Abstract
Mutations in the homologous presenilin 1 (PS1) and presenilin 2 (PS2) genes cause the most common and aggressive form of familial Alzheimer’s disease. Although PS1 function and dysfunction have been extensively studied, little is known about the function of PS2 in vivo. To delineate the relationships of PS2 and PS1 activities and whether PS2 mutations involve gain or loss of function, we generated PS2 homozygous deficient (−/−) and PS1/PS2 double homozygous deficient mice. In contrast to PS1−/− mice, PS2−/− mice are viable and fertile and develop only mild pulmonary fibrosis and hemorrhage with age. Absence of PS2 does not detectably alter processing of amyloid precursor protein and has little or no effect on physiologically important apoptotic processes, indicating that Alzheimer’s disease-causing mutations in PS2, as in PS1, result in gain of function. Although PS1+/− PS2 −/− mice survive in relatively good health, complete deletion of both PS2 and PS1 genes causes a phenotype closely resembling full Notch-1 deficiency. These results demonstrate in vivo that PS1 and PS2 have partially overlapping functions and that PS1 is essential and PS2 is redundant for normal Notch signaling during mammalian embryological development.
The presenilins (PS) are polytransmembrane proteins located in the endoplasmic reticulum and the early Golgi apparatus (1–3). Missense mutations cause familial Alzheimer’s disease (AD) in a dominant fashion (4–6). Although the exact pathogenetic mechanism underlying the disease process remains to be further elucidated, it is fairly established that most PS missense mutations affect the processing of the amyloid precursor protein (APP), resulting in an increased generation of the longer form of the amyloid peptide (7–10). This peptide constitutes the major component of the amyloid plaques in patients. Interestingly, several of the PS mutations appear also to enhance the sensitivity of cells, and in particular neurons, to apoptotic stimuli (11–15). In principle, both mechanisms could contribute to the pathogenesis of AD. Presenilin homologues in Drosophila and Caenorhabditis elegans are involved in the Notch signaling pathway, which controls cell fate decisions during embryogenesis (16–19). In accordance with an important function also in mammalian embryogenesis, inactivation of the PS1 gene in mice results in a severe phenotype (20–23), characterized by late embryonic lethality, disturbed somitogenesis, cranial hemorrhage, underdevelopment of the subventricular zone of the brain, midline closure deficiencies, and a neuronal migration disorder very similar to human lissencephaly type II (22). It has been suggested that many of these lesions, and in particular the disturbed somitogenesis, were a consequence of disturbed Notch signaling in mice as well (20–22). This hypothesis was recently corroborated by the observation that PS1 is needed for the proteolytic processing of the transmembrane domain of Notch. (18, 19, 24). This proteolytic event is an essential step in the Notch signaling cascade and results finally in the nuclear translocation of the cleaved Notch intracellular domain together with members of the CSL family of DNA binding proteins (25). It should, however, be noticed that the phenotype of the PS1-deficient mice differs in several important aspects from the previously described Notch-1 deficient phenotype. For instance, PS1−/− embryos suffer from severe brain hemorrhage and are viable until birth whereas Notch1 deficient animals do not even survive embryonic day 11 (E11) (26, 27). PS1 deficiency affects not only Notch proteolytic processing but also interferes with the γ-secretase-mediated proteolysis of the transmembrane domain of APP (23) and amyloid precursor-like protein 1 (APLP-1) (28). Recently, it has been suggested that PS1 itself is an aspartyl-type proteinase, mediating γ-secretase cleavage of APP. This conclusion was mainly based on the observation that site-directed mutagenesis of either one of the two aspartyl residues in transmembrane domain 6 or 7 of PS1 results in a dominant negative effect on γ-secretase processing of APP (29).
Although PS1 and PS2 share overall 63% amino acid residues at identical positions and have a very similar structure (4, 6), it is not established whether they are functional homologues. The fact that the endogenous PS2 in the PS1-deficient animals does not rescue its lethal phenotype suggests that its activity does not completely overlap with that of PS1 (20–23). On the other hand, we have speculated that the residual amyloid peptide secretion in PS1-deficient neurons could be a consequence of the remaining PS2 in those cells (23). These questions can obviously only be tackled by generating PS2- and PS1/PS2-deficient mice. Those animals could then also be used to investigate the role of PS2 in apoptotic processes in vivo, as mentioned above.
In the current manuscript, we describe the generation of PS2-deficient mice and report on the effects on APP processing and apoptotic processes. Evidence that PS2 and PS1 are functional homologues and both involved in the Notch signaling pathway was obtained in double-deficient mice. These double homozygous animals display a severe embryological lethal phenotype that is similar, if not identical, to that observed in Notch-1-deficient animals (26, 27).
Materials and Methods
Generation of PS2-Deficient Mice.
