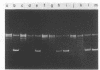
Abstract
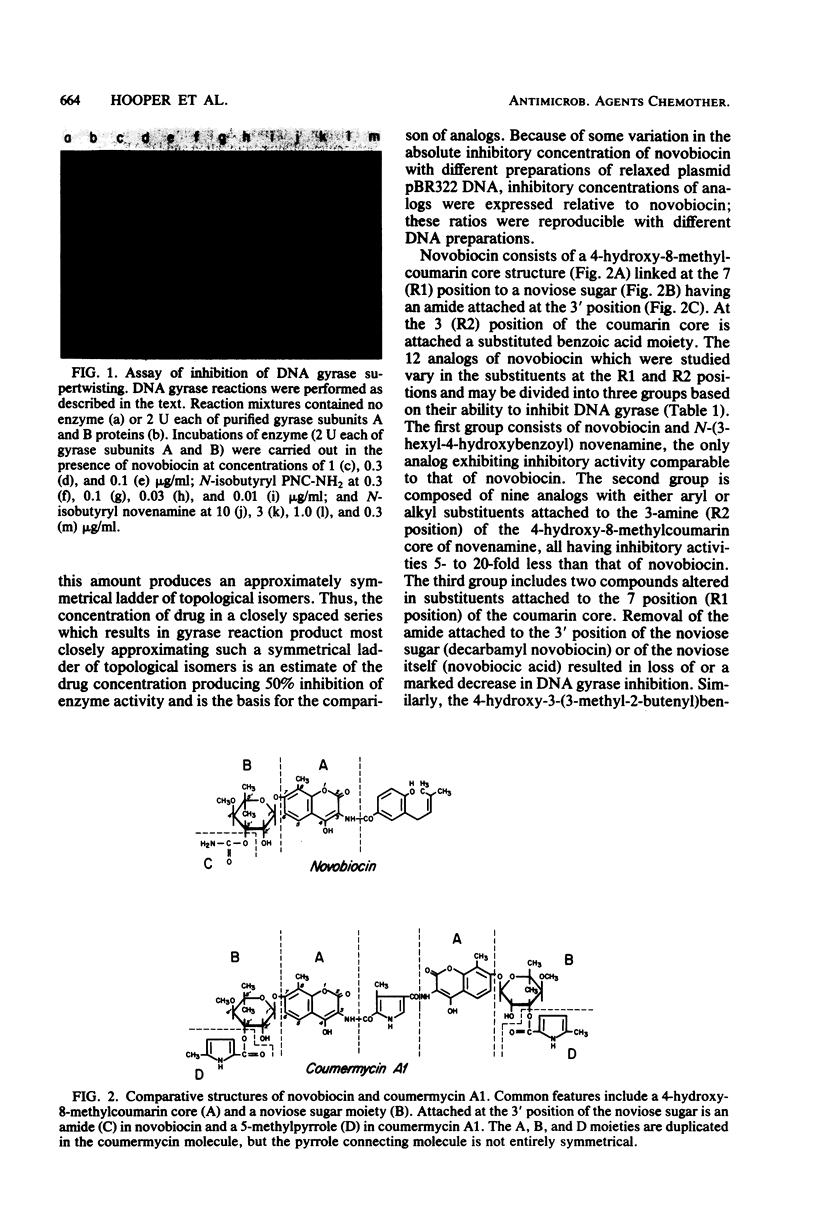
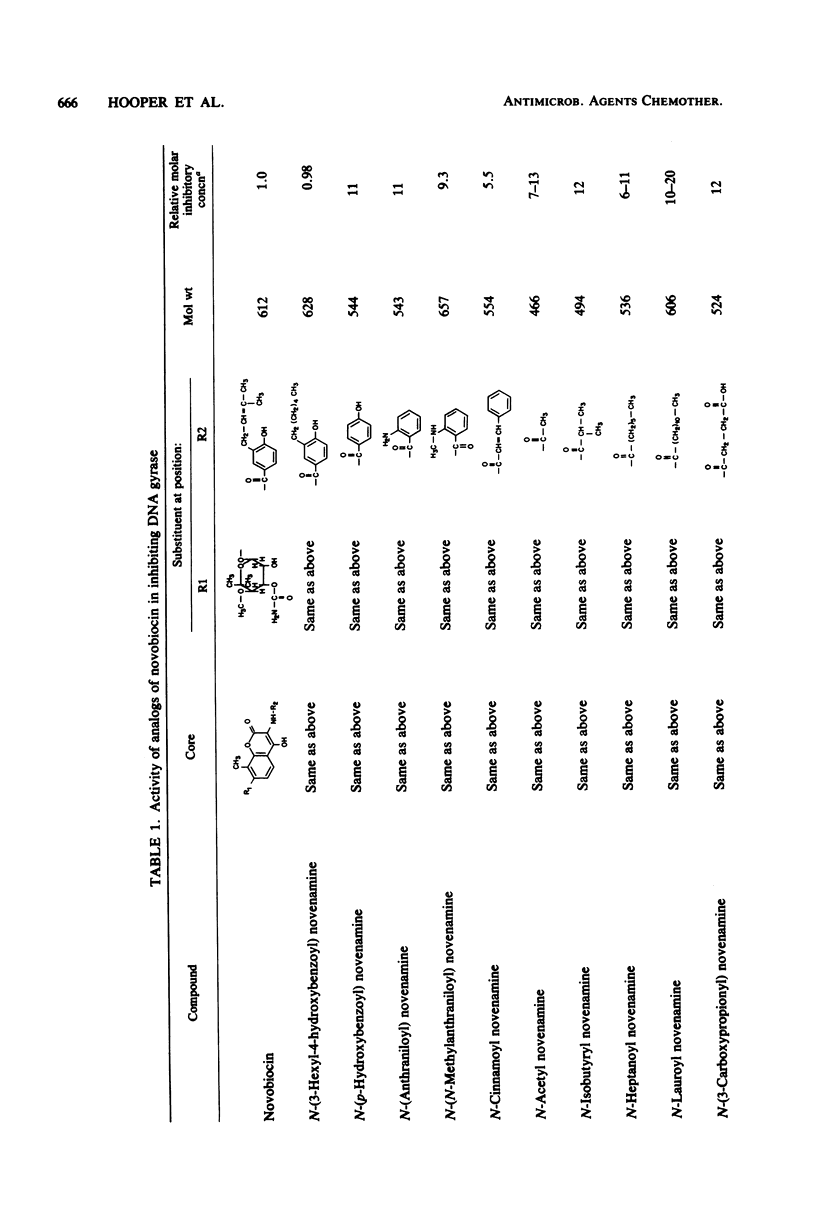
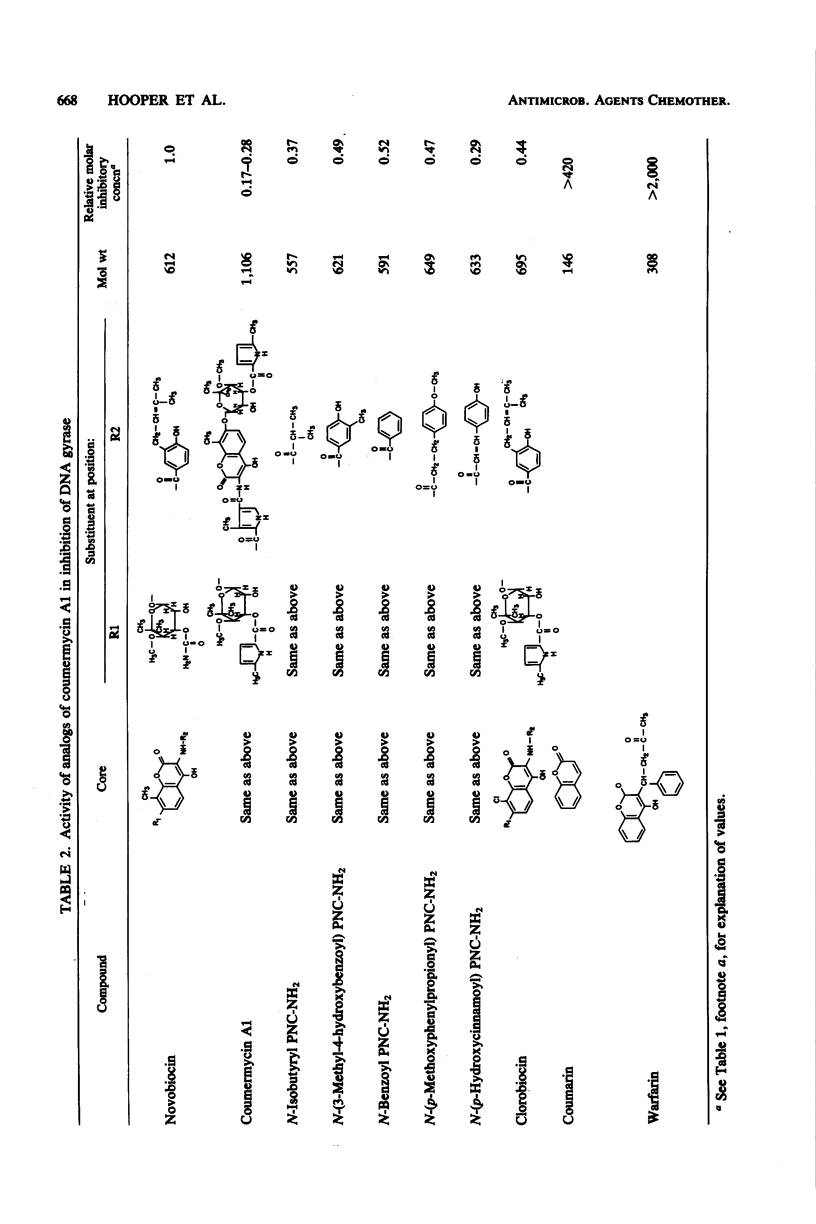
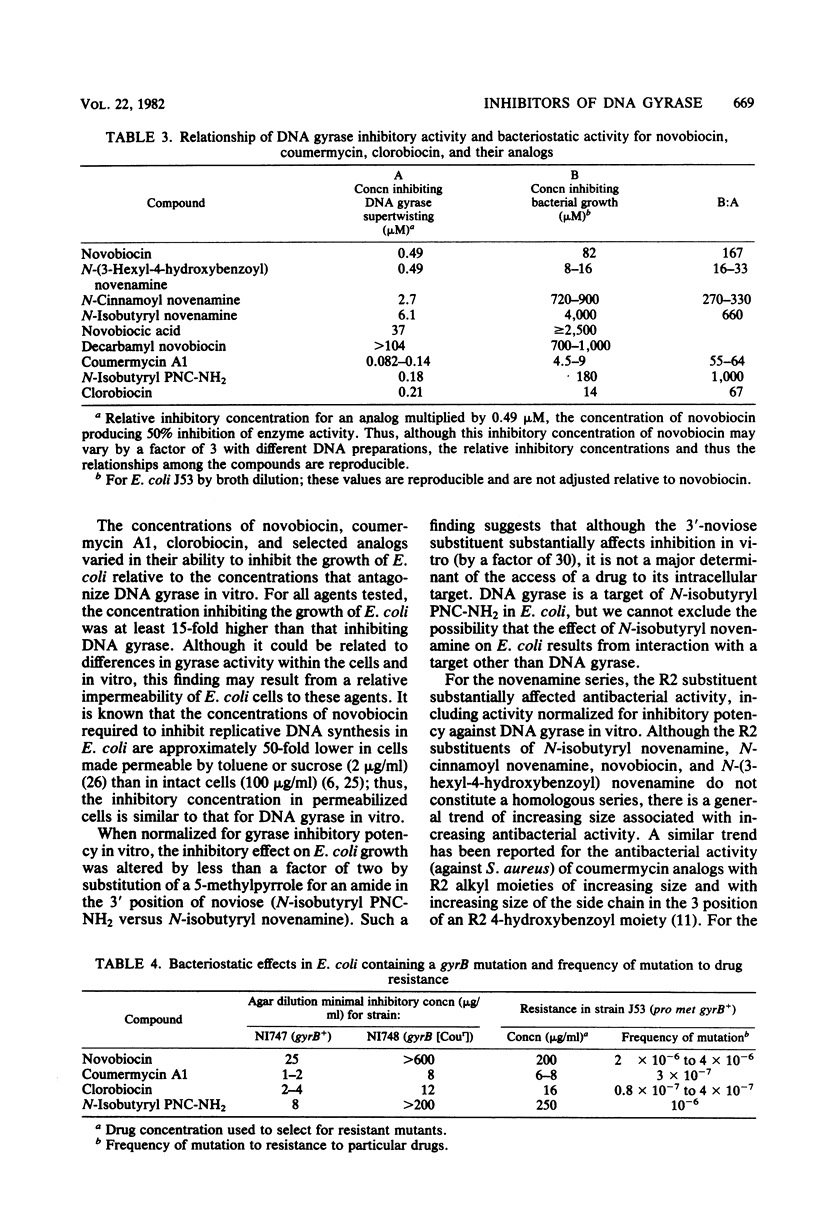
Novobiocin, coumermycin A1, and clorobiocin, structurally related compounds that antagonize the B subunit of the essential bacterial enzyme DNA gyrase, were compared with 18 of their analogs for the inhibition of Escherichia coli DNA gyrase supertwisting activity in vitro and of bacterial multiplication. This family of compounds has a 4-hydroxy-8-methylcoumarin core substituted in the 7 and 3 positions. Important for enzyme inhibition in vitro is a 7 ether linkage to a 3'-substituted noviose sugar. The 3'-ester-linked 5-methylpyrrole, found in the coumermycin series, conferred at least 10-fold more inhibitory activity than did the similarly linked amide, found in the novobiocin series; lack of the pyrrole and amide results in the loss of inhibitory activity. Of many aryl and alkyl substituents linked as an amide at the 3 position, the 4-hydroxyl-3-(3-methyl-2-butenyl)benzoic acid moiety, found in novobiocin and clorobiocin, and the reduplication of the coumarin-noviose-5-methylpyrrole, found in coumermycin A1, were most effective in gyrase inhibition. In vivo, the ability of these compounds to inhibit the growth of E. coli varied greatly. The enhanced inhibition of gyrase in vitro conferred by a 5-methylpyrrole relative to an amide in the 3'-noviose position was reflected in inhibition of bacterial multiplication. Several substitutions at the 3 position of the coumarin core conferring similar antagonism of gyrase in vitro resulted in substantially different inhibitory activities for E. coli, suggesting that these moieties at the 3 position affect drug access to the intracellular target. This target was shown for isobutyryl PNC-NH2 (PNC-NH2 is 3-amino-4-hydroxy-8-methyl-7-[3-O-(5-methyl-2-pyrrolylcarbonyl)noviosyloxy] coumarin) and confirmed for novobiocin, coumermycin A1, and clorobiocin to be in the B subunit of DNA gyrase.