Abstract
Persistent infections by viruses such as HIV-1 and hepatitis B virus can pose long-term health hazards. Because establishment of persistent infections involves close interactions and adjustments in both host and virus, it would be informative to establish a paradigm with which a normally cytolytic viral infection can be easily converted to persistent infection, so that the different stages in developing persistent infection can be examined. Such a model system is described in this paper. Highly cytolytic encephalomyocarditis virus (EMCV) infection was shifted to persistent infection as a result of repressed expression of the double-stranded RNA-dependent protein kinase (PKR) in the promonocytic U937 cells. Because of the apoptogenic potential of PKR, a deficiency of PKR resulted in a delay in virus-induced apoptosis in EMCV-infected U937 cells, allowing the eventual establishment of persistent EMCV infection in these cells (U9K-AV2). That this was a bona fide persistent infection was demonstrated by the ability of infected cells to propagate as long-term virus-shedding cultures; electron microscopy studies showing presence of intracellular EMCV virions and chromatin condensation; detection of virus-induced chromosomal DNA fragmentation and sustained expression of apoptogenic p53 and IL-1β converting enzyme; and demonstration of active EMCV transcription by reverse transcription–PCR. In addition, a host-virus coevolution was observed in U9K-AV2 cultures over time: U9K-AV2 cells exhibited slower growth rates, resistance to viral super-infection, and cessation of IFN-α synthesis, whereas the infectivity of EMCV was drastically attenuated. Finally, data are presented on the suitability of this model to study establishment of persistent infection by other viruses such as Sendai virus and reovirus.
During acute viral infections, the effectiveness with which the host’s immune response can clear viruses dictates that the invading virus must infect new hosts with matching efficiency. Some viruses, such as poliovirus and rotavirus, can survive without a host for some time. Alternatively, viruses may find it advantageous to establish persistent infection with periodic transmission to new hosts. For the host, persistent viral infections can be potentially devastating. Classic examples include clinical progression to AIDS and hepatocarcinoma as a result of chronic infection by HIV-1 and hepatitis B virus, respectively. Thus, it is important to study at the cellular level the molecular mechanisms of how viruses evade the host’s antiviral defenses and are allowed to establish persistent infections in susceptible host cells.
Apoptosis, or programmed cell death, is a self-destruct process whereby unwanted individual cells in metazoan animals undergo a genetically determined program of new protein synthesis and morphological changes, culminating in chromosomal DNA fragmentation and eventual cell death (1). A cell can have more than one apoptotic pathway in its arsenal. Depending on the death signal and the cell type, a cell turns on an appropriate apoptotic pathway with associated death genes, which can either be unique or shared by different apoptotic programs (2). One well-characterized apoptotic pathway important for viral clearance is induced by tumor necrosis factor α (TNF-α). Apparently, there is a cross-talk between the TNF-α and the IFN signaling pathways: a factor with a pivotal role in TNF-α-induced apoptosis is the IFN-inducible, double-stranded RNA-dependent protein kinase (PKR). Initially known for its antiviral action through phosphorylation of the eukaryotic translation initiation factor 2 and subsequent inhibition of protein synthesis (3), PKR has been demonstrated to be essential for activation of the transcription factor NF-κB (4), synthesis of IFN-α (5), TNF-α-induced apoptosis (6), and cellular stress-coping strategies (7).
Viral infection can serve as an apoptotic signal. Apoptosis has been documented to be a cellular response to infection by many viruses including HIV-1, adenovirus, encephalomyocarditis virus (EMCV), cowpox virus, baculovirus, poxvirus, and gammaherpes virus (8). Apoptotic activation in virus-infected cells may limit virus replication in vivo through phagocytosis of apoptosing virus-infected cells. Consequently, some viruses are known to encode antiapoptotic proteins (e.g., E1B in adenovirus) to avoid host-programmed cell death (8). For EMCV, NF-κB-mediated inhibition of apoptosis is required for viral virulence in a mouse model (9). Given the dual role of PKR in inducing an antiviral state and in mediating apoptosis, we investigated whether diminution in PKR expression will affect the outcome of EMCV infection in the human promonocytic U937 cells in vitro. The use of an in vitro system in this study was designed to circumvent the influence of the host immune system so that we could examine directly the host-virus interactions at the cellular level. It was found that PKR was needed for full EMCV virulence in U937 cells in vitro, and that a lack of PKR in a U937 derivative harboring a PKR antisense expression vector actually promoted conversion of cytolytic to persistent infection by EMCV.
