Abstract
The myogenic factor D-MEF2 is required for the proper differentiation of muscle cells during Drosophila embryogenesis and the correct patterning of indirect flight muscles assembled during later metamorphosis. In addition to these essential myogenic functions, mutant D-mef2 adult females are weakly fertile and produce defective eggs. D-MEF2 is expressed in nurse and follicle cells of the wild-type egg chamber. We have analyzed the D-mef2 oogenic phenotype and show that the gene is required for the normal patterning and differentiation of the centripetally migrating follicle cells that are crucial for development of the anterior chorionic structures. D-mef2 alleles exhibit a genetic interaction with a dominant-negative allele of thick veins (tkv), which encodes a type I receptor of the Decapentaplegic-signaling pathway. tkv RNA is overexpressed in D-mef2 mutant egg chambers, and, conversely, forced expression of D-mef2 represses tkv expression. These results indicate a role for D-MEF2 in the regulation of tkv gene expression and Decapentaplegic signal transduction that are essential for proper determination and/or differentiation of the anterior follicle cells. Additionally, they demonstrate a vital function for the D-MEF2 transcription factor in multiple genetic pathways during Drosophila development.
The developing egg chamber of Drosophila is composed of germ-line nurse cells and oocyte surrounded by a layer of somatic follicle cells (reviewed in ref. 1). Inductive interactions between these cells establish polarities in the egg and the developing embryo, initially along the anterior–posterior axis followed by patterning along the dorsal–ventral axis (2–4). These interactions are mediated mainly by the spatially restricted production of Gurken (GRK), a transforming growth factor α-like protein (5), in the oocyte, and the Drosophila epidermal growth factor receptor (EGFR; ref. 6) in the neighboring follicle cells. In addition, subpopulations of about 1,000 follicle cells are responsible for producing the eggshell (chorion), including specialized anterior structures such as the micropile that allows sperm entry, the operculum that forms the hatch exit for the developed embryo, and two dorsal respiratory appendages (1).
These complicated structures reflect a precise developmental program that specifies different cell fates in the follicular epithelium. At stage 9, a group of 6–10 cells (the future border cells) at the anterior pole initiates an inward migration through the nurse cell complex until they reach the anterior center of the oocyte at stage 10A. Also at stage 9, about 950 follicle cells start migrating posteriorly, in step with the enlargement of the oocyte. The migration is complete at stage 10A, when these newly columnar cells become associated with the oocyte. The remaining anterior follicle cells become squamous and are associated with the 15 nurse cells. These are the nurse cell-associated follicle cells (NC-FC). At stage 10B, about 20 of the anterior-most oocyte-associated follicle cells (O-FC) lead the centripetal migration of about 150 cells along the oocyte–nurse cell boundary until the leading centripetally migrating follicle cells (CMFC) reach the border cells. The CMFC eventually form the operculum, and the leading CMFC, together with the border cells, specify the production of the micropile. In addition, NC-FC and CMFC are thought to regulate the dumping of nurse cell contents into the oocyte through a mechanism that is as yet unknown. Both NC-FC and CMFC express the decapentaplegic (dpp) gene (7), which encodes a transforming growth factor β-related signaling molecule (8). Mutants in dpp or in the DPP receptor gene saxophone (sax) produce egg chambers defective in anterior chorion production and nurse cell dumping (7).
Although important information has emerged on cell–cell communication used in egg chamber development, surprisingly little information exists on the transcriptional regulation of signaling pathway members (9). During the course of our studies on the function of the D-MEF2 transcription factor in muscle cell differentiation (10), we observed that adult females harboring hypomorphic alleles of D-mef2 presented an oogenic phenotype. In this report, we characterize the expression of D-MEF2 during egg development, demonstrate D-mef2 interactions with a member of the DPP-signaling pathway, and show the gene’s requirement for the proper formation of anterior eggshell structures. Our results indicate that D-mef2 has important regulatory functions in such diverse differentiation processes as oogenesis and myogenesis during Drosophila development.
Materials and Methods
Fly Stocks and Genetic Crosses.
The genotype of the D-mef2 strains used in this study is dp, cn, D-mef265 or 424, px, sp/CyO (10). dpp and tkv alleles were obtained from the Bloomington Stock Center: dpphr56, cn1, bw1/CyO and tkv7, cn1, bw1, sp1/CyO. The sax allele was provided by L. Raftery (Harvard Medical School, Cambridge, MA): sax1, cn1, bw1, sp1/CyO. The cn1, bw1, sp1 strain was used as the wild-type control in the genetic interaction study described in Fig. 1. Otherwise, Oregon-R was used as the source of wild-type egg chambers and ovarian RNA. The transgenic lines w; P[w+, UAS-D-mef2], w; P[w+, GAL4]55B, and w; P[w+, UAS-lacZ]4–2-4B have been described (11–13).
