Abstract
Optical single-transporter recording, a recently established fluorescence microscopic method, was used to study the selective transport of proteins through single nuclear pore complexes (NPCs) of Xenopus oocytes. Recombinant proteins containing either a nuclear localization signal (import protein) or a nuclear export signal (export protein) were generated as transport substrates. To approximate in vivo conditions as closely as possible, a Xenopus egg extract was applied to the cytosolic side and a Xenopus oocyte nuclear extract to the nuclear side of the NPCs. It was found that protein transport through functionally isolated, “patched” NPCs depended on signal sequences, extracts, and metabolic energy, as in vivo. All NPCs were competent for both import and export. The transport direction was strictly determined by the transport signal, and at none of the conditions explored was the import protein exported or the export protein imported, even when the application sides of the extracts were reversed. The mean transport rates of the single NPC were ≈2 dimers/s for the import protein and ≈4 dimers/s for the export protein (≈15 μM substrate concentration, 22–24°C), in good agreement with in vivo rates estimated for mammalian cells by microinjection experiments. The study shows that optical single-transporter recording permits the analysis of membrane transport processes not previously accessible to single-transporter recording and thus provides additional possibilities for the elucidation of nucleocytoplasmic transport mechanisms.
Single-channel recording (1, 2) has been proven fundamental for the elucidation of membrane transport mechanisms. However, the only method available until recently measures the electrical current passing through a small, functionally isolated membrane patch and therefore is restricted to electrically charged transport substrates. Also, the sensitivity of the electrical patch clamp method requires a flow of >105 charges/s to yield a detectable signal and therefore is most successfully applied to ion channels. We recently have established a method combining fluorescence microphotolysis (3) with confocal scanning microscopy (4) and a novel membrane patching method (5). This optical single-transporter recording (OSTR) is specific and sensitive enough to be applied to any membrane transport system including carriers and pumps provided the transport substrate is fluorescent or can be monitored via a fluorescent indicator. OSTR furthermore permits the simultaneous recording of two or more substrates, the parallel recording of many membrane patches, the generation of very small membrane patches (diameter ≥ 50 nm), and the photochemical generation or removal of substrates, cofactors, or trinucleotides. Recently (6) OSTR has been extended to the nuclear pore complex (NPC).
The NPC is an important control point in the regulatory circuitry between the genetic material in the nucleus and the protein-synthesizing apparatus in the cytoplasm. Consisting of ≈50 different proteins (7) the NPC is a large, approximately cylindrical structure with a diameter of 125 nm, a height of 150–200 nm, and a total mass of 125 MDa (8). Its substructure is highly differentiated, displaying a central granule, eight spokes, inner and outer rings as well as cytoplasmic and nuclear filaments (9). Vertebrate cells frequently contain a few thousand NPCs per nucleus at an area density of ≈1–10 NPCs/μm2. In oocytes, however, the density is much larger, amounting to ≈50 NPCs/μm2 (10). The NPC supports three different transport modes: passive diffusion of small and intermediate molecules (6, 11, 12), selective import into the nucleus of large molecules containing nuclear localization signals (13, 14), and selective export from the nucleus of macromolecules containing nuclear export signals (15, 16). The signal-mediated transport is a multistep process requiring, in addition to signal sequences in the transport substrates, the cooperation of karyopherins, a family of soluble proteins functioning as adapters or receptors (17–20). In addition, the small GTP-ase Ran (21, 22), together with regulating factors, is involved. Important progress has been made recently in the identification and characterization of karyopherins and their molecular interactions with substrates and NPC proteins (for review, see refs. 23–28). The actual mechanisms, however, by which substrates are translocated through the NPC have remained a matter of speculation.
In a preceding study (6) OSTR was used to study the passive permeability of single NPCs. The results suggested that the NPC contains a single diffusion channel with an equivalent radius of 4.4–6.1 nm and an equivalent length of 40–50 nm. The present study demonstrates that OSTR can be used to study processes such as the signal-mediated protein transport through the NPC that previously were not accessible to a single-transporter analysis. To provide a basis for future studies of nucleocytoplasmic transport by OSTR the present study used media approximating the in vivo environment and addressed basic questions concerning the homogeneity, directionality, and rate of nuclear import and export.
Materials and Methods
Solutions, Extracts, and Recombinant Proteins.
