesis.
PNA oligomers were prepared on a MBHA resin by standard procedures (15) and were purified by HPLC. The oligomers were typically obtained in 10–20% yield when isolated. They were homogeneous by HPLC analysis and exhibited one major peak at the expected mass as analyzed by MALDI-TOF mass spectrometry. The latter also confirmed that the S-4-methoxybenzyl group was removed during cleavage from the resin.
Molecular Biology.
Construction and isolation of plasmid DNA, labeling of restriction fragments, and KMnO4 probing experiments were performed as previously described (5, 10, 16). Briefly, plasmids containing the 5′-GTAGATCACT target were constructed by cloning of oligonucleotides 5′-GATCGTAGATCACT and 5′-GATCAGTGATCTAC into the BamHI site of pBluescript KS + (Stratagene) (p206). The plasmids were digested with restriction enzymes HindIII and PvuII and 3′-32P-end-labeled at the HindIII site by using [α-32P]ATP and the Klenow fragment of Escherichia coli DNA polymerase. The small (204-bp) HindIII–PvuII DNA fragment was isolated by polyacrylamide gel electrophoresis and used for the gel-shift and the probing experiments.
Gel-shift experiments were performed by mixing the desired amount of PNA(s) with 40 cps of 32P-labeled DNA fragment in 20 μl of buffer (10 mM sodium phosphate/1 mM EDTA, pH 7.0). The samples were incubated for 16 h at 37°C and subsequently analyzed by gel electrophoresis in 10% polyacrylamide. Radioactive DNA bands were visualized by autoradiography.
Probing experiments with KMnO4 were performed in 100 μl of buffer (10 mM sodium phosphate/1 mM EDTA, pH 7.0) containing approximately 200 cps of 32P-labeled DNA fragment and the desired amount of PNA. After incubation of DNA with the PNA for 16 h at 37°C, the probing reagent was added, the incubation was continued at room temperature for 15 s, and the reactions were finally terminated by the addition of 50 μl of 1 M 2-mercaptoethanol/1.5 M NaOAc, pH 7.0. The samples probed were subsequently treated with piperidine (0.5 M, 90°C, 20 min) prior to gel analysis. The DNA was precipitated by addition of 200 μl of 2% KOAc in 96% EtOH and was analyzed by electrophoresis in 10% polyacrylamide sequencing gels. Radioactive DNA bands were visualized by autoradiography using amplifying screens and Agfa Curix RP1 x-ray films exposed at −70°C.
Job plots were made by mixing various ratios of PNA and oligonucleotide, keeping the total concentration constant. The samples were heated to 90°C and slowly cooled to 20°C, and the absorbance at 260 nm was measured.
Results and Discussion
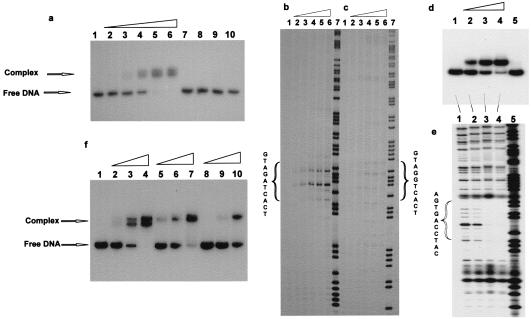
We have previously synthesized PNAs containing 2,6-diaminopurine (D) and found a thermal stabilization of 2–4°C per diaminopurine⋅thymine base pair in PNA⋅DNA duplexes (12). Hence we synthesized a 2-thiouracil (sU) PNA monomer and incorporated this into PNA decamers. Thermal stability (in terms of melting temperature, Tm) measurements on the corresponding PNA⋅DNA duplexes clearly show the expected stabilization upon incorporation of diaminopurine (5–6°C per diaminopurine⋅thymine base pair) (Table 1), and a slight destabilization upon incorporation of thiouracil (2–3°C per thiouracil⋅adenine base pair). Next we synthesized a pair of sequence complementary PNA decamers (GTAGATCACT/AGTGATCTAC) in which all adenines and thymines were replaced by