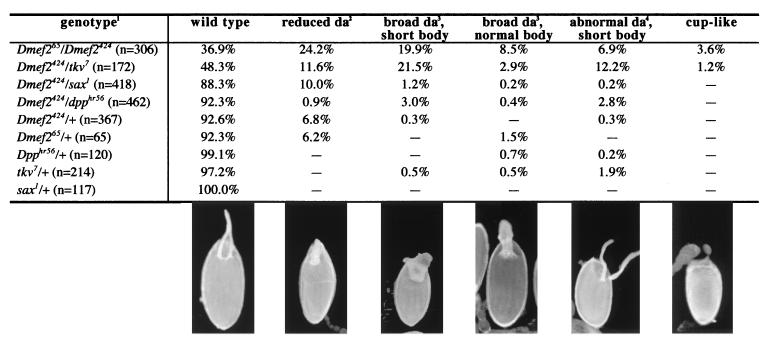
Figure 1.
D-mef2 mutant oogenic phenotypes. (1) The genotypes of the fly strains tested are described in Materials and Methods. The genotype of the “+” chromosome is cn, bw, sp to approximate the other mutant chromosomes. Phenotypes are described in more detail in the text. (2) Reduced da, dorsal appendages are fused and narrowed at the base. (3) Broad da, dorsal appendages do not narrow but the space between the two separate appendages are filled in. (4) Abnormal da, the dorsal appendages are separate but the shape and/or length are abnormal; these eggs are usually shortened as well. In enlarged views of the egg chambers (not shown), the number of follicle cells encompassed by the appendages can be counted and are summarized below: wild type, 14 cells; reduced da, 9 cells; broad da, 14 and 18 cells (in the short and normal body samples, respectively); abnormal da, 14 cells; cup-like, no da.
Egg Chamber Phenotype Analysis.
All flies were grown at 25°C unless noted. Young females were conditioned in the presence of live yeast at 25°C for 3 days except for flies containing the dpphr56 allele and the flies containing the P[w+, GAL4]55B transgene, which were conditioned at 30°C. Ovaries were dissected and mounted in 50% glycerol/1× PBS/0.1% Triton X-100 for microscopic observation.
Gene Expression Analysis.
Protein immunostaining and RNA in situ hybridizations followed established procedures (3, 13, 14). The anti-D-MEF2 polyclonal antibody was provided by H. Nguyen (Albert Einstein Medical College, Bronx, NY). A 1:800-fold dilution of this antibody and a 1:1,000 dilution of the goat anti-rabbit IgG (Vector Laboratories) were used. cDNA probes used for RNA in situ hybridization were as follows: thick veins (tkv), a PCR-generated, 1,565-bp fragment encompassing the entire ORF from a cDNA clone provided by M. Hoffmann (University of Wisconsin, Madison); dpp, a PCR-generated, 457-bp fragment encompassing the central portion of the ORF from a cDNA clone provided by W. Gelbart (Harvard University); argos, a 619-bp fragment encompassing the N terminus of the ORF, generated by reverse transcription–PCR from ovarian total RNA.
S1-Nuclease Protection Assay.
Standard PCR amplification was used to generate double-stranded DNA probes with the antisense primers labeled at the 5′ termini by using T4 polynucleotide kinase (New England Biolabs) and [γ-32P]ATP. The PCR products were gel-purified. The tkv probe was a 205-bp fragment corresponding to the 5′ untranslated region and the first 64 codons. The dpp probe was a 271-bp fragment corresponding to codon 222–312 in the ORF. A 120-bp probe for rp49 (codon 39–81) was used as an internal control.
A mixture of 50 μg of total RNA from wild-type or D-mef265/D-mef2424 ovaries, 1 × 106 cpm of double-stranded DNA probes for either tkv or dpp, and 1 × 106 cpm of the rp49 probe were coprecipitated. The pellet was dissolved in 30 μl of hybridization buffer (40 mM Pipes, pH 6.4/1 mM EDTA, pH 8.0/0.4 M NaCl/80% formamide), denatured at 90°C for 5 min, and then incubated at 59°C for 16 hr. Three hundred microliters of an ice-cold solution containing 800 units of S1-nuclease (Boehringer Mannheim/Roche), 0.1 M NaCl, 60 mM NaOAc (pH 4.5), and 2 mM ZnSO4 was added. S1-nuclease digestion was carried out at 20°C for 0.5 hr and at 4°C for an additional 15 min. The reaction mixtures then were phenol-extracted and ethanol-precipitated. Samples were analyzed on a 6% denaturing polyacrylamide gel.
Results
Animals null for D-mef2 function die during late embryogenesis because of severe muscle defects and a resulting failure to hatch as first instar larvae. However, animals transheterozygous for partial loss-of-function alleles survive into adulthood. We noticed that these females exhibited very low fertility regardless of their male partners, suggesting they may be defective in oogenesis.