Texas red-labeled 70-kDa dextran (TRD70) was from Molecular Probes. Isoporous membrane filters of the type 0.2L were from Infiltec (Speyer, Germany). The ATP-regenerating system (ARS) contained 2 mM ATP, 25 mM phosphocreatine, and 30 units/ml creatine kinase. The extract of Xenopus eggs was prepared according to ref. 29. The nuclear extract was prepared according to ref. 30 by using a buffer containing 140 mM KCl, 3 mM MgCl2, and 10 mM Hepes, pH 7.2. The construction of the import protein (IP), the export protein (EP), and the control protein (CP) (Fig. 1B) was based on the pRSET-B plasmid as provided by Roger Y. Tsien (University of California, San Diego), which encodes the green fluorescent protein mutant GFP(S65T). A plasmid containing the protein kinase A inhibitor α was provided by S. S. Taylor (University of California, San Diego). The nuclear localization signal-containing sequence of the simian virus 40-T antigen has been used previously (31). Plasmids were expressed in the Escherichia coli strain BL21(DE3,pLysS) by using the T7 RNA polymerase-based system (32) and purified by nickel affinity chromatography and gel filtration. By gel chromatography on Sephacryl HR S-200 (Amersham Pharmacia) it was found that the proteins occurred as dimers as expected for glutathione S-transferase proteins. Therefore all concentrations and rates given in this article assume that the recombinant proteins are transported as dimers.
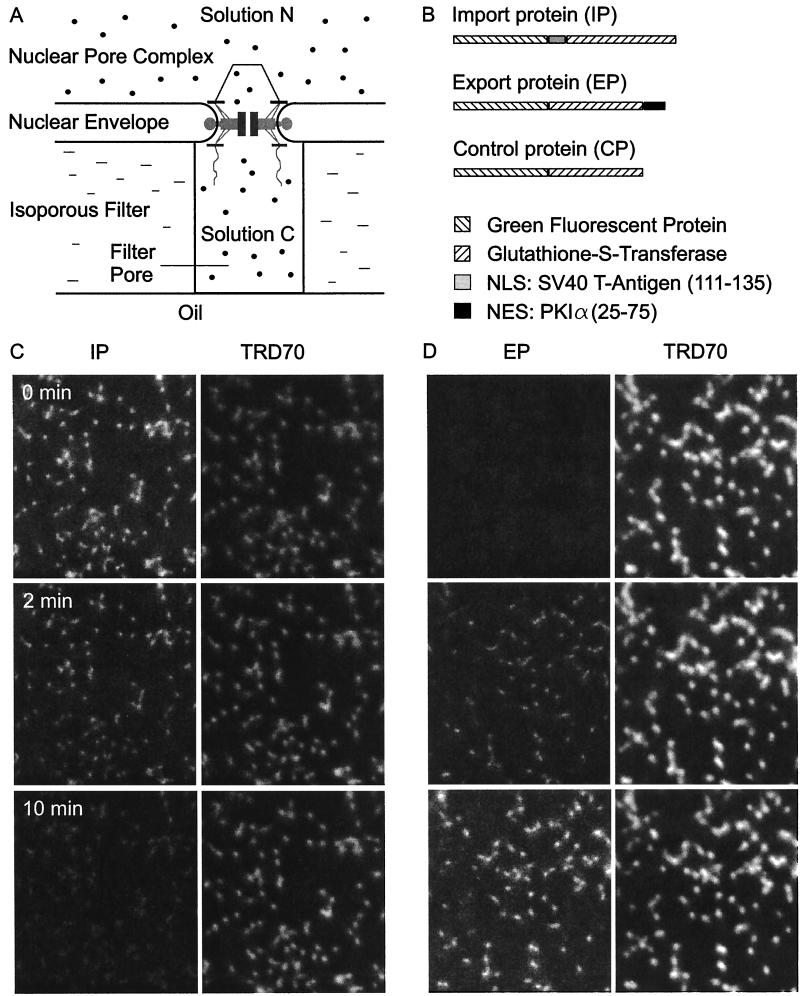
Figure 1.