diaminopurine and thiouracil, respectively. These were found to hybridize better to their DNA complements than did the original PNAs, but did not bind to each other as assayed by Tm measurement or Job plot (data not shown). This pair of pseudocomplementary PNAs was therefore a good candidate for double duplex invasion binding. The gel mobility-shift experiments presented in Fig. 2a clearly show that these two PNAs in combination, and only in combination, bind efficiently in the nanomolar concentration range to a 204-bp double-stranded DNA fragment containing an internal sequence-complementary target for the PNAs. A permanganate chemical probing experiment (Fig. 2b) confirmed these results and, furthermore, demonstrated that binding was indeed taking place exclusively to the target. Very importantly, no specific binding was seen by chemical probing (Fig. 2c) or gel shift analyses (not shown) even at 30-fold higher PNA concentration to a similar DNA fragment in which the 10-mer target contained a single A → G mutation. However, when a set of PNAs complementary to the mutated target (GTAGGTCACT/AGTGACCTAC) was used, binding was restored as detected by a gel shift assay (Fig. 2d), and DNase I footprinting demonstrated that binding was taking place specifically to the dedicated target (Fig. 2e). To address the question how many diaminopurine⋅thiouracil base pairs are required to obtain efficient double duplex invasion with such PNAs, we tested two other pairs of PNAs having only 50% or 40% diaminopurine⋅thiouracil base pairs. The results (Fig. 2f) showed that these PNAs also bind to the target although with considerably lower efficiency (3- and 10-fold) than the fully (60%) substituted PNA. On the basis of these results we predict that any sequence having at least 40% A+T content can be targeted by double duplex invading PNAs, and this would statistically cover more than 83% of all 10-mer targets. In principle, an analogous pseudocomplementary G⋅C base pair that would allow fully sequence unrestricted targeting can be constructed, but it will be a challenge in chemical design and synthesis.
Table 1.
Thermal stabilities (Tm) of PNA·DNA and PNA·PNA complexes
| PNA sequence (PNA number) |
Tm, °C
|
|
|---|---|---|
| Vs. DNA* | Vs. PNA† | |
| H-GTAGATCACT-Lys-NH2 | 51 | 68 |
| H-GTAGDTCACT-Lys-NH2 | 57 | 71 (64)‡ |
| H-GTDGDTCDCT-Lys-NH2 | 67 | 81 (49)§ |
| H-Lys-GsUDGDsUCDCsU-Lys-NH2 (1495) | 58 | 74 |
| H-AGTGATCTAC-Lys-NH2 | 49 | 68 |
| H-AGTGAsUCTAC-Lys-NH2 | 46 | 68 |
| H-AGUsGAsUCsUAC-Lys-NH2 | 44 | 68 |
| H-Lys-DGsUGDsUCsUDC-Lys-NH2 (1496) | 58 | 74 |
| H-Lys-GsUDGDsUCDCT-Lys-NH2 (1674) | 65.5 | |
| H-Lys-AGsUGDsUCsUDC-Lys-NH2 (1672) | 56 | |
| H-Lys-GsUAGDsUCDCT-Lys-NH2 (1675) | 61 | |
| H-Lys-AGsUGDsUCTDC-Lys-NH2 (1673) | 58 | |
| H-Lys-GsUDGGsUCDCsU-Lys-NH2 (1837) | 67 | |
| H-Lys-DGUsGDCCsUDC-Lys-NH2 (1838) | 66 | |
| H-Lys-DCGDCsUCDCsU-Lys-NH2 (1657) | 67 | |
| H-Lys-DGsUGDGsUCGsU-Lys-NH2 (1658) | 68 | |
The complementary DNA oligonucleotides 5′-d(AGTGATCTAC) and 5′-d(GTAGATCACT) were used. Thermal stabilities (Tm) were measured in 10 mM sodium phosphate/100 mM NaCl/0.1 mM EDTA, pH 7.0, at a heating rate of 0.5 °C per step (0.7°C/min) (5–90°C).
The complementary PNA oligomers H-AGTGATCTAC-Lys-NH2 and H-GTAGATCACT-Lys-NH2 were used.
Measured vs. H-AGTGAsUCTAC-Lys-NH2.
Measured vs. H-AGsUGAsUCsUAC-Lys-NH2.
Figure 2.