From our collection of D-mef2 alleles (10), two allelic combinations gave transheterozygous adults: D-mef265/D-mef2424 and D-mef265/D-mef2113. D-mef265 is a nonsense mutation at codon 491 that deletes the C-terminal 25 aa, D-mef2424 is a Ser-to-Pro substitution at codon 186, and D-mef2113 is a splice donor-site mutation resulting in a severe truncation of the protein ORF (10). The viabilities of these transheterozygotes were consistent with the predicted severity of the mutant alleles: D-mef265/D-mef2424 were produced at 20–30% of the expected Mendelian ratio whereas D-mef265/D-mef2113 adults were at about 1% of the expected number. When ovaries were dissected from these transheterozygous females, a large number of egg chambers with various abnormalities were observed. Only the results from D-mef265/D-mef2424 are represented in this report because of the higher number of adult females that could be generated. However, we do note that both classes of transheterozygotes produced similar oogenic phenotypes and D-mef265/D-mef2113 produced fewer wild-type eggs as expected (unpublished observations).
These phenotypes were classified according to the morphology of the dorsal appendages and the shape of the egg chambers (Fig. 1). In one class, the bases of the dorsal appendages fused and encompassed less than 8 follicle cells as compared with the wild-type size of about 14 cells (9). In other groupings the dorsal appendages also fused, but the fusion apparently resulted from overproduction of dorsal appendage material along the dorsal midline because the number of follicle cells encompassed by these broad appendages were either the same as, or slightly larger than, wild type (14–18 cells).
In addition to the appendage phenotypes, a large number of egg chambers were distinctively short, indicating a defect in the dumping of nurse cell contents into the oocyte toward the end of oogenesis (15, 16). The most severely deformed among these short eggs are the cup-like eggs, which lack any anterior structures and are open at the anterior end. These phenotypes can be generalized as defects in patterning of anterior follicle cell fates and are comparable to those observed in dpp-signaling pathway mutants (7).
More than 90% of the egg chambers produced by females heterozygous for D-mef2 or dpp signaling-pathway mutants were phenotypically normal. We therefore sought to examine whether females with transheterozygous combinations of D-mef2 and dpp-signaling mutant alleles produced chorion phenotypes, thus suggesting genetic interactions (Fig. 1). Note that three alleles, D-mef2113, D-mef2424, and D-mef265, were tested and they yielded similar results (unpublished observations). Only the results obtained from D-mef2424 are presented in Fig. 1. None of the D-mef2 alleles tested showed interaction with dpphr56, a dpp allele that has been shown to interact strongly with dppe87 at elevated temperatures (7). A low level of interaction was seen between D-mef2 and sax1. sax1 was completely normal when heterozygous (Fig. 1), but exhibited a strong interaction with an antimorphic tkv allele (tkv7; refs. 7 and 17). Interestingly, D-mef2 showed the strongest interaction with the same tkv7 allele (Fig. 1).
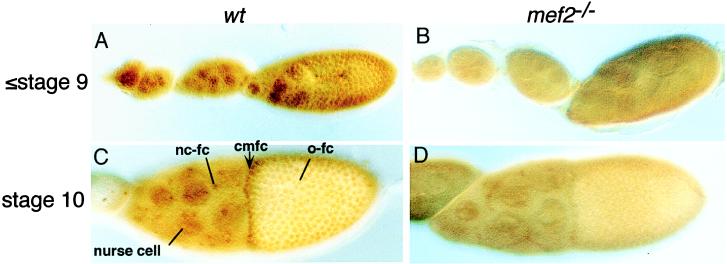
To interpret the D-mef2 phenotypes, we first examined the pattern of D-MEF2 expression during oogenesis. As shown in Fig. 2, D-MEF2 is expressed in both follicle and nurse cells of postgermarium egg chambers. No detectable expression is observed in the oocyte and there is no distinct tissue-specific expression pattern. However, there is a higher level of D-MEF2 in the leading CMFC than in the other follicle cells. Nuclear staining of D-MEF2 is reduced in D-mef2 mutant egg chambers and is accompanied by some general staining in the cytoplasm. This indicates that the missense and truncated D-MEF2 proteins produced by these alleles may impair nuclear localization of the transcription factor (10).
Figure 2.
D-MEF2 expression in developing egg chambers. Ovaries were dissected from either Oregon-R (wt; A and C) or D-mef265/D-mef2424 (mef2−/−; B and D) and stained with an anti-D-MEF2 polyclonal antibody. Anterior is to the left. (A and B) Stage 9 and early eggs. (C and D) Stage 10 eggs. In wild type, D-MEF2 is detected in the nuclei of both nurse and follicle cells up to stage 13 (later stages not shown). Nuclear staining of D-MEF2 is reduced in the mutant background. nc-fc, nurse cell-associated follicle cells; cmfc, centripetally migrating follicle cells; o-fc, oocyte-associated follicle cells.