Material and Methods
Plasmids, Cell Cultures, and Cell Viability.
Construction of PKR antisense expression vector derived from the parental control expression vector pRC-CMV (Invitrogen) as well as isolation of American Type Culture Collection U937 stable derivatives harboring these vectors have been described (5, 6). All U937 cells were maintained at 37°C in suspension cultures in RPMI supplemented with 5% FBS, 400 μg/ml Geneticin (GIBCO/BRL), and 5% CO2. For viral titration studies, L-929 cells were obtained from the American Type Culture Collection and were grown at 37°C as monolayers in DMEM (Mediatech, Herndon, VA) supplemented with 5% FBS and 5% CO2. To determine cell viability, cells were mixed with 0.1 vol of 0.4% trypan blue and were counted with a hemocytometer. Percentage cell viability is defined as the percentage of the number of viable test cells over the number of viable control cells of the same cell line at the end of each experimental treatment.
Viruses and Viral Assays.
All viruses (EMCV, Sendai virus, and reovirus type 3) were obtained from the American Type Culture Collection. For viral infection studies, U937 cells were centrifuged, resuspended in fresh RPMI without FBS, and were incubated with various infecting viral doses at room temperature with constant rocking for 1 hr. The infected cells were diluted with fresh and FBS-supplemented RPMI and further incubated at 37°C for indicated times. The cells were centrifuged; the supernatants and pellets were frozen separately at −70°C until further processing. For viral titration assays, the supernatants and pellets (resuspended in PBS and freeze-thawed three times) were centrifuged before application to titer plates. To determine viral infectivity, 4-fold serial dilutions of samples were added onto L-929 monolayers in 96-well plates and incubated for 48 hr before the cells were stained with 0.05% crystal violet to determine cytopathic effects and the 50% tissue culture infective dose (TCID50).
Microscopy Studies.
For transmission electron microscopy, samples were fixed in Karnovsky solution, followed by cacodylate buffer for ultrastructural examination. The cells were postfixed in 1% osmium tetroxide and dehydrated for staining with uranyl acetate and lead citrate. Thin sections were examined in a JEOL 100 SX electron microscope.
Apoptotic DNA Laddering Assay.
Apoptotic DNA fragments were isolated as described (10). Briefly, cells were extracted twice with lysis buffer (1% NP-40 in 20 mM EDTA, 50 mM Tris⋅HCl, pH 7.5). After centrifugation at 1,600 × g, the combined supernatants were brought to 1% SDS, treated with 5 mg/ml (final concentration) RNase A (Amersham Pharmacia) at 56°C for 2 hr, and followed by digestion with 2.5 mg/ml (final concentration) proteinase K at 37°C for 2 hr. Apoptotic DNA fragments were precipitated and analyzed in 2% agarose gels.
Reverse Transcription–PCR (RT-PCR).
Total RNA extraction and RT-PCR for steady-state RNA levels have been described (6). First-strand cDNA was reverse-transcribed from total RNA by using Random Primers and Moloney murine leukemia virus reverse transcriptase (Promega). In some experiments, total RNA was primed with either 5′ or 3′ EMCV PCR primers before RT (see text). PCRs were performed in a thermal cycler (Coy Laboratory Products, Ann Arbor, MI) for 30 cycles (95°C for 1 min, 55°C for 2 min, and 72°C for 3 min) before arresting the reaction in its logarithmic phase by rapid cooling at 4°C. The PCR products were analyzed on 0.7% agarose gel followed by Southern blotting (11) with gene-specific probes. PCR primer sets used were: (i) EMCV: upstream, 5′-CAAGTCTTCCAACCAGCATA-3′; downstream, 5′-ACCGAAGGCAGTCCAGTGTA-3′; (ii) p53: upstream, 5′-CCTCCTGGCCCCTGTCATCT-3′; downstream, 5′-ACAAACACGCACCTCAAAGC-3′; (iii) IL-1β converting enzyme (ICE): upstream, 5′-AATGCTGCTACAAAATCTGG-3′; downstream, 5′-ATCATCCTCAAACTCTTCTG-3′; and (iv) 18S ribosomal RNA: upstream, 5′-CGCAGCTAGGAATAATGGAA-3′; downstream, 5′-TTATGACCCGCACTTACTGG-3′. For data analysis, radioactivity counts (cpm) for various messages were normalized against those of the corresponding internal control 18S messages in the same sample. In our hands, signals generated from RT-PCR as described above were proportional to the amounts of input cDNA (data not shown).