The mouse PS2 cDNA was kindly provided by L. D’Adamio (National Institutes of Health) (13). An EMBL3-129 SV Mouse genomic phage library (Stratagene) was screened, and one of the obtained phages (phage 7) containing a 15-kilobase (kb) insert including exon 4 was used to generate the targeting construct. In the NotI-HindIII (12.5 kb) fragment, a 2.2-kb NcoI fragment encompassing exon 5 and flanking sequences was replaced by a hygromycin cassette with the PGK promoter (Fig. 1) (30). The plasmid was linearized (HindIII) and electroporated into the embryonic stem cell (ES cell) line E14-1 (30, 31). An external 3′ probe was obtained by HindIII and BglII restriction of phage 7. An external 5′probe was produced by PCR using primers Ne063 (5′-ATG CTC GCA TTC ATG GCC-3′) and Ne064 (5′-CTG GGC AGT GCT CTC TCC-3′). Hygromycin resistant ES colonies were selected after Southern blot analysis of DNA digested with XbaI/SpeI or EcoRV and hybridized with, respectively, the 3′- and the 5′-probe (Fig. 1). Eight ES cell clones displayed the correct 10.5- and 7.8-kb XbaI/SpeI restriction fragments and the 12.5- and 7.5-kb EcoRV restriction fragments. The replacement of exon 5 by the hygromycin cassette results in a frame shift between exons 4 and 6. Mice were genotyped for the PS2 gene mutation by Southern blot analysis or by a hygromycin-specific PCR using primers Ne219 (5′-CAG CGA GAG CCT GAC CTA TTG C-3′) and Ne220 (5′-CGA TTC TGC AAG CTC CGG ATG-3′) and an exon 5-specific PCR using primers Ne257 (5′-AGG TCT GTT TAA CTG GTG A-3′) and Ne204 (5′-CCA GCG GCT CCT CAA CTC-3′). The generation of PS1+/− mice has been described previously (23). All mice used have a C57B6/J black × 129Sv genetic background.
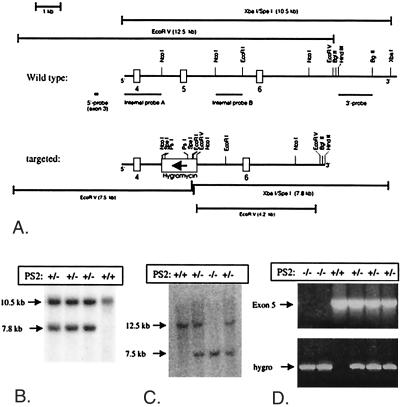
Figure 1.
Generation of PS2-deficient (PS2−/−) mice. (A) Targeting strategy to replace coding exon 5 and flanking sequences of the mouse PS2 gene by a hygromycin cassette. Boxes represent PS2 exons 4, 5, and 6 in the wild-type PS2 gene (upper panel). The lower panel displays the gene locus after targeting. The direction of transcription of the inserted hygromycin cassette is opposite to PS2 transcription (arrow). The predicted lengths of the restriction fragments obtained from the wild-type allele (top) or the targeted allele (bottom) after digestion with EcoRV or combined XbaI and SpeI are shown. (B) Genomic DNA from different ES clones was digested with XbaI and SpeI and was analyzed by Southern blotting using the external 3′probe. (C) Tail genomic DNA from littermate embryos obtained after heterozygote crossings, digested with EcoRV and analyzed by Southern blotting using the external 5′ probe. (D) PCR diagnosis of the PS2 deficient gene in tail genomic DNA.
Northern Blotting.
Total RNA was isolated from tissues with Trizol (GIBCO/BRL). Poly-adenylated RNA was purified from 100 μg of total RNA by using Dynabeads oligo(dT) (Dynal, Oslo) and was separated in formaldehyde agarose gel electrophoresis (32). Filters were hybridized by using the EcoRI-digested PS2cDNA insert as described (32).
Western Blotting.
Rabbit polyclonal antibodies B24.2 to the C-terminal and B22.4 to the N-terminal fragments of PS2 were generated by using the peptides KLDPSSQGALQLPYDPEMEE (amino acids 306–325) or CESTAQWRTQESEED coupled to keyhole limpet hemocyanin. Membrane fractions were prepared from brain or other tissues after homogenization in 0.25 M sucrose, 1 mM EGTA, and 5 mM Tris⋅HCl (pH 7.4) by using a Potter-Elvehjem homogenizer and a low spin centrifugation (450 × g for 10 min). The resulting postnuclear supernatant was ultracentrifuged (136,000 × gmax, 1 hr), yielding a crude membrane pellet. Samples were resuspended in Laemmli buffer, were electrophoresed on 4–20% precast gels (NOVEX, San Diego), and were blotted. Antibodies B22.4 and B24.2 were used at a dilution of 1/10,000 in blocking buffer (blotto: TBS containing 0.1% Tween-20 and 5% nonfat dry milk). PS1 antibodies B17.2 and B19.2 detecting, respectively, the C-terminal and the N-terminal fragment of PS1 have been described previously (2). For detection HRP-coupled secondary antibodies were used, followed by ECL (Amersham Pharmacia).
Histology.
Animals were perfused via the left ventricle with Bouin’s solution diluted 1:4 in PBS. After dissection, individual organs were embedded in low melting point paraffin (Vogel, Giessen, Germany). Serial sections (7 μm) were cut and mounted on glass slides covered with biobond (British Biocell, London). Central sections of each series were stained with hematoxylin and eosin for standard light microscopy. Adjacent sections were used for immunohistological screening by using antibodies to glial fibrillary acid protein (astroglia), F4/80 (microglia/macrophages), CD45 and CD45R (lymphocytes), vimentin (fibroblastic cells), and lectins Solanum tuberosum (endothelia) and RCA-1 (endothelia and activated macrophages). Biotin-labeled secondary antibodies and avidin-biotin or tyramide signal amplification were used. Apoptotic cells in situ were detected by demonstrating tissue type transglutaminase (33) or thermal instability of DNA by using a single strand DNA antibody (34). Tissues were fixed in 4% paraformaldehyde or 80% methanol, respectively, and were embedded in paraffin.