Optical recording of signal-mediated protein transport through single NPCs. (A) The principle. The nuclear envelope of Xenopus oocytes is firmly attached to isoporous filters. Filter pores are sealed by mineral oil. Transport into or out of individual filter pores is measured by confocal laser scanning microscopy. (B) The transport substrates. The recombinant green fluorescent proteins IP, EP, and CP contained either a nuclear localization signal (NLS), nuclear export signal (NES), or no localization signal. (C) Import of IP. Filter pores were filled with a solution containing IP (green fluorescence), TRD70 (red fluorescence), and egg extract while a nuclear extract was applied to the unclear side. Upon addition of ATP (0 min) IP was transported out of the filter pores via the NPCs. Simultaneously, the constant TRD70 fluorescence indicated the tightness of sealing and the integrity of the NPCs. (D) Export no EP. EP was added to nuclear side and IP was omitted on the cytoplasmic side, otherwise analogous to C.
OSTR.
OSTR (5) and its extension to the NPC (6) were as described. The relative fluorescence of the substrates in the filter pores was converted to absolute concentration by calibration curves. These were generated by immersing filter pieces with solution C to which known concentrations of IP, EP, or CP had been added. After sealing by oil the filter pores were imaged and their fluorescence was determined at exactly the same conditions used in transport measurements. A linear relationship between the fluorescence of the filter pores and the concentration of transport substrates was observed.
Microinjection Studies.
Microinjection of NIH 3T3 cells was as described (33).
Results and Discussion
Protein Transport Through Single NPCs Can Be Recorded by OSTR.
The method for recording protein transport through single NPCs is indicated in Fig. 1A. The nuclear envelope of Xenopus oocytes containing a dense array of NPCs (≈50 NPCs/μm2) was firmly attached to 10-μm thick membrane filters containing a homogeneous population of cylindrical pores. Using a filter type with very small pores (0.18 μm diameter) the membrane patches spanning filter pores contained 1.25 NPCs on average and the fractions of membrane patches containing no, one, two, or three NPCs amounted to ≈5%, 70%, 20%, and 3%, respectively (6). One of the recombinant proteins, IP (dimers of 130 kDa), EP (127 kDa), and CP (122 kDa), shown schematically in Fig. 1B, was added at a final concentration of 15 μM to solution C in import experiments and to solution N in export experiments. After starting transport by addition of ARS to solution N the disappearance (import) or appearance (export) of the transport substrate in the filter pores was recorded by confocal sections. In the present study solution C and N contained an extract of Xenopus eggs or Xenopus oocyte nuclei, as specified. Solution C also always contained 30 μM TRD70 to control for the integrity of the NPC and the tightness of sealing between nuclear envelope and filter.
A recording of the import of IP is shown in Fig. 1C. Solution N was made up of IP, egg extract, and TRD70, and solution N consisted of nuclear extract. Through use of the green fluorescence emission of IP, its relative concentration in the filter pores was monitored by confocal sections (Fig. 1C, Left). Simultaneously the relative concentration of the TRD70 was monitored by its red emission (Fig. 1C, Right). After addition of ARS at zero time the concentration of the dextran remained constant (Fig. 1C, Right), indicating that NPCs were intact and the seal was tight. In contrast, the concentration of IP in the filter pores decreased with time (Fig. 1C, Left), indicating that IP was transported through the NPCs in the membrane patches spanning the filter pores. The export of EP is illustrated in Fig. 1D. Here, solution C consisted of egg extract and TRD70, and solution N consisted of nuclear extract and EP. After addition of ARS the concentration of TRD70 remained constant (Fig. 1D, Right) while EP at first appeared in the filter pores and then continuously increased in concentration with time (Fig. 1D, Left).
Protein Transport Through Functionally Isolated, “Patched” NPCs Depends on Signal Sequences, Cofactors, and Metabolic Energy as in Vivo.