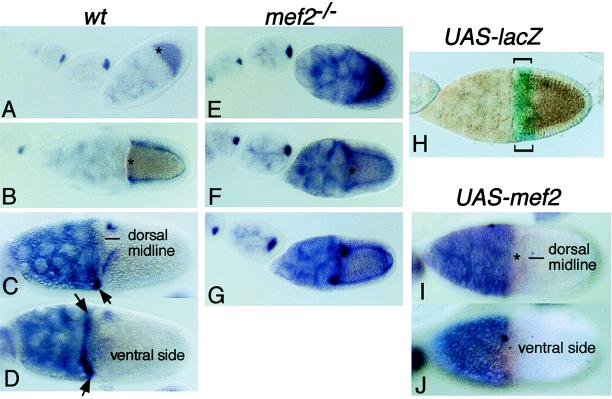
Interaction between D-mef2 and dpp-signaling mutants suggested that D-MEF2 might either regulate the expression of the DPP-signaling components or be regulated by DPP signaling. In tkv7/sax1 transheterozygotes that generated anterior chorion phenotypes, we failed to observe any change in the D-MEF2 expression pattern (data not shown). Because D-mef2 interacts strongly with tkv7, we examined whether or not tkv RNA expression might be affected (Fig. 3). In wild-type egg chambers, tkv RNA is detected in the developing oocyte before stage 8 and the staining starts to fade at stage 9. At stage 10A, the oocyte staining is nondetectable but expression appears in the O-FC, except for a group of cells marking the boundary between the oocyte and nurse cells. These are the leading edge of the future CMFC. However, later at the beginning of stage 10B, tkv expression diminishes in the O-FC but appears in the ventral subpopulation of CMFC and remains in two stripes of O-FC on the dorsal side, separated by the dorsal midline.
Figure 3.
tkv RNA expression is negatively regulated by D-mef2 in the egg chambers. Ovaries were dissected from Oregon-R (wt; A–D), D-mef265/D-mef2424 (mef2−/−; E–G), w; P[w+, GAL4]55B/P[w+, UAS-lacZ]4–2-4B (UAS-lacZ; H), or w; P[w+, GAL4]55B/P[w+, UAS-D-mef2] (UAS-mef2; I and J) and hybridized with digoxigenin-labeled tkv cDNA probes (A–G, and I and J) or stained for the β-galactosidase activity (H). Anterior is to the left. (A and E) Optical cross-sections of stage 9 and early eggs. (B and F) Optical cross-sections of stage 10A eggs. (C and D) Dorsal and ventral surface views of late-stage 10B eggs, respectively. Arrows point to the ventral population of CMFC that express tkv. (G) Optical cross-section of a stage 10B egg. In D-mef−/−, tkv is overexpressed at stage 9 and later. Note that stage 10 eggs from D-mef2−/− are smaller than the wild type. Other features are described in the text. (I and J) Dorsal and ventral surface views, respectively, of a stage 10B egg from a transgenic female w; P[w+, GAL4]55B/P[w+, UAS-D-mef2]. This GAL4/UAS system directs ectopic overexpression in the anterior population of the O-FC as monitored in H by using the lacZ reporter gene (brackets). As a result of D-mef2 overexpression, the tkv expression is diminished in both the dorsal stripes and the ventral CMFC.
In egg chambers from D-mef265/D-mef2424 mutant mothers, tkv RNA expression appears to be normal at stage 8 but is globally overexpressed after stage 9, particularly in the O-FC. Overexpression of tkv in D-mef2 mutants was confirmed further by a quantitative S1-nuclease protection assay (Fig. 4). We observed a 3-fold increase in the tkv RNA level in mutants as compared with wild type. In contrast, there was no change in the level of dpp RNA. In addition, when a wild-type D-mef2 transgene was overexpressed in the anterior population of the O-FC by using the GAL4/UAS binary expression system (refs. 12 and 13; Fig. 3H), tkv RNA expression at stage 10B was abolished (compare Fig. 3 D and E with I and J).
Figure 4.