IFN-α ELISA Assays.
IFN-α was quantified by using an ELISA kit against 13 human IFN-α subtypes (PBL Biomed Laboratories, New Brunswick, NJ).
RNA Dot Blot Hybridization.
After denaturation at 94°C for 1 min and followed by rapid cooling on ice, 10 μg total RNA was dot-blotted onto nitrocellulose membrane and hybridized with appropriate [α-32P]-dATP-random-primed probes specific for EMCV as described (12). Extent of hybridization was determined by scintillation counting of radioactive spots.
Results
EMCV-Induced Apoptosis in U937 Cells Required PKR.
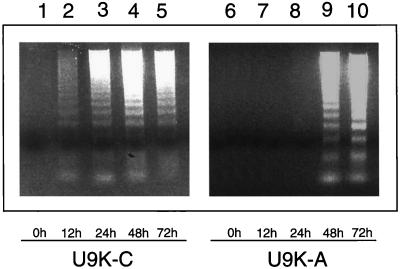
It was demonstrated that PKR is apoptogenic (6) and that some viruses induce apoptotic responses in host cells (8). EMCV is normally cytolytic to host cells. To ascertain whether EMCV induces apoptosis in U937 cells, control U9K-C and PKR-deficient U9K-A cells were infected with EMCV at 0.1 TCID50/cell for up to 72 hr. DNA laddering assays revealed that 198-bp apoptotic DNA fragments appeared in U9K-C control cells as early as 12 hr postinfection (p.i.) (Fig. 1). In contrast, in U9K-A cells in which PKR expression is suppressed (5, 6), apoptotic DNA fragments did not appear until 48 hr p.i., suggesting that a normal level of PKR was required for the EMCV-induced apoptosis in U937 cells.
Figure 1.
Delay of EMCV-induced apoptosis in U9K-A cells. U9K-C and U9K-A cells were infected with EMCV at 0.1 TCID50/cell for up to 72 hr. At indicated times, 5 × 106 cells were harvested from each culture for apoptotic DNA laddering assays.
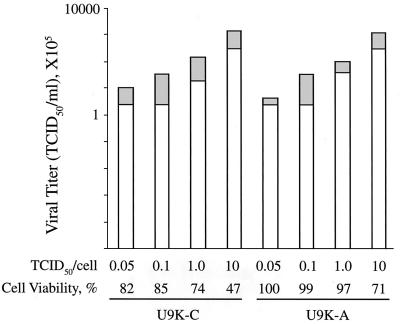
To investigate whether apoptosis was needed for release of EMCV virions from host cells, U937 cells were infected with various EMCV-infecting doses for 12 h to allow for one round of EMCV replication cycle. As expected, increasing infecting doses resulted in increasing levels of cell lysis (Fig. 2). However, U9K-A cells apparently were resistant to EMCV-induced apoptosis unless the infecting dose reached a high 10 TCID50/cell (Fig. 2). This finding correlates with the apoptosis data described above in that EMCV did not efficiently induce apoptosis in U9K-A cells, particularly when the infecting doses were moderate. Despite the differences in the extent of EMCV-induced cell lysis, viral titers present in the supernatant and pellet samples from both EMCV-infected U9K-C and U9K-A cells were quite similar at 12 hr p.i. The release of EMCV in U9K-A cells in the absence of cell lysis argued against apoptosis as a prerequisite for EMCV replication. As in some viral infections, apoptosis induction in EMCV-infected U937 cells was more likely to be a host response to the viral infection (13).
Figure 2.
Cell viability and viral release from EMCV-infected cells. Cells (0.5 × 106) were infected with EMCV with doses from 0.05 to 10 TCID50/cell for 12 hr. After harvesting, viral titers in the cell pellets (freeze-thawed three times) and supernatants were measured by TCID50 bioassays on L-929 cells; cell viability was determined by trypan blue viable cell counts. Viral titers are plotted on a log10 scale. For each stacked bar, the upper and lower portions correspond to EMCV titers in the pellet and supernatant, respectively.
Establishment and Characterization of Persistent EMCV Infection.