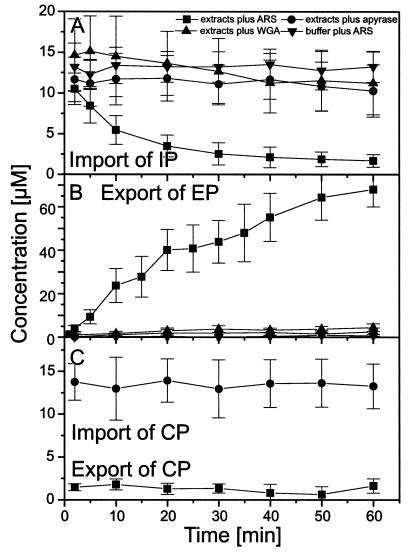
In confocal scans as shown in Fig. 1 C and D the fluorescence intensity of each filter pore was determined individually as a function of time after ARS addition. Averages of the time-dependent fluorescence of large numbers of filter pores are displayed in Fig. 2 A–C. Fig. 2A shows that import of IP had a half-time of ≈10 min (22–25°C) and was observed only in the presence of extracts and ARS, whereas the replacement of extracts by plain buffer, the depletion of extracts from ATP by addition of apyrase, or the addition of the lectin wheat germ agglutinine (WGA) to extracts supplemented with ARS inhibited import completely. The analogous set of experiments for the export of EP is shown in Fig. 2B. The half-time of EP export was ≈5 min (22–25°C). Export of EP also required extracts, and ARS and was completely inhibited by ATP depletion or WGA addition. CP was neither imported nor exported (Fig. 2C). It is well established that signal-mediated protein transport in vivo depends on metabolic energy and can be blocked by ATP depletion or temperature reduction (35, 36). The same is true for signal-mediated protein import into reconstituted nuclei (37) and digitonin-permeabilized cells (17) when extracts are used for providing transport factors. Recently, the energy requirement of protein translocation through the NPC has been determined by using isolated recombinant transport factors instead of extracts (38, 39). It was observed that at least a single translocation of a karyopherin loaded with substrate and the appropriate Ran-trinucleotide complex through the NPC was possible without trinucleotide hydrolysis. At the conditions used in the present study transport had an absolute requirement for trinucleotides. However, whether trinucleotide hydrolysis also was required has yet to be determined.
Figure 2.
Import of IP and export of EP depend on signal sequences, extracts providing soluble transport cofactors, and metabolic energy, and can be inhibited by WGA. (A) From confocal scans as shown in Fig. 1C the time-dependent fluorescence of many filter pores was derived and averaged. Omission of extracts, addition of the ATPase apyrase, or the lectin WGA inhibits transport. (B) Analogous experiments pertaining to the export of EP. (C) CP, containing no localization signal, is neither imported nor exported.
All NPCs Are Competent for Both Protein Import and Export.
From Fig. 1 C and D the conclusion can be drawn that virtually all NPCs are competent for importing IP and exporting EP. Upon starting transport by addition of ARS IP was imported from ≈95% of the filter pores (100% of which could be visualized by means of TRD70 fluorescence). Similarly, EP was exported into ≈95% of filter pores. This was a highly reproducible finding. Previous OSTR measurements (6) showed that ≈95% of the membrane patches spanning filter pores contain at least one NPC. Thus, every patch containing at least one NPC was able to import IP or export EP.
Translocation Through the NPC Is Irreversible, Its Direction Is Determined by Signal Sequences and Its Rate by Soluble Transport Factors.
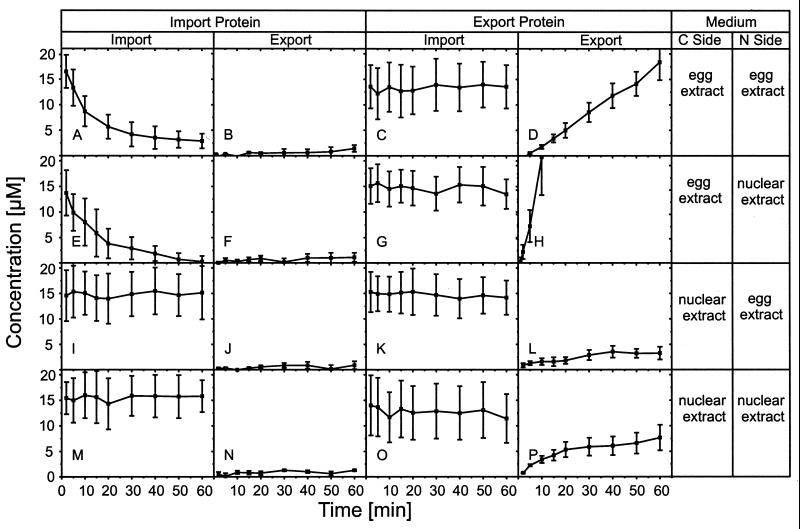
In the experiments described so far the egg extract was applied to the cytoplasmic side and the nuclear extract to the nuclear side of the nuclear envelope. To study which parameters determine the directionality of transport we systematically varied the sides to which transport substrates and extracts were applied, yielding 16 different conditions (Fig. 3).
Figure 3.
