Abstract
The inositol phosphate hydrolyzing activity of human phospholipase Cδ1 (PLCδ1) is markedly inhibited when the enzyme is coexpressed with the human heart Gh/transglutaminase (TG) in human embryonic kidney cells. Because the cotransfection does not affect the amount of PLCδ1 in the cells, the depression of phospholipase activity probably is a result of a direct interaction between the two proteins. An ELISA procedure was employed to document the associations of purified TG preparations from a variety of tissues (human red cells, rabbit lens, guinea pig liver) with PLCδ1. Nucleotides (GTP > GDP > ATP > GMP = ADP, in order of decreasing efficiency) interfered with the formation of the PLCδ1:TG complex. A conformational change in the TG partner, occurring with nucleotide binding, is thought to be responsible for dissociating the two proteins. The structural rearrangement produces a remarkable shift in the anodic mobility of TG in electrophoresis: TGslow + GTP ⇆ [TG:GTP]fast. Altogether, our findings indicate that GTP controls PLCδ1 activity by releasing this protein from an inhibitory association with Gh/transglutaminase.
Enzymes commonly referred to as transglutaminases (TG) (EC 2.3.2.13) are known mostly for activities relating to the posttranslational remodeling of proteins (1–5). They can catalyze the hydrolysis of γ-amides of select glutamine residues in their protein substrates, the incorporation of primary amines (including polyamines) at these same sites, or the formation of Nɛ(γ-glutamyl)lysine crosslinks between protein units. In addition, they can hydrolyze such isopeptide side-chain bridges (6). Recent findings revealed that, apart from these protein modifying capabilities, the cytosolic transglutaminases—found in many tissues—also could function as a component of the signal-transducing G protein complex (7). The cDNA of Ghα, involved in the transmission of adrenergic stimuli, is identical to that of the human endothelial transglutaminase (7).
It has been proposed by Feng et al. (8) that transglutaminases activate the δ1 isoform of phospholipase C (EC 3.1.4.11) (PLCδ1). Though it is known that PLCδ1 is not regulated by interaction with heterotrimeric G proteins or through phosphorylation by any of the receptor-stimulated kinases (9), regulation of this isoform family of PLC is still poorly understood. In vitro, PLCδ1 is sensitive to the composition of both substrate and nonsubstrate phospholipids; it can be stimulated up to 20-fold by the addition of phosphatidylinositol 4,5-bisphosphate (PIP2) and phosphatidylserine (PS; refs. 10 and 11). These lipids act as ligands and bind to the pleckstrin homology and the C2 domain of the enzyme, respectively. The mechanism of activation involves translocation to the plasma membrane, enhancement of Vmax for PIP2, and lowering of the Km for PIP2 (11, 12). There is increasing evidence that PLCδ1 may participate in physiologically relevant protein–protein interactions. The report by Homma and Emori (13) raised the possibility that PLCδ1 was regulated by members of the rho family of small G proteins, whereas the study of Feng et al. (8), mentioned above, led to the notion that PLCδ1 might be activated by the transglutaminase isolated from human heart and referred to as Gh (8). Another report suggests that rhoA acts as an inhibitory factor for PLCδ1 (14). Because all of these proteins can bind and hydrolyze GTP, it may be surmised that guanine nucleotides play an indirect role in the regulation of PLCδ1.
GTP is known to inhibit the amine-incorporating (15–20) as well as the protein-crosslinking activities of TG (20), but the inhibition can be overcome to some extent by raising the concentration of Ca2+ ions. The nucleotide triphosphatase activity of TG resides in the N terminus of the protein (residues 1–185), involving a suggested consensus sequence of residues 165-GFIYQGSVK-173 for the binding of GTP (21, 22). Molecular modeling, based on some degree of sequence homology with the A subunit of factor XIII (22), suggests that the nucleotide-binding domain of TG is distinct from the core that contains the catalytic cysteine (2) and histidine residues (23, 24) as well as an aspartic acid thought to be necessary for amide hydrolysis and exchange. Though the linkage and allosteric communication between these two sites is not yet known, it is reasonable to assume that the binding of GTP to TG is accompanied by significant conformational changes in the enzyme. We find that GTP alters the electrophoretic migration properties of highly purified TG/Gh proteins and affects their binding characteristics to phospholipase Cδ1, suggesting a hormonally controlled pathway by which TG might participate in the regulation of phospholipase C isoforms.