Apoptosis Assay.
Single cells suspensions from spleen and thymus were reacted with monoclonal antibodies to CD45 (clone 30-F12), CD3 (clone 17A2), CD4 (clone GK1.5), and CD8 (clone 53–6.7) and appropriate isotype controls (PharMingen), used at a concentration of 1 μg per 106 cells. The cells were analyzed on a FACScan flowcytometer (Becton Dickinson). The lymphocyte fraction was gated on the basis of forward and side scatter. The rate of apoptotic cells in DNA histograms was determined as described (35).
Results and Discussion
To address the unresolved question of the normal and pathological functions of PS2 in vivo, we generated knock-out mice, in which the PS2 gene was inactivated by homologous recombination (Fig. 1). Exon 5 of the mouse PS2 gene in E14–1 ES cells was replaced by a hygromycin cassette under the control of the PGK promotor (Fig. 1A). Eight of one-hundred and seventy hygromycin-resistant colonies displayed the correct restriction fragments with the external 3′ and 5′ probes in Southern blotting (Fig. 1) (30, 31). Two independently targeted ES clones then were microinjected into C57BL/6J blastocysts, and chimeric males and heterozygous offspring were generated. Genotyping of 131 offspring from heterozygote crosses (Fig. 1) revealed a frequency of 27% for homozygous PS2−/− mice, in agreement with the expected Mendelian frequency (25%). Hence, disruption of the PS2 gene does not result in embryonic lethality, in striking contrast to deleting PS1 (20–22).
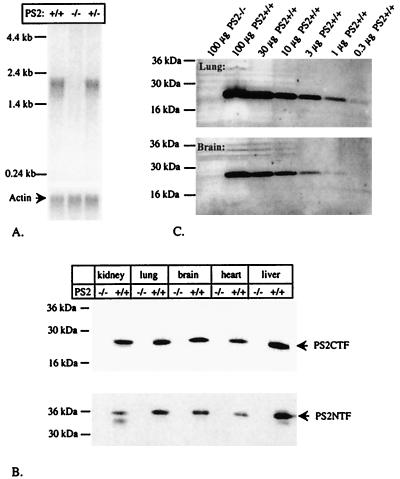
Northern blots on poly-A enriched RNA from brain (Fig. 2A) and Western blots of different tissues (Fig. 2B) using antibodies specific for the N-terminal (PS2NTF) or the C-terminal fragments (PS2CTF) of PS2 confirmed the null mutation of the PS2 gene. Further semiquantitative analyses (Fig. 2C) indicated that PS2CTF fragments would have been detected in as little as 1 μg of brain and 0.3 μg of lung membrane extracts with the approach used.
Figure 2.
Complete absence of PS2 in homozygote PS2−/− mice. (A) Northern blot analysis of PS2 expression. Polyadenylated RNA was isolated from brain from littermate E14 embryos and was hybridized with the EcoRI-digested PS2 cDNA probe. An actin probe was used as positive control. (B) Western blot analysis of PS2 synthesis. Membrane-enriched fractions were prepared from kidney, lung, brain, heart, and liver of 1-week-old animals, and 50 μg of protein was applied on 4–20% SDS/PAGE. Ponceau S staining of the blot confirmed that equal amounts of protein were applied in every lane. The blot was developed by using amino-terminal (NTF) and carboxy-terminal (CTF) specific PS2 antisera. (C) Semiquantitative Western blot analysis of PS2 expression in PS2−/− animals aged 3 months. Membrane material was probed with PS2CTF antiserum. No signal is observed when 100 μg of membrane-enriched material from lung or from brain of PS2−/− animals is applied whereas a signal remains visible in the lanes containing 0.3 μg (lung) or 1 μg (brain) of material derived from wild-type littermates.
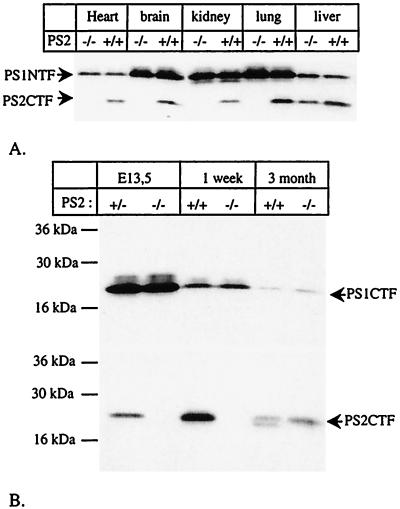
While this work was in progress, Vito et al. reported that a second, shorter transcript is generated from the PS2 gene in adult liver, coding for ALG-3 (36). We confirm by reverse transcription–PCR that, in liver of wild-type and adult PS2−/− mice, such an alternative transcript exists that contains part of the intron sequence between exons 8 and 9 (13) (results not shown). This second message is apparently transcribed from a second, hitherto uncharacterized promoter and translated into ALG-3 starting from methionine 298 or 304 of the PS2 sequence (13, 36). Our carboxyterminal-specific antibody recognizes an epitope carboxyterminal to these methionines and therefore reveals the expression of ALG-3 in the PS2−/− mice. From Fig. 2, it is clear that ALG-3 is not expressed in 1-week-old PS2−/− animals or lung and brain tissue of 3-month-old PS2−/− animals. Further analyses demonstrated that ALG-3 is expressed exclusively in the liver after 3 months (Fig. 3) and therefore does not contribute to the lung phenotype of the PS2−/− animals (see below). Therefore, and because ALG-3 is a dominant negative form of PS2 in apoptosis (13, 36, 37), a complex double-targeting strategy to inactivate the ALG-3 gene in addition to the PS2 gene was not further considered in this study.