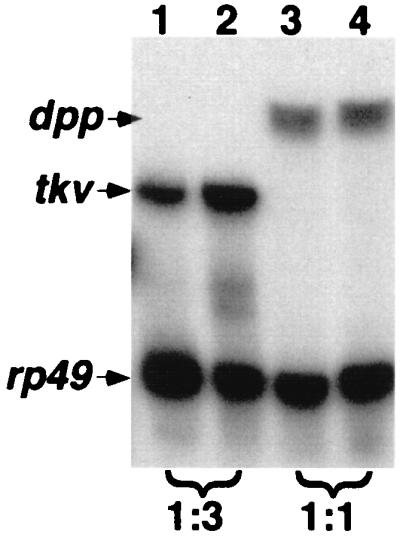
Quantitation of tkv RNA expression. Total RNA was isolated from ovaries of Oregon-R (lanes 1 and 3) or D-mef265/D-mef2424 (lanes 2 and 4) females. S1-nuclease protection assays were performed as described in Materials and Methods. rp49 serves as internal controls. The intensity of the bands were quantitated by using a PhosphorImager. The densities of the tkv and dpp bands were normalized against those of rp49 from the same sample, and the relative values (wild type/D-mef2−/−) are indicated at the bottom.
Although dpp expression levels were not affected in D-mef2 mutant egg chambers, its expression pattern nonetheless provided a valuable marker for the patterning of the anterior follicular epithelium (Fig. 5). In wild type, dpp is expressed in the anterior population of follicle cells at stage 8, before posterior migration. At stage 9, when the posterior migration of follicle cells is in progress, dpp expression remains associated with the anterior population (Fig. 5A; ref. 7). The majority of these dpp-expressing cells are the future NC-FC, but the leading one to two rows of cells appear to be the anterior-most of the future O-FC. These cells mark the future leading edge of the CMFC, as is evident at stage 10A when the posterior migration is complete and centripetal migration is about to commence. At stage 11, these cells can be seen migrating inward along the boundary between the oocyte and the nurse cell complex.
Figure 5.
Follicle cell fates are disrupted in D-mef2 mutant egg chambers. Egg chambers were dissected from either Oregon-R (wt; A, C, and H) or D-mef265/D-mef2424 (mef2−/−; B, D–G, and I) and hybridized with a digoxigenin-labeled dpp cDNA probe. Anterior is to the left. (A) Optical cross-sections of stage 6, 8, and 9 eggs (indicated). Expression in the anterior follicle cells at stage 8 is marked by an arrow. The strongly stained cells at stage 9 are marked by arrows that also denote the direction of follicle cell migration. (B) Markings are the same as in A except that stage 6 and 8 eggs are shown in surface views. (C) Surface view of a stage 10A egg. There is light staining in the NC-FC (slight blue haze over the nurse cells). The strongest staining is seen with the leading CMFC (arrows). (D and E) Dorsal and ventral surface views, respectively, of a stage 10A egg. The presumptive leading CMFC (stained for dpp) are misplaced. (F and G) Two additional samples of stage 10A eggs showing disruption of CMFC determination. (H) Optical cross-section of an early stage 11 egg. The leading CMFC has moved inward (arrows) along the boundary between the oocyte and the nurse cell complex. (I) Optical cross-section of a collapsing stage 11 egg. There is no distinct dpp staining.
In the D-mef2 mutant, the critical leading CMFC defined by dpp expression are severely disrupted. As shown in Fig. 5, there is no disruption of dpp expression before stage 9, but at stage 10 the future leading CMFC become ill-defined. Instead of forming a straight line, they advance too much posteriorly, lag behind, or lose their identities completely (i.e., no distinct dpp expression). These results suggest that the tightly regulated expression of the tkv gene by D-MEF2 may be critical in defining the identity of dpp-expressing follicle cells, which, in turn, elaborate the anterior chorion structures.
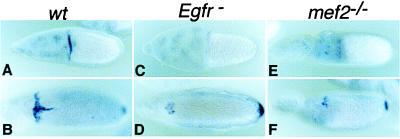
Another interesting aspect of the D-mef2 mutant phenotypes is the appearance of broad and fused dorsal appendages (Fig. 1). Such a phenotype often is attributed to the disruption of a negative-feedback mechanism that reduces the EGFR signaling strength in the dorsal midline O-FC (18, 19). The feedback modulation of the EGFR is carried out by the EGFR blocker Argos (19, 20). argos is expressed in a T-shaped pattern in the dorsal CMFC and in the dorsal midline O-FC. This expression pattern is activated by EGFR signaling itself, presumably at its highest activity level (refs. 19 and 20; Fig. 6). Consistent with the observed phenotype, dorsal midline expression of argos is reduced substantially in the D-mef2 mutant egg chamber (Fig. 6).
Figure 6.
argos expression is reduced in the D-mef2 mutant. Egg chambers were dissected from Oregon-R (wt; A and B), Egfrtop-QY1 (Egfr-; C and D), and D-mef265/D-mef2424 (mef2−/−; E and F) females and hybridized with a digoxigenin-labeled argos cDNA probe. Eggs are shown in dorsal view, and anterior is to the left. In wild type (wt), argos expression starts at stage 10 in the dorsal population of the CMFC (A), whereas dorsal midline expression is added at stage 12 (B). In addition, expression at the posterior polar follicle cells is also detected at stage 12. In the hypomorphic Egfr mutant (C and D), the midline expression is completely abolished and the dorsal CMFC expression is greatly reduced. Note that the posterior polar expression is not affected. Exactly the same reduction pattern is observed in the D-mef2 mutant (E and F).