Materials and Methods
Purification of Enzymes.
Recombinant human PLCδ1 was overexpressed and purified from E. coli as described (10, 25). Tissue TG from human erythrocytes (26), rabbit lens (20), chicken erythrocytes (27), and guinea pig liver (28) were purified as described.
Nucleotide Binding.
The nucleotides GTP, guanosine 5′-O-thiotriphosphate (GTP[γS]), GDP, GMP, ATP, and ADP were obtained from Sigma. The stock solutions of the nucleotides were made in water and stored at −80°C. In binding experiments the nucleotide solutions contained equimolar concentrations of MgCl2. TG was treated with nucleotides/Mg2+ at room temperature for 45 min in reaction volumes of 30 μl of 75 mM imidazole-HCl buffer, pH 7.25, containing 8 μg of TG (≈3.3 μM) and 400 μM of nucleotides, before electrophoresis on agarose under nondenaturing conditions.
Treatment of TG with High Salt.
Approximately 138 μg of human erythrocyte TG in 110 μl of 2.5 M KCl, in 20 mM imidazole-HCl buffer, pH 6.5/10% glycerol, was dialyzed against 500 ml of the same buffer for 18 hr at 4°C followed by dialysis against 2 × 500 ml of the above buffer without KCl for 6 hr at 4°C. Protein concentration was determined by BCA protein assay (Pierce) using BSA (Pierce) as standard.
Electrophoresis.
Nondenaturing electrophoresis was carried out in 2–3% agarose in 75 mM imidazole-HCl buffer/0.5 mM EDTA, pH 7.25, for 2–3 hr at 4°C at 300 V by using the LKB Multiphor Electrophoretic Unit. Samples were applied 2 cm from the cathodic end of the gel. Electrophoresis was carried out until the tracking dye (bromophenol blue) was 1 cm from the anodic end of the gel. Protein staining was performed with Coomassie brilliant blue R (Sigma) in 10% acetic acid and 25% isopropanol, followed by destaining with 10% acetic acid and 20% methanol and densitometric scanning (LKB Ultrascan XL). Alternatively, TG activity was probed with a stain of dansylcadaverine, dimethylated casein, and Ca2+ (29, 30). The fluorescent bands in the gel were visualized by illumination with UV light (λEXC = 360 nm; UVL-56, Ultraviolet Products, San Gabriel, CA).
Electrophoresis in the presence of SDS on 10% acrylamide in the separating gel was carried out by using the discontinuous buffer system of Laemmli (31). Before electrophoresis, the samples were boiled for 5 min in the presence of 40 mM DTT and 2% SDS. The gels were electroblotted (32) onto nitrocellulose (0.2-μM pore size; Schleicher & Schuell) and immunostained with mAb to either human erythrocyte TG (H23.1.2) or PLCδ1.
Antibodies.
Purified human erythrocyte TG was used as antigen to raise hybridomas in mice, and IgG (H23.1.2 and H68.1.1) was purified after subcloning twice according to the procedure described in our earlier report for purification of mAbs to guinea pig liver TG (33). mAbs to human PLCδ1 have been described (25).
Transglutaminase Binding to PLCδ1 by ELISA.
These studies were carried out in 96-well Microtest III assay plates (Falcon 3910; Becton Dickinson Labware) at room temperature on a Titer Plate Shaker (Lab-Line Instruments). The microtiter plate wells were coated with 0.2 μg of PLCδ1 in 100 μl of TBS (50 mM Tris⋅HCl/100 mM NaCl, pH 7.5) for 2 hr. The remaining sites were blocked with 3 × 300 μl/well of 2% BSA (Sigma) in TBS for 30 min. The plates were overlayered with 0.4 μg of wild-type PLCδ1 in 100 μl of 2% BSA, allowed to bind for 2 hr, and washed with 3 × 300 μl/well of 2% BSA for 10 min each. TG was mixed with nucleotide/Mg2+ solution in 2% BSA for 45 min and then overlayered onto PLCδ1-coated wells and allowed to bind for 16–18 hr. The plates were washed with 3 × 300 μl/well of 2% BSA for 5 min each, and monoclonal IgG (H23.1.2 or H68.1.1) to human erythrocyte TG (125 μl of 0.5 μg/ml solution) were added and allowed to bind for 2 hr. After washing with 3 × 300 μl/well of 2% BSA, 125 μl/well alkaline phosphatase-conjugated secondary antibody at 1:5,000 dilution in 2% BSA (anti-mouse IgG developed in rabbit; Sigma) was added and allowed to bind for 2 hr. The plates were washed with 3 × 300 μl/well alkaline phosphatase buffer (100 mM Tris⋅HCl/100 mM NaCl/5 mM MgCl2, pH 9.5) for 5 min each, and 100 μl of 1 mg/ml p-nitrophenyl phosphate disodium (Sigma) in the alkaline phosphatase buffer was added to each well. The color was read in an MR600 microplate reader at 410 nm (Dynatech). For controls, TG was overlayered on BSA-coated wells without PLCδ1.