Figure 3.
Expression of Alg-3 in adult liver. (A) Western blot analysis of material of 3-month-old animals. The blot was stained first with PS2CTF antiserum and afterward with anti-PS1NTF. Notice that PS2CTF are expressed only in liver in the PS2−/− mice. PS1NTF are present at equal levels in PS2 wild-type and knock-out animals. (B) Membrane-enriched material from liver was prepared from E13.5 embryos and 1-week- and 3-month-old animals and was probed with antibodies specific for the carboxyterminus of PS1 (PS1CTF) or PS2 (PS2CTF). Notice that only in 3-month-old PS2−/− animals are PS2CTF fragments expressed.
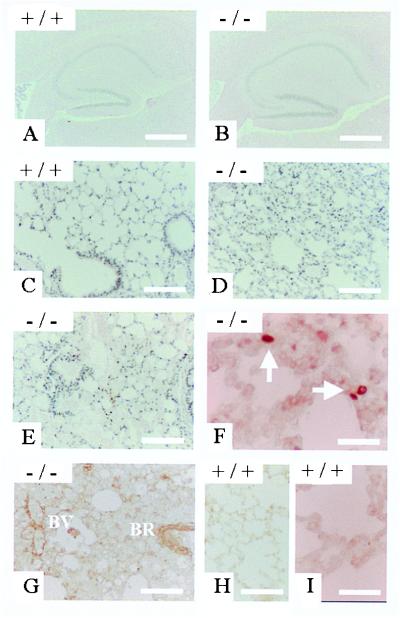
PS2 homozygous mutant, heterozygous, and wild-type mice resulting from heterozygote crosses did not exhibit obvious differences in growth, weight, or health up to age 12 months. PS2-deficient mice were fertile, allowing breeding with PS1+/− animals (see below). Various blood and serum parameters revealed no abnormalities in the PS2−/− mice (not shown). Detailed microscopic examination of brain (shown for the hippocampus as a key target region of Alzheimer’s disease in Fig. 4 A and B), liver, spleen, heart, and skeletal muscle did not reveal any abnormalities in tissue structure or indirect evidence of lesions, such as macrophage activation in peripheral tissues or astrogliosis in the brain. Specifically, none of the salient features of PS1-deficient mice, such as disturbed somitogenesis or neuronal migration disorders, were present in PS2-deficient mice.
Figure 4.
Phenotypical analysis of PS2 knock-out mice. (A and B) Hippocampal formation of control (A) and PS2-deficient (B) mice at age 6 months as seen in routine histological preparation. No differences in tissue architecture can be demonstrated. (C and D) Lung parenchyma of control (C) and PS2-deficient (D) mice at age 3 months. Note the increase in cell mass and the reduction of alveolar space as compared with the control. (E and F) Lung of a PS2-deficient mouse at 6 months exhibiting profuse hemorrhages. Both alveolar spaces and lower conductive airways are virtually filled with extravasated erythrocytes. (F and G) Lung specimens of 6-month-old PS2−/− mice stained for single-stranded DNA after thermal denaturation (F) and tissue transglutaminase type II (G). Both markers for cells undergoing apoptosis label scattered cells within the lung parenchyma as well as in the corresponding blood vessels (BV) and bronchioli (BR). (H and I) Similar experiments carried out on wild-type mice of the same age did not result in any staining either for tissue type transglutaminase (H) or single-stranded DNA (I).
From age 3 months on, however, PS2−/− animals displayed a considerable thickening of alveolar walls, with broad strands of fibrotic tissues sometimes compromising airway patency (Fig. 4 C and D). In animals of 3 and 6 months, we observed hemorrhages within large groups of alveoli and their adjoining airways, suggesting vascular alterations within the lung parenchyma (Fig. 4E). Surprisingly, the transgenic animals suffered very little from these fibrotic alterations, probably because the large physiological reserves in lung capacity suffice to perform all functions needed to survive in the relative quiet environment of an animal facility. Immunohistochemical screening for macrophages (F4/80) and lymphocytes (CD45, CD45R) ruled out the possibility of intercurrent (chronic) infections as a possible nonspecific cause for the observed pulmonal alterations. In situ experiments using antibodies to single-stranded DNA (Fig. 4F) or tissue type transglutaminase (Fig. 4G) revealed cells undergoing programmed cell death scattered throughout the parenchyma and within the bronchial epithelium and also affecting vascular endothelium. For both techniques, control experiments were conducted on PS2+/+ material to rule out any contribution of the physiological apoptotic cell death occurring in postnatal lung development (Fig. 4 H and I).
Because PS2 has been implicated in apoptosis in vitro (11–15), we were surprised to observe no alterations in brain anatomy or digit formation in the PS2−/− mice. Moreover, no deviations in the number of lymphocytes in spleen, thymus, and blood or in the number and distribution of lymphocyte subpopulations defined by the cell surface markers CD45, CD3, CD4, and CD8 were found by flow cytometry (data not shown). The sub-G1 peaks, which correlate with DNA fragmentation during apoptosis (35), were determined in DNA histograms of isolated PS2-deficient and control thymocytes and splenocytes after incorporation of propidium iodide. No differences were found, indicating similar rates of apoptosis in cells derived from PS2-deficient or wild-type mice (not shown). Apparently, PS2 plays only a limited role in the control of apoptotic processes in vivo as detected solely in the lungs. Our in vivo data imply that PS2 is not directly part of the apoptotic execution pathway but may subsequently have an indirect modulating role, i.e., by changing the sensitivity of cells to subsequent apoptotic stimuli.