Discussion
In this report we show that the D-MEF2 transcription factor, an essential regulator of the myogenic program in Drosophila, also functions in oogenesis. Females carrying hypomorphic alleles of D-mef2 exhibit chorion phenotypes similar to those noted in dpp-signaling pathway mutants (7). These morphological defects suggested that D-MEF2 might interact with members of the DPP pathway. The three best-studied components of the DPP pathway have been shown to play important roles during oogenesis: DPP, which is the BMP2/4-like extracellular ligand (8), and SAX and TKV, which are distinct type I receptor molecules (21, 22). Most of the dpp pathway mutants and the D-mef2 alleles are phenotypically normal when heterozygous. This provided a convenient way to test for possible genetic interactions by examining whether or not a significant number of phenocopies could result from transheterozygous combinations. In initial tests, none of the dpp, sax, or tkv hypomorphic alleles showed significant interactions with D-mef2. However, a dominant-negative allele of tkv (tkv7; ref. 17) showed a very strong enhancement effect on D-mef2 (Fig. 1). Subsequent molecular epistatic analyses revealed that D-MEF2 negatively regulates the RNA levels of tkv (Figs. 3 and 4). A negative regulatory role of D-MEF2 on tkv expression is consistent with the observed interactions between D-mef2 and tkv7. Because tkv7 is a dominant-negative allele, overexpression of this altered protein in D-mef2 mutants presumably results in a stronger deleterious effect than with other hypomorphic alleles. sax has been shown to function genetically in the germ line (7), but we observed no change in sax RNA expression in the D-mef2 mutant background (data not shown). Therefore, the low-level genetic interaction we observed between sax1 and D-mef2 (Fig. 1) is likely an indirect effect.
D-MEF2 is expressed in both nurse and follicle cells. We have not observed any defects in the germ line, either the number of germ cells (15 nurse cells and 1 oocyte) or the location of the oocyte within the egg chamber (Figs. 2, 3, and 5). A possible requirement for D-MEF2 in germ-line cells remains to be elucidated. In contrast, D-MEF2 appears to function in the somatic follicle cells, particularly in subpopulations of the O-FC, by negatively regulating tkv RNA levels. We do not know whether D-MEF2 directly represses tkv gene transcription. Curiously, the expression patterns of D-MEF2 and tkv RNA are not mutually exclusive. Whereas D-MEF2 is expressed in all follicle cells, tkv is absent only in different populations of follicle cells at different times. Perhaps subtle changes in D-MEF2 levels can have different effects on tkv expression. For example, at stage 10A, D-MEF2 is more abundant in the leading CMFC than in the other O-FC (Fig. 2), whereas tkv is not expressed in CMFC and expressed at a low level in the rest of the follicle cells (Fig. 3). Alternatively, D-MEF2 may be a constitutive repressor of tkv whereas other tissue-specific factors can counteract D-MEF2 and induce tkv expression.
In wild type, tkv expression is dynamic during oogenesis and appears to highlight a specific group of follicle cells, the leading front of the CMFC. At stage 10A just before the commencement of centripetal migration, these cells form a ring marking the boundary between the oocyte and the nurse cell complex. After stage 10B, this ring of cells migrates inward until they reach the border cells located at the center of the oocyte anterior. At stage 10A, tkv is expressed in O-FC but not in the leading CMFC (Fig. 3). This pattern is opposite to the dpp expression pattern, which is highly expressed in the leading CMFC but not in the rest of the O-FC. It will be of interest to examine whether or not tissue-specific expression of dpp and tkv in the egg chamber is autoregulated by DPP signaling.
At stage 10B, tkv is expressed in the ventral half of the CMFC in addition to two short stripes in the dorsal region of the oocyte-associated follicular epithelium (Fig. 3). This expression pattern appears to be complementary to that of the EGFR blocker argos (refs. 19 and 20; Fig. 6), which forms a T-shaped pattern along the dorsal CMFC and dorsal midline. argos expression is induced by the highest level of EGFR signaling and it, in turn, reduces the signaling strength by blocking the interaction between the receptor and its ligands. Thus, the initial graded distribution of EGFR signaling, extending laterally from the anterodorsal midline of the O-FC, is transformed into two ridges of the EGFR-signaling level just lateral to the dorsal midline. These two ridges define the two lines of O-FC that ultimately produce the two dorsal appendages. Interestingly, argos expression is diminished in the D-mef2 mutant (Fig. 6), consistent with the observed mutant egg chambers possessing broad and fused appendages. Although we favor the notion that argos expression is modulated by D-MEF2 through the action of TKV, it cannot be ruled out that D-MEF2 may directly control the transcription of argos.