Assay of PLCδ1 Activity.
TSA201 cells were transiently transfected with lipofectamine (GIBCO). Twenty-four hours after transfection the cells were labeled with [3H]myo-inositol (4 μCi/ml). The hydrolytic release of soluble inositol phosphates was measured at 48 hr by a modification of the method of Martin (34). Activity was expressed as products of hydrolysis (cpm) per total labeled lipids (as determined from an organic extraction of the cells).
Results and Discussion
Coexpression with Transglutaminase Lowers PLCδ1 Activity.
A recently appreciated and intriguing function of TG is its postulated role in mediating the effects of hormones such as adrenaline and oxytocin (7, 35). It has been reported (36, 37) that the α1-adrenergic receptor (α1-AR) couples to the δ1 isoform of PLC via interaction with a TG (termed Gαh) and another, yet unidentified, 50-kDa protein. Though initially identified by copurification with the α1-AR, Gαh subsequently was sequenced and found to be identical to the human endothelial TG (7). According to this model, agonist binding to α1-AR would lead to the exchange of GDP for GTP on TG. The GTP-bound form of TG then would be able to interact with PLCδ1 and activate it for the hydrolysis of inositol phosphates. The signal would be turned off when the intrinsic GTPase activity of TG hydrolysed GTP to GDP, and the GDP bound form of TG would cease to stimulate PLCδ1 and dissociate from the complex (7).
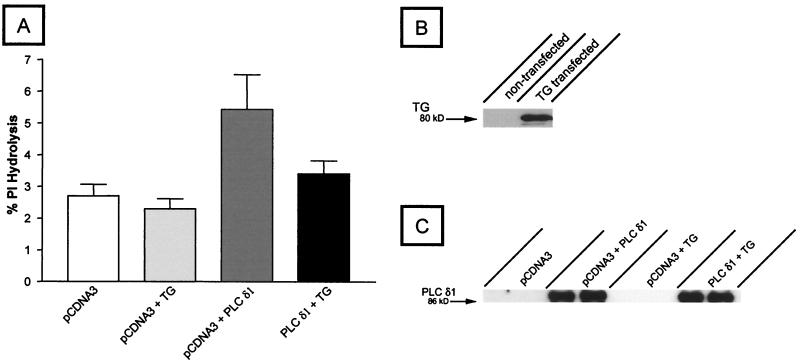
We carried out cotransfection studies to assess the functional consequences of the interaction of TG with PLCδ1. TSA 201 cells, a clone of human embryonic kidney 293 cells stably expressing simian virus 40 large T antigen, were transfected with PLCδ1 and TG from human heart (Gh, courtesy of M. J. Im, The Cleveland Clinic Foundation) (38). PLCδ1 function was measured by assaying for hydrolysis of inositol phospholipids. As shown in Fig. 1, PLCδ1-transfected cells had a 2-fold increase in hydrolysis of inositol phospholipids compared with vector (pCDNA3)-transfected control cells or those transfected with TG alone. However, in sharp contrast to the conclusions reached by Feng et al. (8), in our hands cotransfection of PLCδ1 with TG led to significant inhibition (74%) of PLCδ1 activity. Western blots of transfected cells with antibodies directed against TG showed expression of an ≈80-kDa protein consistent with the size of endothelial TG (39). Western blots probed with a mAb to PLCδ1 showed that expression of this PLC was not affected by cotransfection with TG. PLCδ1 expression remained unchanged, indicating that TG did not cause the reduction of PLCδ1 activity by lowering its level of expression.
Figure 1.