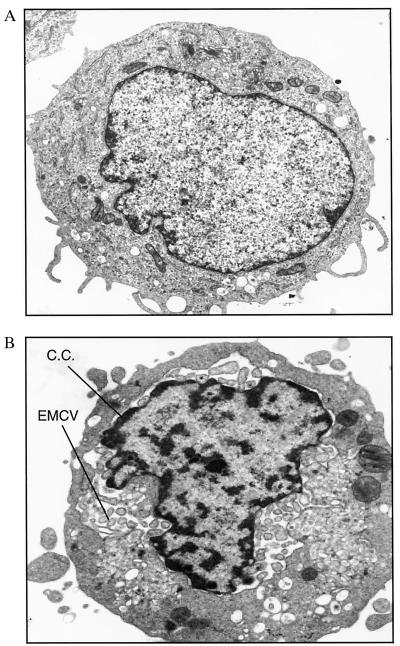
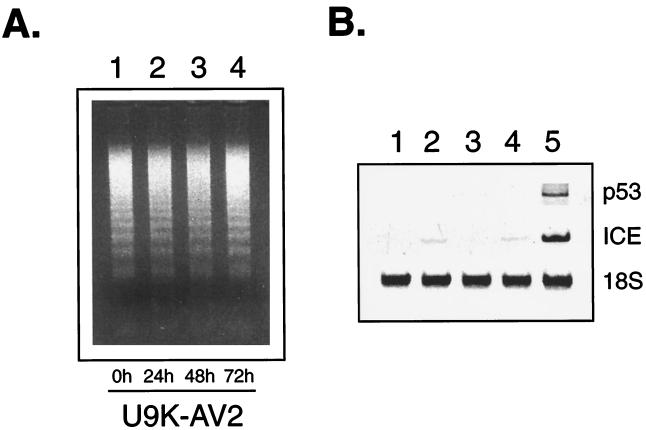
The relative resistance to EMCV-induced apoptosis in U9K-A cells posed the question whether EMCV-infected U9K-A cells would be completely killed with time, particularly at lower infecting doses. Even at 48 hr p.i., viability of EMCV-infected (0.1 TCID50/cell) U9K-A cultures remained at around 40%, whereas EMCV-infected U9K-C cells were all dead (data not shown). If these EMCV-infected U9K-A cells were maintained in culture, they invariably would recover in cell numbers and could be cultured for more than a year. To demonstrate that these cells were persistently infected with EMCV, an EMCV-infected U9K-A line, designated U9K-AV2, was subjected to electron microscopy studies. The presence of viral particles was seen in the cytoplasm of U9K-AV2 cells (Fig. 3). Chromatin condensation along the inner nuclear membrane also was apparent, reflecting the apoptosing nature of the chronically infected culture. This apoptosing nature was corroborated by the presence of apoptotic DNA fragments in U9K-AV2 cells (Fig. 4A) and was further substantiated by the detection of p53 and ICE (Fig. 4B), which are commonly involved in apoptosis. Although ICE messages were detectable in U9K-C and U9K-A cells newly infected with EMCV (Fig. 4B), p53 messages were hard to detect in these cultures. This finding is consistent with the previous observation that p53 expression is extremely low and only transient in U937 cells (14). In contrast, the much higher levels of both p53 and ICE messages in U9K-AV2 cells illustrated the sustained apoptotic response in the persistently infected U9K-AV2 cells (Fig. 4B). Another line of evidence for the apoptotic nature of U9K-AV2 cultures was that by annexin-V (R&D Systems) flow cytometry analysis that detects early apoptosis, persistently infected U9K-AV2 cultures continually grown for 18 months routinely exhibited 10–20% early apoptosis levels; for comparison, the early apoptosis level in U9K-C cells acutely infected with EMCV (0.1 TCID50/cell) for 24 hr was about 20% (unpublished data).
Figure 3.
Electron microscopy of U9K-A cells persistently infected with EMCV. Noninfected U9K-A controls and U9K-AV2 cells persistently infected with EMCV were examined with transmission electron microscopy for the presence of viral particles. (A) Control U9K-A cells. (B) U9K-AV2 cells. Magnification: ×16,000. C.C., chromatin condensation.
Figure 4.
Evidence of apoptosis in U9K-AV2 cells. (A) U9K-AV2 cells (5 × 106) were centrifuged, resuspended in fresh medium, and followed for 3 days. Cells were harvested at indicated times for apoptotic DNA laddering assays. (B) Expression of p53 and ICE in U9K-AV2 cells was assayed by RT-PCR. Lanes 1 and 2, control and EMCV-infected (0.1 TCID50/cell, 24 hr p.i.) U9K-C cells, respectively; lanes 3 and 4, control and EMCV-infected (0.1 TCID50/cell, 24 hr p.i.) U9K-A cells, respectively; lane 5, U9K-AV2 cells.