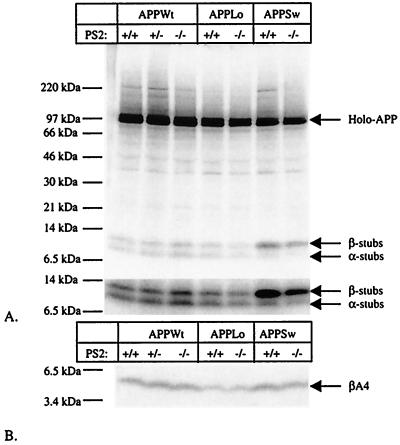
It has been shown that proteolytic processing of APP in neurons derived from PS1−/− animals is markedly inhibited at the level of γ-secretase, albeit not completely (23). It was speculated that the residual secretion of Aβ in these cultures was caused by the presence of PS2 (23). Neurons derived from E14 embryos expressed indeed PS2 whereas their counterparts derived from PS2−/− did not (data not shown). Neurons were cultured and infected with recombinant Semliki Forest Virus coding for human APP, as before (23, 38), and were metabolically labeled with [35S]methionine. Various proteolytic derivatives of APP were immunoprecipitated from cell extracts and conditioned media using a panel of well characterized antibodies (23, 38) and then were analyzed by SDS/PAGE (Fig. 5). Levels of holo-APP, APP carboxyterminal fragments (generated by α- and β-secretase), secreted APPs (generated mainly by α-secretase), and amyloid β-peptide (generated by combined β- and γ-secretase activities) were closely similar in PS2−/− neurons and PS2+/+ neurons in six independent experiments. ELISA experiments (23) to assess amounts of Aβ1–40 (139 ± 22 pg/ml and 166 ± 22 pg/ml in, respectively, PS2+/+ and PS2−/− neurons, n = 8) or Aβ1–42 (49 ± 9 pg/ml and 50 ± 8 pg/ml in, respectively, PS2+/+ and PS2−/− neurons, n = 9) in the medium confirmed that the absence of PS2 does not significantly alter APP metabolism. This result implies that PS2 mutations causing AD must act via a gain-of-function mechanism.
Figure 5.
APP processing in neurons derived from E14 PS2−/− embryos. (A) Analysis of APP expression and generation of α- and β-secretase-generated APP carboxyterminal stubs (23, 38). Neurons were infected with recombinant Semliki Forest Virus driving expression of human wild-type APP (APPWt) or APP containing the London (APPLo) or the Swedish (APPSw) type of AD-causing mutations. After metabolic labeling, antibody B12/4 recognizing the 20 carboxyterminal amino acid residues of APP was used to immunoprecipitate holo-APP and α- and β-secretase-generated carboxyterminal stubs from the cell extracts. The material was analyzed on a Tris⋅tricine 10–20% gel. The lower part of the figure shows a stronger exposure of the bottom part of the gel to better reveal the APP carboxyterminal fragments. (B) Analysis of Aβ secretion into the medium (23, 38). The conditioned medium of the same neurons examined in A was immunoprecipitated with the Aβ antibody B7/6. This antibody recognizes an epitope between the α- and the β-secretase sites and therefore does not precipitate the p3 fragment. Material was analyzed on a Tris⋅tricine 10–20% gel.
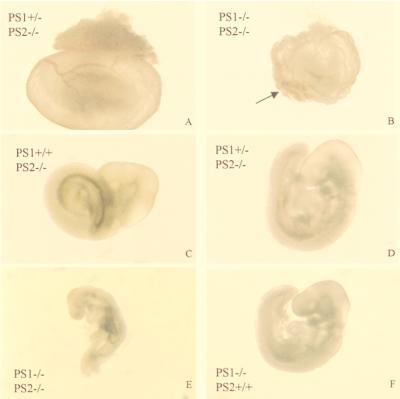
Given the strikingly different phenotype of PS2−/− mice versus PS1−/− mice (20–23), the question arose as to whether PS2 is actually a functional homologue of PS1. We therefore crossed PS2−/− mice with PS1+/− mice to obtain PS1+/−PS2+/− double heterozygous mice (Table 1, first row). Double heterozygotes then were crossed with PS2−/− homozygous mice to obtain PS1+/−PS2−/− mice. Even with only one active PS1 gene left, these mice remain completely viable and fertile (Table 1, third row). We failed to detect liveborn double homozygous offspring from these heterozygous intercrosses. At E9.5, we could recover homozygous PS1−/−PS2−/− embryos in a nearly Mendelian distribution (15/66), but embryos were developmentally retarded by approximately half a day when compared with heterozygous littermates (Fig. 6). Vasculogenesis of the yolk sac was delayed in most of the mutants (Fig. 6B). Although an initial vascular plexus and primitive red blood cells had formed, organization into a discrete network of vitelline vessels was always lacking. Furthermore, yolk sacs did not expand properly and often had a blistered appearance. A severely impaired yolk sac is shown in Fig. 6B. The embryo itself was always devoid of blood circulation and appeared posteriorly truncated. Heart development was largely unaffected, with the exception of an occasional enlarged pericardial sac (not shown). Somitogenesis had begun, and turning occurred in the majority of the mutants (Fig. 6E). The optic and otic vesicle, the first branchial arch, and the forelimb buds were visible. Mutants had a vestigial fore- and hindbrain, and fusion of headfolds was delayed. The neural tube often had a kinked appearance, which may be secondary to the circulation problems. This phenotype of the double-deficient embryos is clearly different from that of PS2−/− embryos, which appear normal (Fig. 6C), and PS1−/− embryos, which are only marginally retarded at E9.5 (Fig. 6F) (20, 21, 22).