The PLCδ1-catalyzed hydrolysis of inositol phospholipids is inhibited by expression of transglutaminase in transfected TSA201 cells. TSA201 cells were transiently transfected with cDNA encoding the human PLCδ1 and human heart tissue transglutaminase (Gαh). PLC activity was assessed by isolating the soluble products of phospholipid hydrolysis (IP1, IP2, and IP3) by using anionic-exchange chromatography according to the method of Martin (A; ref. 34). Products of hydrolysis are expressed as a percentage of total labeled lipid. B and C represent Western blots of transfected cells probed with a mAb for transglutaminase or PLCδ1, respectively. Approximately 3 μg of total protein was applied to each lane. Molecular mass markers (for TG, 80 kDa, and for PLCδ1, 86 kDa) are shown on the left. The data in A represent three separate experiments, and the error bars have been calculated as mean ± SD.
Binding of Transglutaminase to PLCδ1 Is Regulated by GTP.
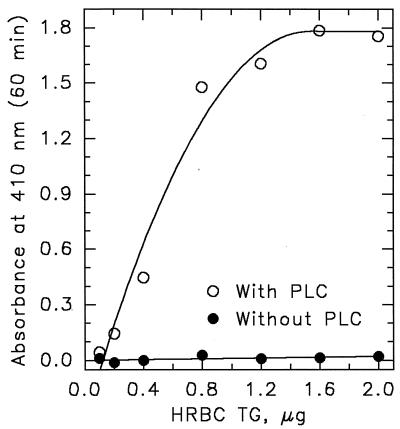
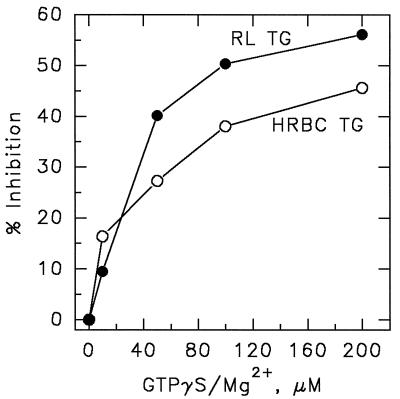
We developed an ELISA to demonstrate that cytoplasmic transglutaminases, isolated from various tissues, could, in fact, associate with PLCδ1. TG from human red blood cells, rabbit lens, and guinea pig liver all bound with relatively high affinity to plated human PLCδ1. As illustrated for red cell TG in Fig. 2, binding was both concentration-dependent and saturable. In this case, again in contrast to the observations of Im and coworkers (40), GTP proved to be a strong inhibitor, blocking the association between PLCδ1 and TG (Fig. 3). The rank order for potency of inhibition was GTP > GDP > ATP > GMP = ADP, and the IC50 for GTP[γS] was between 40 and 100 μM. Because the nucleotides have no effect on PLCδ1 function and because prior incubation of PLCδ1 with GTP[γS] did not inhibit TG binding (data not shown), it can be assumed that the GTP effect is mediated by the binding of the nucleotide to TG and not to PLC.
Figure 2.
Transglutaminase binds to immobilized PLCδ1. Human erythrocyte TG (0.05–2 μg/well) was allowed to bind to wells coated with two layers of PLCδ1 (0.2 μg/well) or to BSA-coated wells. Bound TG was monitored by ELISA by using a mAb to TG (H68.1.1) and an alkaline phosphatase-conjugated secondary antibody.
Figure 3.
GTP[γS] inhibits the binding of TG to immobilized PLCδ1. TG from human erythrocyte (HRBC) and rabbit lens (RL) were mixed with GTP[γS]/Mg2+ (45 min, room temperature) and then allowed to bind to ELISA plate wells coated with PLCδ1 as in Fig. 2. Bound TG was monitored by ELISA by using a mAb to TG (H68.1.1) and an alkaline phosphatase-conjugated secondary antibody.
GTP-Induced Changes in Transglutaminase.
TG obtained from different sources (guinea pig liver, rabbit lens, and human and chicken red cells) vary somewhat in sensitivities to GTP inhibition (20), but the transamidating activities of all of these enzymes is negatively regulated by the nucleotide. GTP has been shown to lower the affinity of TG for Ca2+ (17), which prevents the unmasking of its CySH active center. However, important details of the allosteric communication between the GTP and Ca2+-binding sites of TG and the catalytic center required for transamidation are yet to be worked out. In chicken red blood cell TG, the putative GTP-binding consensus sequence GFIYQGSVK (22) is replaced by GLIYMGSRD (27); yet, GTP is a potent inhibitor of the chicken enzyme, too (unpublished results).