Directionality of transport. The application of the recombinant green fluorescent proteins IP and EP and of egg and nuclear extract to the cytoplasmic and nuclear side of the NPC was permutated to obtain clues for the parameters determining the directionality of transport. See text for details.
Fig. 3 reveals that at no condition IP was exported (Fig. 3 B, F, J, and N) nor EP imported (Fig. 3 C, G, K, and O), implying that the signal sequence strictly determined the transport direction and that translocation was irreversible, at least on the time scale investigated. The transport rates were highest when both transport substrates and extracts were applied to the “correct” sides, i.e., IP and egg extract to the cytoplasmic side, EP and nuclear extract to the nuclear side (Fig. 3 E and H). When the extracts were applied to the “wrong” sides import of IP was completely abolished (Fig. 3I) whereas export of EP was reduced to a low rate (Fig. 3L). The same was true when nuclear extract was applied to both sides (Fig. 3 M and P). When egg extract was applied to both sides (Fig. 3 A and D) both import of IP and export of EP proceeded at reduced rates. The data suggest that all three components involved in the transport process, i.e., the signal sequences, extracts, and NPC, cooperate to uniquely determine the transport direction, to make translocation irreversible, and to optimize the transport rate. However, the signal sequence has priority because it strictly determines the transport direction while soluble factors in the media bathing the nuclear envelope optimize the transport rate. At certain conditions deviating from normal (Fig. 3 A, D, and L) the blocking of transport was not complete. This, however, may be simply attributed to the fact that the egg extract, made from laid and activated eggs (29), was not a purely cytosolic preparation but contained nuclear contents.
The Mean Rate of Protein Translocation Amounts to ≈2 Dimers/s per NPC for the Import Protein and to ≈4 Dimers/s per NPC for the Export Protein, in Agreement with Estimates for Living Cells.
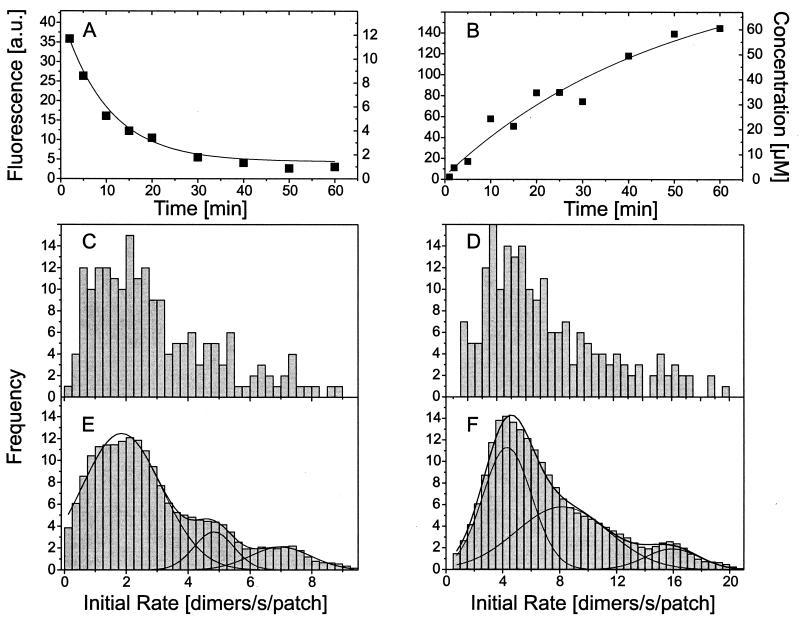
Confocal scans as shown in Fig. 1 C and D were used to determine the fluorescence intensity of individual filter pores as a function of time after ARS addition (Fig. 4 A and B). By means of calibration curves (see Materials and Methods) fluorescence (Fig. 4 A and B, left ordinate) was converted into concentration (Fig. 4 A and B, right ordinate). After conversion the experimental data were fitted, as first approximation, by a simple exponential (lines in Fig. 4 A and B), and taking the volume of the filter pores (0.25 fl) into account, the fit was used to determine the initial transport rate in terms of dimers/s. Populations of initial transport rates pertaining to given experimental conditions were plotted as frequency diagrams (Fig. 4 C-F). Herein the frequency at which a given initial rate occurs within a population is plotted versus the initial rate. Raw data pertaining to the import of IP and the export EP are shown in Fig. 4 C and D, respectively, while corresponding smoothed data are given in Fig. 4 D and F. In the frequency diagrams a number of equidistantly spaced peaks can be recognized. These represent subpopulations of membrane patches with 1,2,3,… transporters (5, 7, 40). The decomposition into subpopulations by Gaussians (lines) is shown in Fig. 4 E and F. From these Gaussians the mean rate of the single NPC for IP import and EP export are determined to be 1.75 dimers/s and 4.25 dimers/s, respectively.