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Camerini-Otero R. D., Felsenfeld G. Supercoiling energy and nucleosome formation: the role of the arginine-rich histone kernel. Nucleic Acids Res. 1977;4(5):1159–1181. doi: 10.1093/nar/4.5.1159-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crumplin G. C. The involvement of DNA topoisomerases in DNA repair and mutagenesis. Carcinogenesis. 1981;2(2):157–160. doi: 10.1093/carcin/2.2.157. [DOI] [PubMed] [Google Scholar]
- FINLAND M., NICHOLS R. L. Novobiocin. Antibiot Chemother. 1957;4:209–392. doi: 10.1159/000386637. [DOI] [PubMed] [Google Scholar]
- Fairweather N. F., Orr E., Holland I. B. Inhibition of deoxyribonucleic acid gyrase: effects on nucleic acid synthesis and cell division in Escherichia coli K-12. J Bacteriol. 1980 Apr;142(1):153–161. doi: 10.1128/jb.142.1.153-161.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellert M., Fisher L. M., O'Dea M. H. DNA gyrase: purification and catalytic properties of a fragment of gyrase B protein. Proc Natl Acad Sci U S A. 1979 Dec;76(12):6289–6293. doi: 10.1073/pnas.76.12.6289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellert M., Mizuuchi K., O'Dea M. H., Itoh T., Tomizawa J. I. Nalidixic acid resistance: a second genetic character involved in DNA gyrase activity. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4772–4776. doi: 10.1073/pnas.74.11.4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellert M., Mizuuchi K., O'Dea M. H., Nash H. A. DNA gyrase: an enzyme that introduces superhelical turns into DNA. Proc Natl Acad Sci U S A. 1976 Nov;73(11):3872–3876. doi: 10.1073/pnas.73.11.