Fourier-transformed infrared spectroscopy (41) failed to detect changes in the secondary structure of human erythrocyte TG upon binding GTP. Yet, the nucleotide was found to stabilize the enzyme against heat denaturation (41) and against proteolytic degradation (15, 17). In addition, the immunoreactivity of a C-terminal fragment (residues 666–680) of rat brain TG—which, incidentally, includes a proposed PLC-binding site (8)—is markedly altered in the presence of GTP (42). This latter study would seem to suggest that the binding of GTP is accompanied by significant movements and rearrangements of the protein structure.
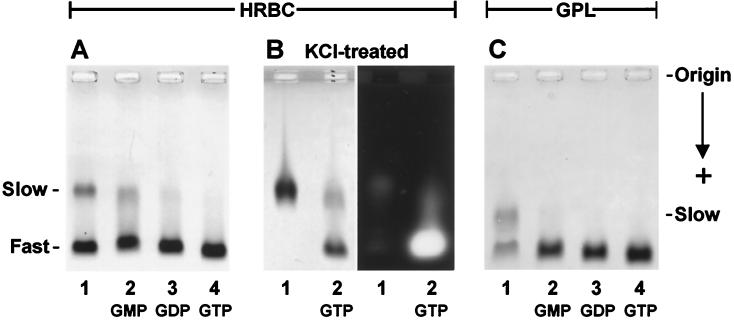
Electrophoresis under nondenaturing conditions often reveals a heterogeneity in TG preparations that seem to be homogeneous by criteria of gel filtration and SDS/PAGE (29, 30). As shown in Fig. 4, the human red cell and the guinea pig liver enzymes both comprise a slow (s) and fast (f) migrating form of TG. Respective mobilities of the s and f forms of the two enzymes are different, and the relative proportions of s and f can vary from preparation to preparation and might also be affected by storage. What is highly significant, however, is that admixture of nucleotides: GMP, GDP, GTP, or GTP[γS] just before electrophoresis promotes varying degrees of s → f transformation, with GTP or GTP[γS] being the most effective in this regard (Fig. 4 A and C, lanes 2–4). If the migration distance of the fast-moving [HRBC TG:GTP] band (Fig. 4A, lane 4) is taken for unity, the relative mobilities of the other fast forms of [HRBC TG:GMP] and [HRBC TG:GDP] are 0.97 (Fig. 4A, lane 2) and 0.98 (Fig. 4A, lane 3), whereas the fast species in the original preparation migrates as 0.98 (Fig. 4A, lane 1). The relative migration distance of the slow component (Fig. 4A, lane 1) was only 0.65. The slow form in GPL TG (Fig. 4C, lane 1) migrated as 0.78 in relation to the GTP complex of the same TG in Fig. 4C, lane 4. These findings indicate that the purified preparations are really mixtures of a slow, nucleotide-free and a fast, nucleotide-bound form of the TG protein. In the HRBC preparation (Fig. 4A, lane 1), the fast form migrated as [TG:GDP] (Fig. 4A, lane 3), and in the GPL material (Fig. 4C, lane 1), migration of the fast band was similar to that of [TG:GMP] (Fig. 4C, lane 2).
Figure 4.
Shift in the electrophoretic mobility of tissue transglutaminases after binding to nucleotides. Purified TG isolated from human erythrocytes (A and B) and from guinea pig liver (C) were examined by electrophoresis in agarose (3% in A and B; 2% in C) with (lanes 2, 3, and 4) and without (lane 1) admixture (45 min at room temperature) of nucleotides/Mg2+ (0.4 mM of GMP, GDP, or GTP). The proteins were prepared at concentrations of 3.3 μM in a buffer of 75 mM imidazole-HCl, pH 7.2, containing 0.5 mM EDTA. Samples of 30 μl (i.e., 8 μg protein per lane) were applied to the gel, and electrophoresis was carried out for 2 hr at 4°C in the above buffer. The gels with light backgrounds in the picture were developed with Coomassie blue; the segment shown in B Right, probing the electrophoretic mobility of KCl-stripped red cell TG, was photographed under UV light after a transamidase activity-specific staining procedure, based on the incorporation of fluorescent dansylcadaverine into dimethylated casein (29, 30).