Binding of pseudocomplementary PNAs to double-stranded DNA. (a) PNAs 1495 and 1496 (see Table 1) were incubated for 16 hr at 37°C in 10 mM sodium phosphate/1 mM EDTA buffer, pH 7.0, with a 204-bp DNA fragment (HindIII/PvuII fragment of p206 3′-32P-end-labeled at the HindIII site) containing the 10-bp PNA target. The following concentrations were used: lanes 1–6, 0, 5, 10, 20, 40, and 80 nM each of PNAs 1495 and 1496; lanes 7 and 9, 40 and 80 nM, respectively, of PNA 1495; and lanes 8 and 10, 40 and 80 nM, respectively, of PNA 1496. The presence of two shifted complexes is not understood. HPLC and MALDI-TOF analyses of the PNAs did not indicate heterogeneity, but it could be that there are two slowly exchanging “structural isomers” of this very “crowded” double duplex invasion complex. (b and c) Sequence specificity of the PNA binding. PNAs 1495 and 1496 were bound to the HindIII/PvuII fragment of p206 (b) or p259 (one mismatch target) (c) as described above. Subsequently the complexes were probed with KMnO4 and the samples were analyzed by electrophoresis in polyacrylamide sequencing gels followed by autoradiography. The PNA concentrations in lanes 1–7 were 0, 60, 200, and 600 nM and 2 mM and 6 mM. Lane 7 is an A/G sequence marker. The 10-mer PNA targets are indicated. (d) Experiment as in a except that the PNA pairs 1837/1838 (H-Lys-GsUDGGsUCDCsU-Lys-NH2/H-Lys-DGsUGDCCsUDC-Lys-NH2) and the plasmid p259 were used. The PNA concentrations were as follows: lane 1, control without PNA; lanes 2–5, 30 nM PNA 1838 and 100 nM, 150 nM, 200 nM, and 0 nM of PNA 1837, respectively. (e) DNase I probing (after adjusting the buffer to 50 mM Tris⋅HCl, pH 7.5/1 mM Mg2Cl) of samples 1–4 as described for d; lane 5 is an A/G sequence marker. (f) Experiment as in a except that the PNA pairs 1495/1496 (60% AT modification) (lanes 2–4), 1674/1672 (H-Lys-GsUDGDsUCDCT-Lys-NH2/H-Lys-AGsUGDsUCsUDC-Lys-NH2) (50% AT modification) (lanes 5–7), and 1675/1673 (H-Lys-GsUAGDsUCDCT-Lys-NH2/H-Lys-AGsUGDsUCTDC-Lys-NH2) (40% AT modification) (lanes 8–10) were used at concentrations of 5 nM (lane 2), 15 nM (lanes 3, 5, and 8), 45 nM (lanes 4, 6, and 9), or 150 nM (lanes 7 and 10). Lane 1 is a control without PNA.
As has previously been found for (10-mer) PNA triplex invasion complexes (17, 18), preliminary results indicate that the binding of the presently investigated double duplex invasion PNAs is kinetically controlled—i.e., the binding efficiency is reflecting the on-rate binding constants, and the complexes do not dissociate measurably during the experiment (results not shown). The double duplex invasion reaction also resembles that of the triplex invasion in terms of ionic strength dependence (15, 19), exhibiting significantly decreased binding with increasing ionic strength. Further studies are required to determine the kinetic and mechanistic details of this PNA binding mode.
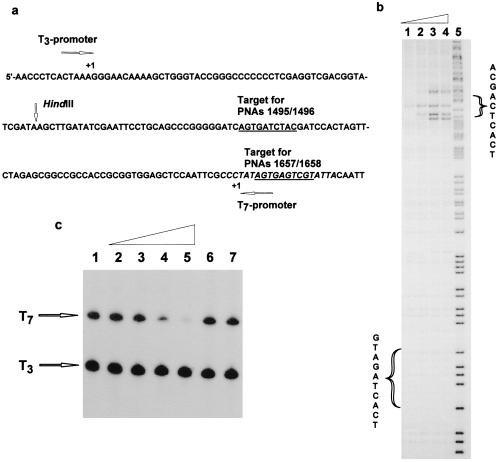
Because our PNA target was positioned in the polylinker region of the Bluescript plasmid (Fig. 3a), we could test the effect of the double duplex PNA invasion complexes had on transcription elongation by T3 and T7 RNA polymerases. In contrast to PNA triplex invasion complexes (20, 21), no inhibition of transcription elongation could be obtained with PNAs 1495 and 1496, which bind downstream from the transcription initiation (data not shown). We therefore synthesized a new set of pseudocomplementary PNAs targeted to the −3/−13 region of the T7 promoter (Fig. 3a). These PNAs (1657/1658) also bound sequence specifically and efficiently to their target as assayed by gel mobility-shift electrophoresis and permanganate probing (Fig. 3b), and efficient inhibition of T7 transcription was seen in this case (Fig. 3c). Thus these 10-mer double duplex invasion PNA complexes are efficiently blocking the access of an enzyme such as RNA polymerase to the DNA, but in contrast to PNA triplex invasion complexes, they are not capable of arresting the elongating phage RNA polymerase. It should be stressed that these experiments were performed with PNA–double-stranded DNA invasion complexes preformed at low ionic strength followed by buffer adjustment and transcription reaction because the buffer conditions required for transcription are highly inhibitory to PNA invasion binding. However, the complexes appear kinetically stable under the transcription conditions, and it is therefore likely that it is the helicase activity of the RNA polymerase that is able to disrupt the PNA⋅DNA duplex. However, it is quite likely that eukaryotic RNA polymerases (such as Pol II) are significantly more sensitive to a double duplex “PNA block” as compared with the much more robust phage polymerases.