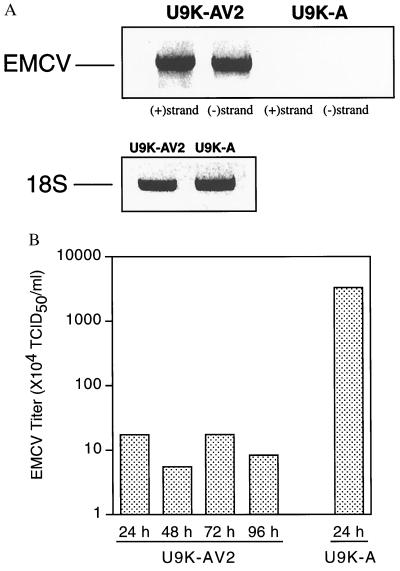
Similar to other picornaviruses, EMCV contains a positive-strand RNA genome that is transcribed into complementary negative RNA strands during viral replication (15). To show that U9K-AV2 cells carried replicating rather than latent EMCV, RT-PCR was performed on total RNA from U9K-AV2 cells to detect EMCV negative-strand RNAs. In this assay, the positive-strand genomic RNA was specifically primed by the EMCV-specific downstream PCR primer (see Methods and Materials), whereas the complementary negative-strand RNAs were specifically primed by the corresponding upstream EMCV-specific PCR primer. Subsequent PCRs on these selectively primed cDNAs yielded exclusively positive- or negative-strand amplified products, respectively. Indicative of ongoing EMCV replication, messages corresponding to the EMCV negative-strand RNAs were readily detected in U9K-AV2 cells (Fig. 5A). As further supporting evidence, EMCV titers in U9K-AV2 cells remained constant but low over time, which was in sharp contrast to the case with control U9K-A cells newly infected with wild-type EMCV (Fig. 5B). This finding is in agreement with the von Magnus effect, in which the constant competition between wild-type and defective interfering (DI) particles for the host cellular machinery to replicate results in a continual, steady-state release of measurable low titers in a cell culture persistently infected with virus in vitro (16).
Figure 5.
EMCV replication in U9K-AV2 cells. (A) Total RNA was extracted from noninfected U9K-A controls and persistently infected U9K-AV2 cells to detect steady-state EMCV RNA by RT-PCR. To detect positive- and negative-strand EMCV RNA, cDNA synthesis was achieved by priming total RNA with either 5′ or 3′ EMCV PCR primer. RT-PCR detection of 18S RNA was performed on cDNA from random-primed total RNA. (B) A U9K-AV2 culture was maintained for 4 days. Aliquots were taken at indicated times, freeze-thawed three times, and centrifuged, and the supernatants were titered for EMCV. For comparison, a typical viral yield from U9K-A cells newly infected with EMCV at 0.1 TCID50/cell for 24 hr is included. EMCV titers are plotted on a log10 scale.
Host and Virus Coevolution.
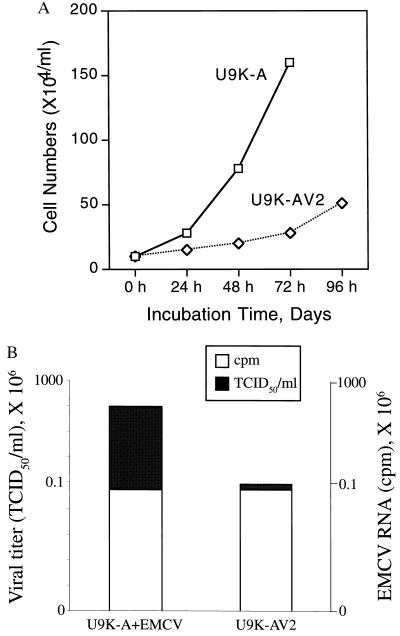
Establishment of persistent infection often results in changes in both host and virus with time (17, 18). Some of the data reported above suggested marked changes in U9K-AV2 cultures. These included apoptotic morphological changes and up-regulation of p53 and ICE. Similarly, the following observations attest to the host cell changes induced by persistent EMCV infection. First, U9K-AV2 cultures exhibited growth characteristics slower than the parental’s (Fig. 6A). Second, U9K-AV2 cells could not be super-infected by wild-type EMCV (normal infecting doses, 0.1 TCID50/cell; data not shown). Third, by constant viral induction, 6-month-old U9K-AV2 cells could synthesize up to 40 units/ml IFN-α; however, older U9K-AV2 cultures (e.g., EMCV persistently infected for over a year) ceased to synthesize IFN-α (data not shown). Fourth, for some cell lines that we were able to subclone by limiting dilution of U9K-AV2 cells grew about 30–40% slower than the parental U9K-A cells (data not shown). These subcloned cells were free of EMCV as demonstrated by RT-PCR and were susceptible to EMCV only at much higher infecting doses (above 10 TCID50/cell; data not shown).