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellert M., O'Dea M. H., Itoh T., Tomizawa J. Novobiocin and coumermycin inhibit DNA supercoiling catalyzed by DNA gyrase. Proc Natl Acad Sci U S A. 1976 Dec;73(12):4474–4478. doi: 10.1073/pnas.73.12.4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hane M. W., Wood T. H. Escherichia coli K-12 mutants resistant to nalidixic acid: genetic mapping and dominance studies. J Bacteriol. 1969 Jul;99(1):238–241. doi: 10.1128/jb.99.1.238-241.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hays J. B., Boehmer S. Antagonists of DNA gyrase inhibit repair and recombination of UV-irradiated phage lambda. Proc Natl Acad Sci U S A. 1978 Sep;75(9):4125–4129. doi: 10.1073/pnas.75.9.4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue S., Ohue T., Yamagishi J., Nakamura S., Shimizu M. Mode of incomplete cross-resistance among pipemidic, piromidic, and nalidixic acids. Antimicrob Agents Chemother. 1978 Aug;14(2):240–245. doi: 10.1128/aac.14.2.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreuzer K. N., Cozzarelli N. R. Escherichia coli mutants thermosensitive for deoxyribonucleic acid gyrase subunit A: effects on deoxyribonucleic acid replication, transcription, and bacteriophage growth. J Bacteriol. 1979 Nov;140(2):424–435. doi: 10.1128/jb.140.2.424-435.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuuchi K., Fisher L. M., O'Dea M. H., Gellert M. DNA gyrase action involves the introduction of transient double-strand breaks into DNA. Proc Natl Acad Sci U S A. 1980 Apr;77(4):1847–1851. doi: 10.1073/pnas.77.4.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuuchi K., Gellert M., Nash H. A. Involement of supertwisted DNA in integrative recombination of bacteriophage lambda. J Mol Biol. 1978 May 25;121(3):375–392. doi: 10.1016/0022-2836(78)90370-4. [DOI] [PubMed] [Google Scholar]