In addition to regulating the expression pattern of argos, D-mef2 may play a more general role in modulating the EGFR-signaling level. This is suggested by the presence of D-mef2 mutant egg cambers with reduced and fused dorsal appendages, a phenotype typical of hypomorphic EGFR-signaling pathway mutants (4). Indeed, we have observed reduced expression of EGFR-signaling components such as rhomboid in D-mef2 mutants (data not shown). More detailed and expansive studies are needed to elucidate the possible interaction between the DPP- and EGFR-signaling pathways with D-MEF2 as a potential mediator.
On the other hand, this report does demonstrate that the dpp-expressing CMFC are poorly defined in D-mef2 mutant egg chambers (Fig. 5). CMFC is responsible for forming the operculum and, together with the border cells, specifying the construction of the micropile. Formation of these structures is also essential to closing the anterior end of the egg chamber. Because DPP is critical for specifying anterior chorion production, the disrupted patterning of CMFC in the D-mef2 mutant may explain, at least in part, the chorion phenotypes we have observed.
Earlier studies have shown that D-mef2 is a late-acting component of the genetic network controlling embryonic myogenesis (10, 11, 23). In the cardiac lineage, D-mef2 is a direct target of Tinman (Tin; refs. 26 and 27), a homeodomain transcription factor essential for heart formation (28, 29). tin function is required for the specification of cardiac precursor cells within the dorsal mesoderm, and its expression within this domain is induced by DPP, produced by cells of the dorsal ectoderm (28, 29). Additionally, D-mef2 expression in a broader dorsal mesodermal domain is controlled by DPP and the transcription factor MEDEA (30). Thus, D-mef2 can be considered a downstream regulator within the DPP-signaling pathway needed for cardiogenesis in the fly.
In the current study, we show that D-MEF2 modulates the dissemination of a DPP signal in the egg chamber through its control of tkv expression levels. Because multifunctional proteins often are members of conserved gene cassettes that function in different developmental processes (31), it is possible that D-MEF2 provides an additional critical function for DPP signaling in the mesoderm. tkv is expressed during and required for inductive events occurring in the dorsal mesoderm (32). Likewise, D-MEF2 is present throughout the mesoderm at this stage and may have a comparable function in regulating tkv expression in mesodermal cells as has been elucidated in follicle cells. This could occur through a possible feedback regulatory loop from D-mef2 to tkv. It will be important to investigate potential interactions of the two genes in the mesoderm, perhaps providing new insights into the specificity of DPP signaling during Drosophila development.
Acknowledgments
We are grateful to S. Abmayr, M. Hoffmann, L. Raftery, and the Bloomington Stock Center for sending fly strains, H. Nguyen for providing D-MEF2 antibody, and W. Gelbart and M. Hoffmann for sending dpp and tkv cDNA clones. This research was supported by grants from the Medical University of South Carolina Internal Research Funds (T.H.), Department of Energy (T.H.), Muscular Dystrophy Association (R.A.S.), National Science Foundation (R.A.S.), and National Institutes of Health (T.H. and R.A.S.).
Abbreviations
- CMFC
centripetally migrating follicle cells
- NC-FC
nurse cell-associated follicle cells
- O-FC
oocyte-associated follicle cells
- dpp
decapentaplegic
- D-mef2
Drosophila myocyte enhancer factor 2
- tkv
thick veins
- EGFR
epidermal growth factor receptor
References
- 1.Spradling A C. The Development of Drosophila melanogaster. Plainview, NY: Cold Spring Harbor Lab. Press; 1993. pp. 1–70. [Google Scholar]
- 2.Gonzalez-Reyes A, Elliot H, St. Johnston D. Nature (London) 1995;375:654–658. doi: 10.1038/375654a0. [DOI] [PubMed] [Google Scholar]
- 3.Roth S, Neuman-Silberberg F S, Barcelo G, Schüpbach T. Cell. 1995;81:967–978. doi: 10.1016/0092-8674(95)90016-0. [DOI] [PubMed] [Google Scholar]