Figure 3.
Binding of anti-T7-promoter PNAs 1657/1658 (H-Lys-DCGDCsUCDCsU-Lys-NH2/H-Lys-DGsUGDGsUCGsU-Lys-NH2) to their target and effect on T7-transciption. (a) Sequence showing the T7 promoter and the PNA targets. (b) KMnO4 probing of the binding of PNAs 1657/1658 to their target. The experiment was performed as described for Fig. 2c. The PNA concentrations in lanes 1–4 were 30 nM, 90 nM, 3 mM, and 9 mM. Lane 5 is an A/G sequence marker. (c) Effect on T7 transcription of the binding of promoter-directed PNAs 1657/1658. The PNAs were prebound to the DNA (p206 cut with HindIII) for 16 hr at 37°C in 10 mM Tris⋅HCl/1 mM EDTA buffer, pH 7.4. Subsequently polymerase buffer, NTP mix containing [32P]UTP and T7 and T3 RNA polymerases were added and transcription was allowed to proceed for 1 min at 37°C. The transcripts were analyzed by PAGE and autoradiography. The following concentrations (mM,) of PNAs were used (PNA 1657/PNA 1658): lane 1, 0/0; lane 2, 0.03/0.03; lane 3, 0.1/0.1; lane 4, 0.3/0.3; lane 5, 1/1; lane 6, 0.3/0; lane 7, 0/0.3. As an internal control, transcription with RNA polymerase T3, for which the promoter does not contain the PNA-binding site, was performed simultaneously.
The present results show that double-stranded DNA can be efficiently and sequence specifically targeted at low ionic strength in a digital way (base pair by base pair readout) exploiting simple Watson–Crick base pair type recognition. Although the binding mechanism is not known, it most likely involves dynamic base pair breathing of the double-stranded DNA, by which the PNA may trap the nucleobases in the open state. It may be argued that the ionic conditions existing in the cell nucleus could constitute an insurmountable obstacle for any eventual use of PNA as gene therapeutic agents. However, recent reports have already indicated that PNAs may indeed be able to bind their targets in the cell nucleus (22, 23), and in vitro experiments have shown that biological events taking place in the nucleus such as transcription and the resulting negative supercoiling of the DNA greatly facilitate strand displacement binding (24, 25). Also, the construction of covalently linked pseudocomplementary bis-PNAs could enhance the performance. Thus additional experiments are required to evaluate the potential of exploiting the double duplex invasion principle in drug development. The principle can, however, readily be used for various gene mapping (26, 27) and sample preparation techniques because these procedures can easily be adapted to low ionic strength binding conditions. Therefore these techniques should no longer be limited to homopurine targets.
Acknowledgments
The expert technical assistance of Karin Frederiksen, Annette Jørgensen, and Margit Jørgensen is gratefully acknowledged. This work was supported by the Danish National Research Foundation and the Lundbeck Foundation.