- Mizuuchi K., O'Dea M. H., Gellert M. DNA gyrase: subunit structure and ATPase activity of the purified enzyme. Proc Natl Acad Sci U S A. 1978 Dec;75(12):5960–5963. doi: 10.1073/pnas.75.12.5960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr E., Fairweather N. F., Holland I. B., Pritchard R. H. Isolation and characterisation of a strain carrying a conditional lethal mutation in the cou gene of Escherichia coli K12. Mol Gen Genet. 1979;177(1):103–112. doi: 10.1007/BF00267259. [DOI] [PubMed] [Google Scholar]
- Orr E., Staudenbauer W. L. An Escherichia coli mutant thermosensitive in the B subunit of DNA gyrase: effect on the structure and replication of the colicin E1 plasmid in vitro. Mol Gen Genet. 1981;181(1):52–56. doi: 10.1007/BF00339004. [DOI] [PubMed] [Google Scholar]
- Sanzey B. Modulation of gene expression by drugs affecting deoxyribonucleic acid gyrase. J Bacteriol. 1979 Apr;138(1):40–47. doi: 10.1128/jb.138.1.40-47.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. L., Kubo M., Imamoto F. Promoter-specific inhibition of transcription by antibiotics which act on DNA gyrase. Nature. 1978 Oct 5;275(5679):420–423. doi: 10.1038/275420a0. [DOI] [PubMed] [Google Scholar]
- Smith D. H., Davis B. D. Mode of action of novobiocin in Escherichia coli. J Bacteriol. 1967 Jan;93(1):71–79. doi: 10.1128/jb.93.1.71-79.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staudenbauer W. L. Letters to the editor: Novobiocin-a specific inhibitor of semiconservative DNA replication in permeabilized Escherichia coli cells. J Mol Biol. 1975 Jul 25;96(1):201–205. doi: 10.1016/0022-2836(75)90191-6. [DOI] [PubMed] [Google Scholar]