Figure 6.
Growth characteristics and attenuated EMCV infectivity in U9K-AV2 cells. (A) U9K-A and U9K-AV2 cultures (each with a starting cell density of 106 cells in 10 ml of medium) were followed for up to 96 hr for growth curve determination (trypan blue viable cell counting). (B) Cells (5 × 106) of EMCV-infected U9K-A (48 hr p.i.) and U9K-AV2 cells were harvested for viral titration and total RNA extraction. Ten micrograms of total RNA was dot-blotted onto nitrocellulose membrane and hybridized with 33P-random-primed EMCV-specific probes. Both viral titers and EMCV RNA axes are plotted on log10 scales.
Changes also were apparent for the EMCV in U9K-AV2 cultures. As described earlier, supernatant from U9K-AV2 cultures routinely yielded 3 logs lower TCID50/ml than parental U9K-A cells newly infected with EMCV (Fig. 5). To ascertain whether this lower viral titer was caused by lower viral replication rates or attenuated viral infectivity, total RNA extracted from U9K-AV2 and newly EMCV-infected (0.1 TCID50/cell for 48 hr) U9K-A cells was hybridized with EMCV-specific probes. Fig. 6B shows that whereas the two cultures had similar EMCV RNA contents, the viral infectivities of the two cultures differed by at least 3 logs, suggesting, but not proving, that EMCV in U9K-AV2 cells was less infectious than its wild-type counterpart, and that selection of EMCV DI particles probably had occurred in U9K-AV2 cultures. Interestingly, despite the attenuation of EMCV in U9K-AV2 cultures, no subclones harboring EMCV could ever be isolated by limiting dilution of U9K-AV2 cells, suggesting that even attenuated EMCV in these cultures ultimately would lyse the host cells. It follows then continuation of the viral persistence state in U9K-V2 cultures would partly depend on new infection of the noninfected subpopulation of U9K-AV2 cells described above.
Infection of U9K-A Cells with Sendai Virus and Reovirus.
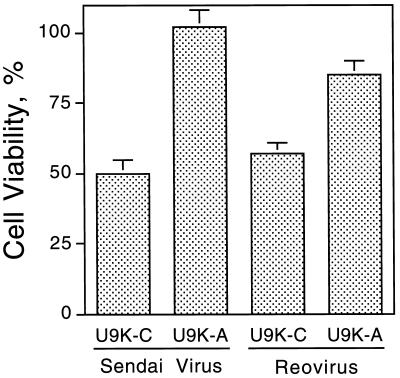
To investigate whether U9K-A cells were similarly resistant to cytolysis by other RNA viruses, U9K-A cells were infected with Sendai virus (0.5 TCID50/cell) and reovirus type 3 (5 TCID50/cell). At 48 hr p.i., U9K-A cells exhibited 50% and 25% more cell viability than U9K-C cells when infected with Sendai virus and reovirus, respectively (Fig. 7).
Figure 7.
Viability of Sendai- and reovirus-infected U937 cells. U9K-C and U9K-A cells were infected with either Sendai (0.5 TCID50/cell) or reovirus (5 TCID50/cell) for 48 hr before cell viability was determined by trypan blue viable cell counting.
Discussion
From the previous finding that PKR is apoptogenic (6) and the observation in this work that the U9K-A cells deficient in PKR showed a delay in EMCV-induced apoptosis, a potentially important role for the multifunctional PKR emerges: assurance of apoptotic cell death after viral infection. As in most altruistic apoptotic processes, this commitment to self-suicide in virally infected cells serves to limit the extent of viral infection in the host organism; noninflammatory clearance of apoptotic cells by phagocytic cells usually confines toxic cellular components (virus in this case) within the apoptotic bodies, effectively preventing the spread of potentially harmful substances (viruses) to neighboring tissues (19). It is not surprising that some viruses have evolved strategies to combat the host apoptotic responses in virally infected cells (8). In this regard, it is noted that inhibition of apoptosis enhances EMCV virulence in a mouse model (9). Inhibition of apoptosis in adenovirus-infected HeLa cells similarly increases virus production in vitro (20). Thus, the apoptogenic nature of PKR is crucial in the overall antiviral repertoire in the host.