Abbreviations
- PNA
peptide nucleic acid
- HBTU O-benzotriazole-1-yl-N,N,N′,N′-tetramethyluronium hexafluorophosphate
MALDI-TOF, matrix-assisted laser desorption ionization time-of-flight
- D
2,6-diaminopurine
- sU
2-thiouracil
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Nielsen P E. Chem Eur J. 1997;3:505–508. [Google Scholar]
- 2.Denison C, Kodadek T. Chem Biol. 1998;5:R129–R145. doi: 10.1016/s1074-5521(98)90167-3. [DOI] [PubMed] [Google Scholar]
- 3.Hélène C. Current Opin Biotech. 1993;4:29–36. doi: 10.1016/0958-1669(93)90028-u. [DOI] [PubMed] [Google Scholar]
- 4.Giovannangeli C, Hélène C. Antisense Nucleic Acid Drug Dev. 1997;7:413–421. doi: 10.1089/oli.1.1997.7.413. [DOI] [PubMed] [Google Scholar]
- 5.Nielsen P E, Egholm M, Berg R H, Buchardt O. Science. 1991;254:1497–1500. doi: 10.1126/science.1962210. [DOI] [PubMed] [Google Scholar]
- 6.Good L, Nielsen P E. Antisense Nucleic Acid Drug Dev. 1997;7:431–437. doi: 10.1089/oli.1.1997.7.431. [DOI] [PubMed] [Google Scholar]
- 7.Gottesfeld J M, Neely L, Trauger J W, Baird E E, Dervan P B. Nature (London) 1997;387:202–205. doi: 10.1038/387202a0. [DOI] [PubMed] [Google Scholar]
- 8.White S, Szewczyk J W, Turner J M, Dervan P B. Nature (London) 1998;391:468–471. doi: 10.1038/35106. [DOI] [PubMed] [Google Scholar]
- 9.Frank-Kamenetskii M D, Mirkin S M. Annu Rev Biochem. 1995;64:65–95. doi: 10.1146/annurev.bi.64.070195.000433. [DOI] [PubMed] [Google Scholar]
- 10.Nielsen P E, Christensen L. J Am Chem Soc. 1996;118:2287–2288. [Google Scholar]
- 11.Kutyavin I V, Rhinehart R L, Lukhtanov E A, Gorn V V, Meyer R B, Jr, Gamper H B., Jr Biochemistry. 1996;35:11170–11176. doi: 10.1021/bi960626v. [DOI] [PubMed] [Google Scholar]
- 12.Haaima G, Hansen H F, Christensen L, Dahl O, Nielsen P E. Nucleic Acids Res. 1997;25:4639–4643. doi: 10.1093/nar/25.22.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McKinnon M D, Chauhan S M. Can J Chem. 1978;56:725–729. [Google Scholar]
- 14.Dueholm K, Egholm M, Buchardt O. Org Prep Proced Int. 1993;25:457–461. [Google Scholar]
- 15.Christensen L, Fitzpatrick R, Gildea B, Petersen K H, Hansen H F, Koch T, Egholm M, Buchardt O, Nielsen P E, Coull J, Berg R. J Peptide Sci. 1995;3:175–183. doi: 10.1002/psc.310010304. [DOI] [PubMed] [Google Scholar]
- 16.Nielsen P E, Egholm M, Buchardt O. J Mol Recognit. 1994;7:165–170. doi: 10.1002/jmr.300070303. [DOI] [PubMed] [Google Scholar]
- 17.Demidov V V, Yavnilovich M V, Belotserkovskii B P, Frank-Kamenetskii M D, Nielsen P E. Proc Natl Acad Sci USA. 1995;92:2637–2641. doi: 10.1073/pnas.92.7.2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuhn H, Demidov V, Frank-Kamenetskii M D, Nielsen P E. Nucleic Acids Res. 1997;26:582–587. doi: 10.1093/nar/26.2.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peffer N J, Hanvey J C, Bisi J E, Thomson S A, Hassman F C, Noble S A, Babiss L E. Proc Natl Acad Sci USA. 1993;90:10648–10652. doi: 10.1073/pnas.90.22.10648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nielsen P E, Egholm M, Buchardt O. Gene. 1994;149:139–145. doi: 10.1016/0378-1119(94)90422-7. [DOI] [PubMed] [Google Scholar]
- 21.Hanvey J C, Peffer N C, Bisi J E, Thomson S A, Cadilla R, Josey J A, Ricca D J, Hassman C F, Bonham M A, Au K G, et al. Science. 1992;258:1481–1485. doi: 10.1126/science.1279811. [DOI] [PubMed] [Google Scholar]
- 22.Boffa L C, Carpaneto E M, Mariani M R, Louissaint M, Allfrey V G. Oncol Res. 1997;9:41–51. [PubMed] [Google Scholar]
- 23.Faruqi A F, Egholm M, Glazer P M. Proc Natl Acad Sci USA. 1998;95:1398–1403. doi: 10.1073/pnas.95.4.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bentin T, Nielsen P E. Biochemistry. 1996;35:8863–8869. doi: 10.1021/bi960436k. [DOI] [PubMed] [Google Scholar]
- 25.Larsen H J, Nielsen P E. Nucleic Acids Res. 1996;24:458–463. doi: 10.1093/nar/24.3.458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Veselkov A G, Demidov V V, Nielsen P E, Frank-Kamenetskii M. Nucleic Acids Res. 1996;24:2483–2487. doi: 10.1093/nar/24.13.2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bukanov N O, Demidov V V, Nielsen P E, Frank-Kamenetskii M D. Proc Natl Acad Sci USA. 1998;95:5516–5520. doi: 10.1073/pnas.95.10.5516. [DOI] [PMC free article] [PubMed] [Google Scholar]