Figure 4.
Protein transport rate of the NPC. (A and B) From confocal scans as shown in Fig. 1 C and D the time-dependent fluorescence of single-filter pores was derived. Fluorescence was converted into concentration and fitted by a simple exponential. The fit was used to derive the initial rate and, taking the volume of the filter pore into account, expressed in molecules/s per patch. (C–F) Populations of transport rates were plotted as frequency diagrams where C and D are the raw data, and E and F are smoothed data. The peaks that are equidistantly spaced on the abscissa represent membrane patches with one, two, or three NPCs.
To compare the transport rates of the patched NPC with its rates in vivo, IP, EP, or CP were injected together with TRD70 into the cytoplasm or nucleus of 3T3 cells (data not shown). The time development of the intracellular fluorescence was assessed by confocal scans. IP was found to be only imported and EP to be only exported. From the scans the mean nuclear fluorescence was determined. As in OSTR measurements fluorescence was converted into concentration by means of calibration curves (not shown), although in this case the conversion can be considered as rough approximation only. From the time dependence of intranuclear concentration the initial transport rates were determined for each injected cell individually. Taking the mean nuclear volume of 3T3 cells as 420 fl and the mean number of NPCs per nucleus as 3,300 (34), the import and export rates were estimated in terms of transported dimers/s per NPC and plotted as a function of concentration. Interestingly, the rates of both import and export depended linearly on concentration without indication of saturation. Thus, the rate of IP import increased from 0.0 to 1.0 dimer/s per NPC and for EP export from 0.0 to 5.0 dimers/s per NPC for a substrate concentration range of 0 to 10 μM. Extrapolated to the concentrations used in OSTR measurements (15 μM) the in vivo rates for IP import and EP export amounted to 1.6 and 7.6 dimers/s per NPC, respectively, resembling protein transport rates of the patched NPC.
Franke (41) suggested in 1970 that the “nuclear pore flow rate” is a parameter characteristic of the functional state of the pore complex. He and many others (most recently in ref. 27) arrived at rates of ≈1–2 ribosomal protein molecules/s per NPC or 0.05 ribosomal subunits/s per NPC. Goldfarb et al. (42) studied the import of nuclear localization signal-conjugated BSA into nuclei of Xenopus oocytes by microinjection and reported a maximal rate of 3.5 molecules/s per NPC. From the data of Fischer et al. (16) for the export of a nuclear export signal-BSA conjugate from microinjected Xenopus oocyte nuclei an export rate of 0.2 molecules/s per NPC can be derived. Thus the rate of ≈2 molecules/s per NPC for the import of IP and 4 molecules/s per NPC for the export of EP as determined in this study are at the upper limit of previous estimates and also agree very well with our own semiquantitative determination pertaining to living 3T3 cells. Although the transport rates determined by us hold for only specific conditions they shed some light on the efficiency of the NPC. Ion channels can accommodate large transport rates such as 106 ions/s per channel, but active transport by ion pumps in which a high specificity is achieved by tight binding of the substrate to the transporter and in which transport against electrochemical gradients is achieved by input of metabolic energy proceeds at much smaller rates. For instance, both the sodium-potassium pump and the calcium pump (43) operate at approximately 100 cycles/s. Taking into consideration that protein transport through the NPC involves substrates very much larger than inorganic ions and is highly specific and strictly directional the rates determined in this study appear to be large. Apparently, transport through the NPC is an optimized, fast, and accurate process, consistent with the important role the NPC assumes in cell regulation.
Concluding Remarks
Many membrane transport processes cannot be analyzed by the electrical patch clamp method, for instance because the rate is too small, the electrical background noise too large, or the transporter density too high. The present study shows that such transport processes can be analyzed by OSTR provided the transport substrate is fluorescent or can be monitored by means of a fluorescent indicator and the patching method is applicable. Independently of the single-transporter analysis OSTR has another property particularly relevant for nucleocytoplasmic transport: the media bathing the NPC on its cytoplasmic and nuclear sides can be freely determined. This feature provides access to the analysis of import and export and the determinants of directionality. The present study yielded a basis for the analysis of nucleocytoplasmic transport by OSTR. It will be highly instructive to use now defined mixtures of recombinant transport factors instead of cell extracts.