Table 1.
Genotyping of offspring of various crossings at 3 weeks of age
| Crossings
|
Pups expected
|
Pups observed, no. | |||||
|---|---|---|---|---|---|---|---|
| PS1 | PS2 | × | PS1 PS2 | PS1 | PS2 | ||
| +/+ | −/− | +/− +/+ | +/− | +/− | (50%) | 28 (42%) | |
| +/+ | +/− | (50%) | 39 (58%) | ||||
| 67 (100%) | |||||||
| +/− | +/− | +/+ −/− | +/+ | +/− | (25%) | 40 (30%) | |
| +/+ | −/− | (25%) | 30 (22%) | ||||
| +/− | −/− | (25%) | 26 (20%) | ||||
| +/− | +/− | (25%) | 39 (29%) | ||||
| 135 (100%) | |||||||
| +/− | −/− | +/− −/− | +/+ | −/− | (25%) | 12 (32%) | |
| +/− | −/− | (50%) | 26 (68%) | ||||
| −/− | −/− | (25%) | 0 (0%) | ||||
| 38 (100%) | |||||||
Figure 6.
PS1−/−PS2−/− embryos at E9.5 display severe growth retardation. The two upper panels display the yolk sac of E9.5 PS1+/−PS2−/− (A) and PS1−/−PS2−/− (B) littermates (identical magnification). The arrowhead points to the vascular plexus. The four lower panels display E9.5 embryos (identical magnification). See text for description of the phenotype.
Models have been proposed (18, 23, 24, 29) in which PS1 mediates the proteolytic cleavage of APP and Notch in their respective transmembrane domains. These models suggest that inhibition of PS1 function could be a potential therapeutic target for AD (23, 24, 29). On the other hand, concerns have been raised with respect to the potential severe side effects of this approach (24) as a consequence of the inhibition of the Notch signaling pathway, which controls cell fate and affects differentiation and proliferation during development. Although the phenotype of PS1−/− mice only marginally mimicked the phenotype of Notch1-deficient mice, the similarity of PS1−/−PS2−/− mice (above) with Notch-1-null mutants is striking. In the latter, an analogous deficit in posterior development as found for the PS1−/−PS2−/− occurs at E9.0 (26, 27), and the early lethal phenotype of our double homozygous null embryos is reminiscent of what has been observed in the Notch-1 knock-out mice (26, 27) and contrasts with the PS1−/− single-deficient mice, which can survive until birth. Our results therefore further establish the absolute requirement for presenilins in Notch signaling (18, 19, 24) and provide functional evidence in vivo for the hypothesis that PS2 has a similar molecular function as PS1. Interestingly, the aspartates in the transmembrane domain 6 and 7 of PS1 that are critically involved in γ-secretase activity (29) are conserved in PS2. Recent data indicate that mutagenesis of these aspartate residues in mammalian PS2 also results in a dominant negative effect on APP processing and Notch processing (39). Although PS1 can apparently completely compensate for the putative role(s) of PS2 during development, the reverse is not true. This may be explained by the fact that PS2 is less abundantly expressed than PS1 during embryological development (ref. 40 and our unpublished results).
Enhancement of PS function as a consequence of AD-causing missense mutations results in increased release of the Aβ1–42-peptide (7–10). Because enhanced production of this peptide is believed to be the central problem in sporadic as well as familial AD (41), we suggest that therapeutic strategies aimed at lowering PS1 and -2 activity partially should be considered for the treatment of sporadic AD. Given the need for PS in Notch signaling, for instance, in hematopoiesis (42), marked inhibition of PS activity as a therapy for AD would likely be toxic. On the other hand, PS1+/−PS2−/− mice appear to maintain a remarkably healthy condition. It remains to be investigated to what degree γ-secretase activity toward APP is down-regulated in these mice. Our in vivo results help define the biological limits of such a therapeutic strategy and constitute in combination with previous work elucidating the molecular function of PS1 (26, 28, 30, 34) a basis for considering the PS as the currently best defined molecular target for treating and preventing AD.
Acknowledgments
The BARN (Bayer Alzheimer Research Network), the Fonds voor Wetenschappelijk Onderzoek (FWO-Vlaanderen), the Human Frontier of Science Program (HFSP), the Flanders Interuniversity Institute for Biotechnology (VIB), the Interuniversitaire Attractiepolen (IUAP), International Alzheimer’s Research Foundation (IARF) Belgium, and the K.U.Leuven supported this work. B.D.S., W.A., P.C., and K.C. are researchers of the FWO. P.S. and D.H. are supported by the Deutsche Forschungsgemeinschaft (DFG). R.D. was supported by the Lower Saxony/Israel joint project 210.3-70631/9-99-19/96. We would like to thank Dr. L. D’Adamio for the mouse PS2 cDNA and Dr. E. Günther and Dr. D. Selkoe for helpful discussion.