As a further illustration of this critical role of PKR, a consequence of a cell having nonfunctional or limited PKR activity would be the risk of establishing persistent infection in some viral infections. This risk was exemplified in this study in which normally cytolytic infection of U937 cells by EMCV was converted to persistent infection in cells deficient in PKR. Moreover, as long as there was a deficiency in PKR, establishment of this persistent infection by EMCV cells was apparently not a difficult feat for the virus; we could routinely isolate U9K-A lines persistently infected with EMCV even when the infecting EMCV dose used was as high as 10 TCID50/cell (data not shown). Hence, another interpretation of the PKR role in virus-induced apoptosis is inhibition of viral-persistent infection.
Like most picornaviruses of which EMCV is a member, EMCV infection is highly cytolytic. The ease with which persistent EMCV infection could be established in U9K-A cells in this study presents a paradigm for studying the molecular mechanisms of viral-persistent infections and the host-virus interactions at the cellular level. The lack of sufficient PKR activity to execute apoptosis in U9K-A cells (6) probably provided some undefined initial intracellular environment conducive to the development and selection of less cytolytic variants and defective DI particles, which then would compete with wild-type EMCV virions for the cellular machinery to replicate. This view is supported by the establishment of persistent EMCV infection in the human erythroleukemic K562 cells, which do not support EMCV growth and hence present favorable conditions for establishing persistent EMCV infection (21). In addition, the likely presence of DI particles in U9K-AV2 cells (Fig. 6B) is consistent with the idea of variant selection and propagation, leading eventually to persistent infection. On the other hand, the reduced capacity to synthesize IFN-α in U9K-A cells (5) also may be a contributing, albeit not necessarily the defining, factor in developing persistent EMCV infection. U9K-A cells still can synthesize other IFNs (5), which may explain why there were no viral titer surges in U9K-A cells as compared with U9K-C control cells (Fig. 2). Besides, there are residual PKR activities in U9K-A cells (6) that would allow some IFN-α synthesis (see below) as well as the delayed EMCV-induced apoptosis in newly infected U9K-A cells. In any event, these considerations further emphasize the importance of the apoptogenic function of PKR in establishing persistent infection in U9K-A cells as discussed above.
As with many viral persistent infections, changes in both host cells and virus often accompany and stabilize the development of persistent infection. This host-virus coevolution was apparent in U9K-AV2 cultures; drastic changes in both U9K-AV2 cells and EMCV were observed (see Results). In particular, EMCV in U9K-AV2 cells was substantially attenuated. These observations strengthen the suitability of U9K-A cells as a model for elucidating the different stages of persistent viral infections. More in-depth analyses of U9K-AV2 cells, as well as the genomes of EMCV from U9K-AV2 cells that have been in culture for different times, should further delineate the interactions and changes in both virus and host cells that are necessary for the development and maintenance of EMCV-persistent infection in U937 cells. In any event, data described in this paper would support a model in which the EMCV present in U9K-AV2 cultures was attenuated to such an extent that it could no longer efficiently and overwhelmingly infect and lyse the still noninfected U9K-A subpopulation, which also had altered physiologically in terms of growth rates and resistance to EMCV infection. The dynamic balance of these two subpopulations of infected and noninfected cells allows the continuance of the persistence nature of the U9K-AV2 culture as a whole.
Even though no attempts were made to isolate cultures persistently infected with Sendai virus and reovirus in this study, the initial data of U9K-A cells being similarly resistant to infection by these viruses suggested the applicability of our model to study persistent infection by other viruses as well. A case in point is the ability of HIV-1 to persistently infect host cells (22). It is interesting to note that the tat protein of HIV-1 can be both a substrate and an inhibitor of PKR (23). It would be informative to study a possible establishment of persistent HIV-1 infection in the promonocytic U9K-A cells possessing only residual PKR activities. Finally, Vero cells deficient in PKR also exhibited resistance to EMCV-induced apoptosis (unpublished data), lending further support to the role of PKR in EMCV-induced apoptosis in at least one other cell type as well.
Acknowledgments
We thank Dr. Jay Tureen for critical comments on the manuscript. We also thank Amy Narvaez and Ron Gascon for their assistance with flow cytometry. This work was supported in part by a grant from the Rhone-Poulenc Group-Pasteur Merieux Connaught, France, and from the Department of Pediatrics, University of California, San Francisco.