Acknowledgments
We thank Dr. S. S. Taylor (Department of Chemistry and Biochemistry, University of California, San Diego) and Dr. R. Y. Tsien (Howard Hughes Medical Institute, University of California, San Diego) for providing plasmids and Dr. M. Madeja (Physiologisches Institute, Universität Münster) for proving Xenopus oocytes. The study was supported by the Deutsche Forschungsgemeinschaft.
Abbreviations
- ARS
ATP-regenerating system
- CP
control protein
- EP
export protein
- IP
import protein
- NPC
nuclear pore complex
- OSTR
optical single-transporter recording
- TRD70
Texas red-labeled dextran of 70 kDa
- WGA
wheat germ agglutinine
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Neher E, Sakman B. Nature (London) 1976;260:770–802. [Google Scholar]
- 2.Sakmann B, Neher E. Single-Channel Recording. 2nd Ed. New York: Plenum; 1995. [Google Scholar]
- 3.Peters R, Peters J, Tews K H, Bähr W. Biochim Biophys Acta. 1974;367:282–294. doi: 10.1016/0005-2736(74)90085-6. [DOI] [PubMed] [Google Scholar]
- 4.Wedekind P, Kubitscheck U, Peters R. J Microsc. 1994;176:23–33. doi: 10.1111/j.1365-2818.1994.tb03496.x. [DOI] [PubMed] [Google Scholar]
- 5.Tschödrich-Rotter M, Peters R. J Microsc. 1998;192:114–125. doi: 10.1046/j.1365-2818.1998.00411.x. [DOI] [PubMed] [Google Scholar]
- 6.Keminer O, Peters R. Biophys J. 1999;77:217–228. doi: 10.1016/S0006-3495(99)76883-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rout M P, Blobel G. J Cell Biol. 1993;123:771–783. doi: 10.1083/jcb.123.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reichelt R, Holzenburg A, Buhle E L, Jr, Jarnik M, Engel A, Aebi U. J Cell Biol. 1990;110:883–894. doi: 10.1083/jcb.110.4.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Akey C W, Radermacher M. J Cell Biol. 1993;122:1–119. doi: 10.1083/jcb.122.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maul G G. Int Rev Cytol. 1977;6,Suppl.:75–186. [PubMed] [Google Scholar]
- 11.Paine P L, Moore L C, Horowitz S B. Nature (London) 1975;254:109–114. doi: 10.1038/254109a0. [DOI] [PubMed] [Google Scholar]
- 12.Peters R. EMBO J. 1984;3:1831–1836. doi: 10.1002/j.1460-2075.1984.tb02055.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dingwall C, Sharnik S V, Laskey R A. Cell. 1982;30:449–458. doi: 10.1016/0092-8674(82)90242-2. [DOI] [PubMed] [Google Scholar]
- 14.Kalderon D, Roberts B L, Richardson W D, Smith A E. Cell. 1984;39:499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- 15.Wen W, Meinkoth J L, Tsien R Y, Taylor S S. Cell. 1995;82:463–473. doi: 10.1016/0092-8674(95)90435-2. [DOI] [PubMed] [Google Scholar]
- 16.Fischer U, Huber J, Boelens W C, Mattaj I W, Lührmann R. Cell. 1995;82:475–483. doi: 10.1016/0092-8674(95)90436-0. [DOI] [PubMed] [Google Scholar]
- 17.Adam S A, Gerace L. Cell. 1991;66:837–847. doi: 10.1016/0092-8674(91)90431-w. [DOI] [PubMed] [Google Scholar]
- 18.Adam E J H, Adam S A. J Cell Biol. 1994;125:547–555. doi: 10.1083/jcb.125.3.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Görlich D, Prehn S, Laskey R A, Hartmann E. Cell. 1994;79:767–778. doi: 10.1016/0092-8674(94)90067-1. [DOI] [PubMed] [Google Scholar]