Abbreviations
- PS1
presenilin 1
- AD
Alzheimer’s disease
- APP
amyloid precursor protein
- kb
kilobase
- ES cell
embryonic stem cell
- E11
embryonic day 11
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Doan A, Thinakaran G, Borchelt D R, Slunt H H, Ratovitsky T, Podlisny M, Selkoe D J, Seeger M, Gandy S E, Price D L, Sisodia S S. Neuron. 1996;17:1023–1030. doi: 10.1016/s0896-6273(00)80232-9. [DOI] [PubMed] [Google Scholar]
- 2.De Strooper B, Beullens M, Contreras B, Levesque L, Craessaerts K, Cordell B, Moechars D, Bollen M, Fraser P, George-Hyslop P S, Van Leuven F. J Biol Chem. 1997;272:3590–3598. doi: 10.1074/jbc.272.6.3590. [DOI] [PubMed] [Google Scholar]
- 3.Walter J, Capell A, Grunberg J, Pesold B, Schindzielorz A, Prior R, Podlisny M B, Fraser P, Hyslop P S, Selkoe D J, Haass C. Mol Med. 1996;2:673–691. [PMC free article] [PubMed] [Google Scholar]
- 4.Rogaev E I, Sherrington R, Rogaeva E A, Levesque G, Ikeda M, Liang Y, Chi H, Lin C, Holman K, Tsuda T, et al. Nature (London) 1995;376:775–778. doi: 10.1038/376775a0. [DOI] [PubMed] [Google Scholar]
- 5.Sherrington R, Rogaev E I, Liang Y, Rogaeva E A, Levesque G, Ikeda M, Chi H, Lin C, Li G, Holman K, et al. Nature (London) 1995;375:754–760. doi: 10.1038/376775a0. [DOI] [PubMed] [Google Scholar]
- 6.Levy-Lahad E, Wasco W, Poorkaj P, Romano D M, Oshima J, Pettingell W H, Yu C E, Jondro P D, Schmidt S D, Wang K, et al. Science. 1995;269:973–977. doi: 10.1126/science.7638622. [DOI] [PubMed] [Google Scholar]
- 7.Xia W, Zhang J, Kholodenko D, Citron M, Podlisny M B, Teplow D B, Haass C, Seubert P, Koo E H, Selkoe D J. J Biol Chem. 1997;272:7977–7982. doi: 10.1074/jbc.272.12.7977. [DOI] [PubMed] [Google Scholar]
- 8.Tomita T, Maruyama K, Saido T C, Kume H, Shinozaki K, Tokuhiro S, Capell A, Walter J, Grunberg J, Haass C, et al. Proc Natl Acad Sci USA. 1997;94:2025–2030. doi: 10.1073/pnas.94.5.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duff K, Eckman C, Zehr C, Yu X, Prada C M, Perez-tur J, Hutton M, Buee L, Harigaya Y, Yager D, et al. Nature (London) 1996;383:710–713. doi: 10.1038/383710a0. [DOI] [PubMed] [Google Scholar]
- 10.Lemere C A, Lopera F, Kosik K S, Lendon C L, Ossa J, Saido T C, Yamaguchi H, Ruiz A, Martinez A, Madrigal L, et al. Nat Med. 1996;2:1146–1150. doi: 10.1038/nm1096-1146. [DOI] [PubMed] [Google Scholar]
- 11.Deng G, Pike C J, Cotman C W. FEBS Lett. 1996;397:50–54. doi: 10.1016/s0014-5793(96)01142-8. [DOI] [PubMed] [Google Scholar]
- 12.Wolozin B, Iwasaki K, Vito P, Ganjei J K, Lacana E, Sunderland T, Zhao B, Kusiak J W, Wasco W, D’Adamio L. Science. 1996;274:1710–1713. doi: 10.1126/science.274.5293.1710. [DOI] [PubMed] [Google Scholar]
- 13.Vito P, Ghayur T, L D A. J Biol Chem. 1997;272:28315–28320. doi: 10.1074/jbc.272.45.28315. [DOI] [PubMed] [Google Scholar]
- 14.Guo Q, Sopher B L, Furukawa K, Pham D G, Robinson N, Martin G M, Mattson M P. J Neurosci. 1997;17:4212–4222. doi: 10.1523/JNEUROSCI.17-11-04212.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Janicki S, Monteiro M J. J Cell Biol. 1997;139:485–495. doi: 10.1083/jcb.139.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levitan D, Greenwald I. Nature (London) 1995;377:351–354. doi: 10.1038/377351a0. [DOI] [PubMed] [Google Scholar]
- 17.Baumeister R, Leimer U, Zweckbronner I, Jakubek C, Grunberg J, Haass C. Genes Funct. 1997;1:149–159. doi: 10.1046/j.1365-4624.1997.00012.x. [DOI] [PubMed] [Google Scholar]
- 18.Struhl G, Greenwald I. Nature (London) 1999;398:522–525. doi: 10.1038/19091. [DOI] [PubMed] [Google Scholar]
- 19.Ye Y, Lulinova N, Fortini M E. Nature (London) 1999;398:525–529. doi: 10.1038/19096. [DOI] [PubMed] [Google Scholar]
- 20.Shen J, Bronson R T, Chen D F, Xia W, Selkoe D J, Tonegawa S. Cell. 1997;89:629–639. doi: 10.1016/s0092-8674(00)80244-5. [DOI] [PubMed] [Google Scholar]