As illustrated for the HRBC preparation in Fig. 4B, after treatment with 2.5 M KCl for removal of bound nucleotides, only the slow electrophoretic form of TG could be seen (Fig. 4B, lane 1). UV absorption of the preparation, measured as a ratio at 251 and 278 nm, also changed with the KCl treatment from 0.65 to 0.53. Admixture of GTP to the KCl-stripped TG caused nearly a complete (86%) conversion of the slow form back to the fast one (Fig. 4B, lane 2). Developing a similar gel for transamidase activity through the incorporation of fluorescent dansylcadaverine into a casein substrate (29, 30) showed that the KCl-stripped protein had no appreciable enzymatic activity (Fig. 4B Right, lane 1), but activity was regained after exposure to GTP and conversion to the fast electrophoretic form (Fig. 4B Right, lane 2).
The differences in the relative mobilities of TG with GMP, GDP, or GTP as ligands (i.e., by a stepwise increase of excess negative charges) are very modest (0.97 → 1.0; Fig. 4 A and C, lanes 2–4) in comparison with those between the slow and fast forms of TG (0.65 → 1.0 for the HRBC and 0.78 → 1.0 for the GPL protein). The large, GTP-induced anodic shift of the s → f conversion is surely a sign of a major reorganization in the secondary/tertiary structure of the protein upon binding the nucleotide. Interestingly (as seen in Fig. 4 A and C, lanes 2 and 3), GMP and GDP also appear to be somewhat effective in stabilizing a fast-moving configuration of the TG protein. As a corollary to the binding of nucleotide, the rearrangement of amino acid side chains in TG could result in the burying of positive charges or might bring negatively charged residues to the surface, or both; also, there might be a decrease in the frictional coefficient (Stoke’s radius) of the TG molecule upon binding the nucleotide ligand. Considering that the various nucleotide-bound forms of TG can survive the lengthy procedures of purification and electrophoresis without any free ligand in the buffer, it must be assumed that the binding constants for these equilibria and, particularly, for TGslow + GTP ⇆ [TG:GTP]fast are quite high.
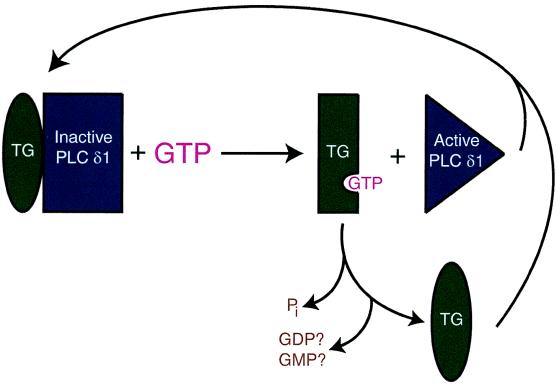
Current models for the regulation of PLCδ1 propose its activation by TG (8, 22, 40), whereas our findings suggest an inhibitory mechanism that could be modulated through GTP (Fig. 5). The activity of PLCδ1 is suppressed by interaction with TG. However, the association between the two proteins would be negated by the binding of GTP to TG, which, in turn, would cause the activation of PLCδ1. Thus, TG could be an important player in the signaling pathway that regulates calcium homeostasis and modulates physiological processes such as vascular smooth muscle tone (i.e., blood pressure) and neurotransmitter release.
Figure 5.
Model of PLCδ1 regulation by TG and GTP. The activity of PLCδ1 is negatively regulated by interaction with TG, but PLCδ1 is released from this inhibitory association when GTP binds to TG. It is not yet known whether the TG molecule in the PLCδ1 complex contains a nucleotide (e.g., GDP or GMP) ligand.
Because inositol phosphate substrates of PLCδ1 are present in the plasma membrane and because this phospholipase is cytosolic in most cell types, to bind and hydrolyse its substrates, PLCδ1 must shuttle between the membrane and the cytoplasm. The N-terminal pleckstrin homology domain is largely responsible for enabling the enzyme to fulfill this “interfacial” role, and mutations in this domain severely impair its function (10). It is reasonable to suggest that the observed PLCδ1 inhibition by cotransfection with the cytosolic TG (Fig. 1) is due to a sequestering of the [PLCδ1:TG] complex in the cytoplasm away from the plasma membrane.