- 4.Ray R P, Schüpbach T. Genes Dev. 1996;10:1711–1723. doi: 10.1101/gad.10.14.1711. [DOI] [PubMed] [Google Scholar]
- 5.Neuman-Silberberg F S, Schüpbach T. Cell. 1993;75:165–174. [PubMed] [Google Scholar]
- 6.Schejter E D, Shilo B-Z. Cell. 1989;56:1093–1104. doi: 10.1016/0092-8674(89)90642-9. [DOI] [PubMed] [Google Scholar]
- 7.Twombly V, Blackman R K, Jin H, Graff J M, Padgett R W, Gelbart W M. Development. 1996;122:1555–1565. doi: 10.1242/dev.122.5.1555. [DOI] [PubMed] [Google Scholar]
- 8.Padgett R W, St. Johnston D, Gelbart W M. Nature (London) 1987;325:81–84. doi: 10.1038/325081a0. [DOI] [PubMed] [Google Scholar]
- 9.Hsu T, Bagni C, Sutherland J D, Kafatos F C. Genes Dev. 1996;10:1411–1421. doi: 10.1101/gad.10.11.1411. [DOI] [PubMed] [Google Scholar]
- 10.Ranganayakulu G, Zhao B, Dokidis A, Molkentin J D, Olson E N, Schulz R A. Dev Biol. 1995;171:169–181. doi: 10.1006/dbio.1995.1269. [DOI] [PubMed] [Google Scholar]
- 11.Bour B A, O’Brien M A, Lockwood W L, Goldstein E S, Bodmer R, Taghert P H, Abmayr S M, Nguyen H. Genes Dev. 1995;9:730–741. doi: 10.1101/gad.9.6.730. [DOI] [PubMed] [Google Scholar]
- 12.Brand A H, Perrimon N. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 13.Mantrova E Y, Hsu T. Genes Dev. 1998;12:1166–1175. doi: 10.1101/gad.12.8.1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsu T, King D L, LaBonne C, Kafatos F C. Proc Natl Acad Sci USA. 1993;90:6488–6492. doi: 10.1073/pnas.90.14.6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gutzeit H O. J Cell Sci. 1986;80:159–169. doi: 10.1242/jcs.80.1.159. [DOI] [PubMed] [Google Scholar]
- 16.Cant K, Cooley L. Genetics. 1996;143:249–258. doi: 10.1093/genetics/143.1.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tearle R G, Nüsslein-Volhard C. Dros Inform Serv. 1987;66:209–269. [Google Scholar]
- 18.Morimoto A M, Jordan K C, Tietzer K, Britton J S, O’Neill E M, Ruohola-Baker H. Development. 1996;122:3745–3754. doi: 10.1242/dev.122.12.3745. [DOI] [PubMed] [Google Scholar]
- 19.Wasserman J D, Freeman M. Cell. 1998;95:355–364. doi: 10.1016/s0092-8674(00)81767-5. [DOI] [PubMed] [Google Scholar]
- 20.Queenan A M, Ghabrial A, Schüpbach T. Development. 1997;124:3871–3880. doi: 10.1242/dev.124.19.3871. [DOI] [PubMed] [Google Scholar]
- 21.Letsou A, Arora K, Wrana J L, Simin K, Twombly V, Jamal J, Staehling-Hampton K, Hoffmann F M, Gelbart W M, Massagué J, et al. Cell. 1995;80:899–908. doi: 10.1016/0092-8674(95)90293-7. [DOI] [PubMed] [Google Scholar]
- 22.Ruberte E, Marty T, Nellen D, Affolter M, Basler K. Cell. 1995;80:889–897. doi: 10.1016/0092-8674(95)90292-9. [DOI] [PubMed] [Google Scholar]
- 23.Lilly B, Zhao B, Ranganayakulu G, Paterson B M, Schulz R A, Olson E N. Science. 1995;267:688–693. doi: 10.1126/science.7839146. [DOI] [PubMed] [Google Scholar]
- 24.Gajewski K, Kim Y, Lee Y M, Olson E N, Schulz R A. EMBO J. 1997;16:515–522. doi: 10.1093/emboj/16.3.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gajewski K, Kim Y, Choi C Y, Schulz R A. Dev Genes Evol. 1998;208:382–392. doi: 10.1007/s004270050194. [DOI] [PubMed] [Google Scholar]
- 26.Azpiazu N, Frasch M. Genes Dev. 1993;7:1325–1340. doi: 10.1101/gad.7.7b.1325. [DOI] [PubMed] [Google Scholar]
- 27.Bodmer R. Development. 1993;118:719–729. doi: 10.1242/dev.118.3.719. [DOI] [PubMed] [Google Scholar]
- 28.Frasch M. Nature (London) 1995;374:464–467. doi: 10.1038/374464a0. [DOI] [PubMed] [Google Scholar]
- 29.Xu X, Yin Z, Hudson J B, Ferguson E L, Frasch M. Genes Dev. 1998;12:2354–2370. doi: 10.1101/gad.12.15.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nguyen H T, Xu X. Dev Biol. 1998;204:550–566. doi: 10.1006/dbio.1998.9081. [DOI] [PubMed] [Google Scholar]
- 31.Ruohola-Baker H, Jan L Y, Jan Y N. Trends Genet. 1994;10:89–94. doi: 10.1016/0168-9525(94)90231-3. [DOI] [PubMed] [Google Scholar]
- 32.Yin Z, Frasch M. Dev Genet. 1998;22:187–200. doi: 10.1002/(SICI)1520-6408(1998)22:3<187::AID-DVG2>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]