- 21.Wong P C, Zheng H, Chen H, Becher M W, Sirinathsinghji D J, Trumbauer M E, Chen H Y, Price D L, Van der Ploeg L H, Sisodia S S. Nature (London) 1997;387:288–292. doi: 10.1038/387288a0. [DOI] [PubMed] [Google Scholar]
- 22.Hartmann D, Strooper B D, Saftig P. Curr Biol. 1999;9:719–727. doi: 10.1016/s0960-9822(99)80331-5. [DOI] [PubMed] [Google Scholar]
- 23.De Strooper B, Saftig P, Craessaerts K, Vanderstichele H, Guhde G, Annaert W, Von Figura K, Van Leuven F. Nature (London) 1998;391:387–390. doi: 10.1038/34910. [DOI] [PubMed] [Google Scholar]
- 24.De Strooper B, Annaert W, Cupers P, Saftig P, Craessaerts K, Mumm J S, Schroeter E H, Schrijvers V, Wolfe M S, Ray W J, et al. Nature (London) 1999;398:518–522. doi: 10.1038/19083. [DOI] [PubMed] [Google Scholar]
- 25.Schroeter E H, Kisslinger J A, Kopan R. Nature (London) 1998;393:382–386. doi: 10.1038/30756. [DOI] [PubMed] [Google Scholar]
- 26.Swiatek P J, Lindsell C E, del Amo F F, Weinmaster G, Gridley T. Genes Dev. 1994;8:707–719. doi: 10.1101/gad.8.6.707. [DOI] [PubMed] [Google Scholar]
- 27.Conlon R A, Reaume A G, Rossant J. Development (Cambridge, UK) 1995;121:1533–1545. doi: 10.1242/dev.121.5.1533. [DOI] [PubMed] [Google Scholar]
- 28.Naruse S, Thinakaran G, Luo J J, Kusiak J W, Tomita T, Iwatsubo T, Qian X, Ginty D D, Price D L, Borchelt D R, et al. Neuron. 1998;21:1213–1221. doi: 10.1016/s0896-6273(00)80637-6. [DOI] [PubMed] [Google Scholar]
- 29.Wolfe M S, Xia W, Ostaszewski B L, Diehl T S, Kimberly W T, Selkoe D J. Nature (London) 1999;398:513–517. doi: 10.1038/19077. [DOI] [PubMed] [Google Scholar]
- 30.Te Riele H, Robanus Maandag E, Clarke A, Hooper M, Berns A. Nature (London) 1990;348:649–651. doi: 10.1038/348649a0. [DOI] [PubMed] [Google Scholar]
- 31.Hooper M, Hardy K, Handyside A, Hunter S, Monk M. Nature (London) 1987;326:292–295. doi: 10.1038/326292a0. [DOI] [PubMed] [Google Scholar]
- 32.Lorent K, Overbergh L, Delabie J, Van Leuven F, Van den Berghe H. Differentiation (Berlin) 1994;55:213–223. doi: 10.1046/j.1432-0436.1994.5530213.x. [DOI] [PubMed] [Google Scholar]
- 33.Melino G, Piacentini M. FEBS Lett. 1998;430:59–63. doi: 10.1016/s0014-5793(98)00521-3. [DOI] [PubMed] [Google Scholar]
- 34.Frankfurt O S, Robb J A, Sugarbaker E V, Villa L. Exp Cell Res. 1996;226:387–397. doi: 10.1006/excr.1996.0240. [DOI] [PubMed] [Google Scholar]
- 35.Ormerod M G, Collins M K, Rodriguez-Tarduchy G, Robertson D. J Immunol Methods. 1992;153:57–65. doi: 10.1016/0022-1759(92)90305-d. [DOI] [PubMed] [Google Scholar]
- 36.Vito P, Lacana E, D’Adamio L. Science. 1996;271:521–525. doi: 10.1126/science.271.5248.521. [DOI] [PubMed] [Google Scholar]
- 37.Vito P, Wolozin B, Ganjei J K, Iwasaki K, Lacana E, D’Adamio L. J Biol Chem. 1996;271:31025–31028. doi: 10.1074/jbc.271.49.31025. [DOI] [PubMed] [Google Scholar]
- 38.De Strooper B, Simons M, Multhaup G, Van Leuven F, Beyreuther K, Dotti C G. EMBO J. 1995;14:4932–4938. doi: 10.1002/j.1460-2075.1995.tb00176.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Steiner, H., Duff, K., Capell, A., Romig, H., Grim, M., Lincoln, S., Hardy, J., Yu, X., Picciano, M., Fechteler, K., et al. (1999) J. Biol. Chem., in press. [DOI] [PubMed]
- 40.Lee M K, Slunt H H, Martin L J, Thinakaran G, Kim G, Gandy S E, Seeger M, Koo E, Price D L, Sisodia S S. J Neurosci. 1996;16:7513–7525. doi: 10.1523/JNEUROSCI.16-23-07513.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Selkoe D J. Science. 1997;275:630–631. doi: 10.1126/science.275.5300.630. [DOI] [PubMed] [Google Scholar]
- 42.Varnum-Finney B, Purton L E, Yu M, Brashem-Stein C, Flowers D, Staats S, Moore K A, Le Roux I, Mann R, Gray G, et al. Blood. 1998;91:4084–4091. [PubMed] [Google Scholar]