Acknowledgments
We are grateful to Dr. M. J. Im of The Cleveland Clinic Foundation for providing us the cDNA coding for transglutaminase, Gh. This research was supported by grants from the National Institutes of Health (HL-02212 and EY-03942 to L.L. and HL-55591 and HL-03961 to J.W.L).
Abbreviations
- TG
transglutaminase(s)
- PLCδ1
δ1 isoform of phospholipase C
References
- 1.Clark D D, Mycek M J, Neidle A, Waelsch H. Arch Biochem Biophys. 1959;79:338–354. doi: 10.1016/0003-9861(59)90613-7. [DOI] [PubMed] [Google Scholar]
- 2.Folk J E. Annu Rev Biochem. 1980;49:517–531. doi: 10.1146/annurev.bi.49.070180.002505. [DOI] [PubMed] [Google Scholar]
- 3.Lorand L, Conrad S M. Mol Cell Biochem. 1984;58:9–35. doi: 10.1007/BF00240602. [DOI] [PubMed] [Google Scholar]
- 4.Greenberg C S, Birckbichler P J, Rice R H. FASEB J. 1991;5:3071–3077. doi: 10.1096/fasebj.5.15.1683845. [DOI] [PubMed] [Google Scholar]
- 5.Aeschlimann D, Paulsson M. Thromb Haemostasis. 1994;71:402–415. [PubMed] [Google Scholar]
- 6.Parameswaran K N, Cheng X F, Chen E C, Velasco P T, Wilson J H, Lorand L. J Biol Chem. 1997;272:10311–10317. doi: 10.1074/jbc.272.15.10311. [DOI] [PubMed] [Google Scholar]
- 7.Nakaoka H, Perez D M, Baek K J, Das T, Husain A, Misono K, Im M J, Graham R M. Science. 1994;264:1593–1596. doi: 10.1126/science.7911253. [DOI] [PubMed] [Google Scholar]
- 8.Feng J F, Rhee S G, Im M J. J Biol Chem. 1996;271:16451–16454. doi: 10.1074/jbc.271.28.16451. [DOI] [PubMed] [Google Scholar]
- 9.Rhee S G, Bae Y S. J Biol Chem. 1997;272:15045–15048. doi: 10.1074/jbc.272.24.15045. [DOI] [PubMed] [Google Scholar]
- 10.Lomasney J W, Cheng H F, Wang L P, Kuan Y S, Liu S M, Fesik S W, King K. J Biol Chem. 1996;271:25316–25326. doi: 10.1074/jbc.271.41.25316. [DOI] [PubMed] [Google Scholar]
- 11.Lomasney J W, Cheng H F, Roffler S R, King K. J Biol Chem. 1999;274:21995–22001. doi: 10.1074/jbc.274.31.21995. [DOI] [PubMed] [Google Scholar]
- 12.Bromann P A, Boetticher E E, Lomasney J W. J Biol Chem. 1997;272:16240–16246. doi: 10.1074/jbc.272.26.16240. [DOI] [PubMed] [Google Scholar]
- 13.Homma Y, Emori Y. EMBO J. 1995;14:286–291. doi: 10.1002/j.1460-2075.1995.tb07002.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hodson E A, Ashley C C, Hughes A D, Lymn J S. Biochim Biophys Acta. 1998;1403:97–101. doi: 10.1016/s0167-4889(98)00028-7. [DOI] [PubMed] [Google Scholar]
- 15.Achyuthan K E, Greenberg C S. J Biol Chem. 1987;262:1901–1906. [PubMed] [Google Scholar]
- 16.Bergamini C M, Signorini M, Poltronieri L. Biochem Biophys Acta. 1987;916:149–151. doi: 10.1016/0167-4838(87)90222-6. [DOI] [PubMed] [Google Scholar]
- 17.Bergamini C M. FEBS Lett. 1988;239:255–258. doi: 10.1016/0014-5793(88)80928-1. [DOI] [PubMed] [Google Scholar]
- 18.Lee K N, Birckbichler P J, Patterson M K., Jr Biochem Biophys Res Commun. 1989;162:1370–1375. doi: 10.1016/0006-291x(89)90825-5. [DOI] [PubMed] [Google Scholar]
- 19.Kawashima S. Experientia. 1991;47:709–712. doi: 10.1007/BF01958822. [DOI] [PubMed] [Google Scholar]
- 20.Murthy S N P, Velasco P T, Lorand L. Exp Eye Res. 1998;67:273–281. doi: 10.1006/exer.1998.0509. [DOI] [PubMed] [Google Scholar]