- Staudenbauer W. L., Orr E. DNA gyrase: affinity chromatography on novobiocin-Sepharose and catalytic properties. Nucleic Acids Res. 1981 Aug 11;9(15):3589–3603. doi: 10.1093/nar/9.15.3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugino A., Bott K. F. Bacillus subtilis deoxyribonucleic acid gyrase. J Bacteriol. 1980 Mar;141(3):1331–1339. doi: 10.1128/jb.141.3.1331-1339.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugino A., Cozzarelli N. R. The intrinsic ATPase of DNA gyrase. J Biol Chem. 1980 Jul 10;255(13):6299–6306. [PubMed] [Google Scholar]
- Sugino A., Higgins N. P., Brown P. O., Peebles C. L., Cozzarelli N. R. Energy coupling in DNA gyrase and the mechanism of action of novobiocin. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4838–4842. doi: 10.1073/pnas.75.10.4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugino A., Peebles C. L., Kreuzer K. N., Cozzarelli N. R. Mechanism of action of nalidixic acid: purification of Escherichia coli nalA gene product and its relationship to DNA gyrase and a novel nicking-closing enzyme. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4767–4771. doi: 10.1073/pnas.74.11.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi J., Furutani Y., Inoue S., Ohue T., Nakamura S., Shimizu M. New nalidixic acid resistance mutations related to deoxyribonucleic acid gyrase activity. J Bacteriol. 1981 Nov;148(2):450–458. doi: 10.1128/jb.148.2.450-458.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Wright A., Bridges B. A. Effect of gyrB-mediated changes in chromosome structure on killing of Escherichia coli by ultraviolet light: experiments with strains differing in deoxyribonucleic acid repair capacity. J Bacteriol. 1981 Apr;146(1):18–23. doi: 10.1128/jb.146.1.18-23.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]