Abbreviations
- DI
defective interfering
- EMCV
encephalomyocarditis virus
- ICE
IL-1β converting enzyme
- p.i.
postinfection
- PKR
double-stranded RNA-dependent protein kinase
- U9K-A
U937 derivative deficient in PKR
- U9K-C
control U937 derivative
- RT-PCR
reverse transcription–PCR
- TCID50
50% tissue culture infective dose
Footnotes
See commentary on page 11693.
References
- 1.Evan G, Littlewood T. Science. 1998;281:1317–1322. doi: 10.1126/science.281.5381.1317. [DOI] [PubMed] [Google Scholar]
- 2.Ashkenazi A, Dixit V M. Science. 1998;281:1305–1308. doi: 10.1126/science.281.5381.1305. [DOI] [PubMed] [Google Scholar]
- 3.Hershey J W. Annu Rev Biochem. 1991;60:717–755. doi: 10.1146/annurev.bi.60.070191.003441. [DOI] [PubMed] [Google Scholar]
- 4.Kumar A, Haque J, Lacoste J, Hiscott J, Williams B R. Proc Natl Acad Sci USA. 1994;91:6288–6292. doi: 10.1073/pnas.91.14.6288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Der S D, Lau A S. Proc Natl Acad Sci USA. 1995;92:8841–8845. doi: 10.1073/pnas.92.19.8841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yeung M C, Liu J, Lau A S. Proc Natl Acad Sci USA. 1996;93:12451–12455. doi: 10.1073/pnas.93.22.12451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Der S D, Yang Y L, Weissmann C, Williams B R. Proc Natl Acad Sci USA. 1997;94:3279–3283. doi: 10.1073/pnas.94.7.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Razvi E S, Welsh R M. Adv Virus Res. 1995;45:1–60. doi: 10.1016/s0065-3527(08)60057-3. [DOI] [PubMed] [Google Scholar]
- 9.Schwarz E M, Badorff C, Hiura T S, Wessely R, Badorff A, Verma I M, Knowlton K U. J Virol. 1998;72:5654–5660. doi: 10.1128/jvi.72.7.5654-5660.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Herrmann M, Lorenz H M, Voll R, Grunke M, Woith W, Kalden J R. Nucleic Acids Res. 1994;22:5506–5507. doi: 10.1093/nar/22.24.5506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Southern E M. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 12.Church G M, Gilbert W. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gooding L R. Cell. 1992;71:5–7. doi: 10.1016/0092-8674(92)90259-f. [DOI] [PubMed] [Google Scholar]
- 14.Yeung M C, Lau A S. J Biol Chem. 1998;273:25198–25202. doi: 10.1074/jbc.273.39.25198. [DOI] [PubMed] [Google Scholar]
- 15.Rueckert R R. In: Virology. Fields B N, Knipe D M, Howley P M, editors. Philadelphia: Lippincott-Raven; 1996. pp. 609–654. [Google Scholar]
- 16.Magnus P V. Arch Gesamte Virusforsch. 1965;17:414–423. doi: 10.1007/BF01241196. [DOI] [PubMed] [Google Scholar]
- 17.Ahmed R, Canning W M, Kauffman R S, Sharpe A H, Hallum J V, Fields B N. Cell. 1981;25:325–332. doi: 10.1016/0092-8674(81)90050-7. [DOI] [PubMed] [Google Scholar]
- 18.Ahmed R, Morrison L A, Knipe D M. In: Virology. Fields B N, Knipe P M, Howley P M, editors. Philadelphia: Lippincott-Raven; 1996. pp. 219–249. [Google Scholar]
- 19.Duvall E, Wyllie A H, Morris R G. Immunology. 1985;56:351–358. [PMC free article] [PubMed] [Google Scholar]
- 20.Chiou S K, White E. Virology. 1998;244:108–118. doi: 10.1006/viro.1998.9077. [DOI] [PubMed] [Google Scholar]
- 21.Pardoe I U, Grewal K K, Baldeh M P, Hamid J, Burness A T. J Virol. 1990;64:6040–6044. doi: 10.1128/jvi.64.12.6040-6044.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Antoni B A, Sabbatini P, Rabson A B, White E. J Virol. 1995;69:2384–2392. doi: 10.1128/jvi.69.4.2384-2392.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brand S R, Kobayashi R, Mathews M B. J Biol Chem. 1997;272:8388–8395. doi: 10.1074/jbc.272.13.8388. [DOI] [PubMed] [Google Scholar]