- 21.Lai T S, Slaughter T F, Koropchak C M, Haroon Z A, Greenberg C S. J Biol Chem. 1996;271:31191–31195. doi: 10.1074/jbc.271.49.31191. [DOI] [PubMed] [Google Scholar]
- 22.Iismaa S E, Chung L, Wu M J, Teller D C, Yee V C, Graham R M. Biochemistry. 1997;36:11655–11664. doi: 10.1021/bi970545e. [DOI] [PubMed] [Google Scholar]
- 23.Parameswaran K N, Lorand L. Biochemistry. 1981;20:3703–3711. doi: 10.1021/bi00516a006. [DOI] [PubMed] [Google Scholar]
- 24.Micanovic R, Procyk R, Lin W, Matsueda G R. J Biol Chem. 1994;269:9190–9194. [PubMed] [Google Scholar]
- 25.Cheng H F, Jiang M J, Chen C L, Liu S M, Wong L P, Lomasney J W, King K. J Biol Chem. 1995;270:5495–5505. doi: 10.1074/jbc.270.10.5495. [DOI] [PubMed] [Google Scholar]
- 26.Radek J T, Jeong J M, Murthy S N P, Ingham K C, Lorand L. Proc Natl Acad Sci USA. 1993;90:3152–3156. doi: 10.1073/pnas.90.8.3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weraarchakul-Boonmark N, Jeong J M, Murthy S N P, Engel J D, Lorand L. Proc Natl Acad Sci USA. 1992;89:9804–9808. doi: 10.1073/pnas.89.20.9804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jeong J M, Murthy S N P, Radek J T, Lorand L. J Biol Chem. 1995;270:5654–5658. doi: 10.1074/jbc.270.10.5654. [DOI] [PubMed] [Google Scholar]
- 29.Lorand L, Siefring G E, Jr, Tong Y S, Bruner-Lorand J, Gray A J., Jr Anal Biochem. 1979;93:453–458. doi: 10.1016/s0003-2697(79)80178-5. [DOI] [PubMed] [Google Scholar]
- 30.Lorand L, Dailey J E, Turner P M. Proc Natl Acad Sci USA. 1988;85:1057–1059. doi: 10.1073/pnas.85.4.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 32.Towbin H, Stahelin T, Gordon J. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trejo-Skalli A V, Velasco P T, Murthy S N P, Lorand L, Goldman R D. Proc Natl Acad Sci USA. 1995;92:8940–8944. doi: 10.1073/pnas.92.19.8940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martin T F. J Biol Chem. 1983;258:14816–14822. [PubMed] [Google Scholar]
- 35.Baek K J, Kwon N S, Lee H S, Kim M S, Muralidhar P, Im M J. Biochem J. 1996;315:739–744. [PMC free article] [PubMed] [Google Scholar]
- 36.Im M J, Riek R P, Graham R M. J Biol Chem. 1990;265:18952–18960. [PubMed] [Google Scholar]
- 37.Baek J B, Das T, Gray C D, Desai S, Hwang K C, Gacchui R, Ludwig M, Im M J. Biochemistry. 1996;35:2651–2657. doi: 10.1021/bi9522965. [DOI] [PubMed] [Google Scholar]
- 38.Wu D, Katz A, Simon M I. Proc Natl Acad Sci USA. 1993;90:5297–5301. doi: 10.1073/pnas.90.11.5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gentile V, Saydak M, Chiocca A, Akande O, Birckbichler P J, Lee K N, Stein J P, Davies P J A. J Biol Chem. 1991;266:478–483. [PubMed] [Google Scholar]
- 40.Hwang K C, Gray C D, Sivasubramanian N, Im M J. J Biol Chem. 1995;270:27058–27062. doi: 10.1074/jbc.270.45.27058. [DOI] [PubMed] [Google Scholar]
- 41.Tanfani F, Bertoli E, Signorini M, Bergamini C M. Eur J Biochem. 1993;218:499–505. doi: 10.1111/j.1432-1033.1993.tb18402.x. [DOI] [PubMed] [Google Scholar]
- 42.Monsonego A, Friedmann I, Shani Y, Eisenstein M, Schwartz M. J Mol Biol. 1998;282:713–720. doi: 10.1006/jmbi.1998.2052. [DOI] [PubMed] [Google Scholar]