- 20.Moroianu J, Blobel G, Radu A. Proc Natl Acad Sci USA. 1995;92:2008–2011. doi: 10.1073/pnas.92.6.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moore M S, Blobel G. Nature (London) 1993;365:661–663. doi: 10.1038/365661a0. [DOI] [PubMed] [Google Scholar]
- 22.Melchior F, Paschak B, Evans E, Gerace L. J Cell Biol. 1993;123:1649–1659. doi: 10.1083/jcb.123.6.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doyé V, Hurt E. Curr Opin Cell Biol. 1997;9:401–411. doi: 10.1016/s0955-0674(97)80014-2. [DOI] [PubMed] [Google Scholar]
- 24.Daneholt B. Cell. 1997;88:585–588. doi: 10.1016/s0092-8674(00)81900-5. [DOI] [PubMed] [Google Scholar]
- 25.Pemberton L F, Blobel G, Rosenblum J S. Curr Opin Cell Biol. 1998;10:392–399. doi: 10.1016/s0955-0674(98)80016-1. [DOI] [PubMed] [Google Scholar]
- 26.Mattaj I, J, Englmeier L. Annu Rev Biochem. 1998;67:265–306. doi: 10.1146/annurev.biochem.67.1.265. [DOI] [PubMed] [Google Scholar]
- 27.Görlich D, Mattaj I W. Science. 1996;271:1513–1518. doi: 10.1126/science.271.5255.1513. [DOI] [PubMed] [Google Scholar]
- 28.Wozniak R W, Rout M P, Atchinson J D. Trends Cell Biol. 1998;8:184–188. doi: 10.1016/s0962-8924(98)01248-3. [DOI] [PubMed] [Google Scholar]
- 29.Newmeyer D D, Wilson K L. Methods Cell Biol. 1991;36:607–635. doi: 10.1016/s0091-679x(08)60299-x. [DOI] [PubMed] [Google Scholar]
- 30.Lehman C W, Carroll D. Proc Natl Acad Sci USA. 1991;88:10840–10844. doi: 10.1073/pnas.88.23.10840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rihs H-P, Peters R. EMBO J. 1989;8:1479–1484. doi: 10.1002/j.1460-2075.1989.tb03531.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 33.Middeler G, Zerf K, Jenovai S, Thülig A, Tschödrich-Rotter M, Kubitscheck U, Peters R. Oncogene. 1997;14:1407–1417. doi: 10.1038/sj.onc.1200949. [DOI] [PubMed] [Google Scholar]
- 34.Kubitscheck U, Wedekind P, Zeidler O, Grote M, Peters R. Biophys J. 1996;70:2067–2077. doi: 10.1016/S0006-3495(96)79811-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Newmeyer D D, Forbes D J. Cell. 1988;52:641–653. doi: 10.1016/0092-8674(88)90402-3. [DOI] [PubMed] [Google Scholar]
- 36.Richardson W D, Mills A D, Dilworth S M, Laskey R A, Dingwall C. Cell. 1988;52:655–664. doi: 10.1016/0092-8674(88)90403-5. [DOI] [PubMed] [Google Scholar]
- 37.Newmeyer D D, Lucocq J M, Bürglin T R, DeRobertis E M. EMBO J. 1986;5:5001–5010. doi: 10.1002/j.1460-2075.1986.tb04239.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schwoebel E D, Talcott B, Cushman I, Moore M S. J Biol Chem. 1998;52:35170–35175. doi: 10.1074/jbc.273.52.35170. [DOI] [PubMed] [Google Scholar]
- 39.Englmeier L, Olivio J-C, Mattaj I W. Curr Biol. 1999;9:30–41. doi: 10.1016/s0960-9822(99)80044-x. [DOI] [PubMed] [Google Scholar]
- 40.Peters R, Sauer H, Tschopp J, Fritsch G. EMBO J. 1990;9:2447–2451. doi: 10.1002/j.1460-2075.1990.tb07421.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Franke W W. Naturwissenschaften. 1970;57:44–45. doi: 10.1007/BF00593568. [DOI] [PubMed] [Google Scholar]
- 42.Goldfarb D S, Gariepy J, Schoolnik G, Kornberg R D. Nature (London) 1988;322:641–644. doi: 10.1038/322641a0. [DOI] [PubMed] [Google Scholar]
- 43.Kubitscheck U, Pratsch L, Passow H, Peters R. Biophys J. 1995;69:30–41. doi: 10.1016/S0006-3495(95)79875-7. [DOI] [PMC free article] [PubMed] [Google Scholar]