Abstract
The aim of this study was to clarify the role of Fgfr2 during later stages of embryonic development. Of two previously reported gene-targeting experiments, the more extensive Fgfr2 deletion was lethal shortly after implantation, because of trophoblast defects, whereas the less extensive one survived until midgestation with placental insufficiency and defective limb outgrowth [Xu, X., Weinstein, M., Li, C., Naski, M., Cohen, R. I., Ornitz, D. M., Leder, P. & Deng, C. (1998) Development (Cambridge, U.K.) 125, 753–765]. Fgfr2 in the early embryo is expressed in the trophectoderm, and this extra-embryonic localization persists into mid- and late gestation, when Fgfr2 also is expressed in multiple developing organs. To gain insight into the later functions of Fgfr2, fusion chimeras were constructed from homozygous mutant embryonic stem cells and wild-type tetraploid embryos. This allowed survival until term and revealed that Fgfr2 is required for both limb outgrowth and branching lung morphogenesis. The use of fusion chimeras demonstrated that early lethality was indeed because of trophectoderm defects and indicated that in the embryonic cell lineages Fgfr2 activity manifests in limb and lung development. Highly similar lung and limb phenotypes were detected recently in the loss of function mutation of Fgf10, a ligand of Fgfr2. It is likely, therefore, that whereas during early development Fgfr2 interacts with Fgf4, in limb and lung development interactions between Fgf10 and Fgfr2 may be required. Possible epithelial–mesenchymal interactions between the splicing alternatives of Fgfr2 and their specific ligands will be discussed.
Fibroblast growth factors (FGF) contribute to numerous developmental processes throughout embryogenesis. They are the main mediators of limb outgrowth, as shown by ectopic limb bud formation induced by FGF beads transplanted into the flank of chicken embryos (1–3) or by Fgf4 overexpression in transgenic mice (4). Additional roles were suggested for FGF4 and FGF8 in the maintenance of the progress zone and the zone of polarizing activity (ZPA) (for review, see ref. 5). Which of the 18 FGF isotypes is responsible for limb outgrowth remained, however, undefined until targeted null mutations of Fgf10 recently were reported. Targeted mutations of Fgf10 displayed complete abrogation of limb outgrowth coupled with a loss of lung branching morphogenesis. This suggested a crucial role for a single growth factor in the development of these two unrelated organs (6, 7).
Involvement of Fgf10 in limb outgrowth and lung morphogenesis raised the question: Which receptor or receptors transmit its signals in the developing limb and lung? Chimera experiments with homozygous mutant embryonic stem (ES) cells suggested a role for Fgfr1 in limb and central nervous system development (8). Involvement of Fgfr2 in limb outgrowth was indicated by a targeted mutation that displayed no limb buds but, because of placental insufficiency, did not survive beyond early limb outgrowth (9) Questions arose, therefore, of whether one or more Fgf receptors are required for limb outgrowth and whether loss of Fgfr2 function also can lead, beyond retardation, to a complete loss of limb development.
The FGF receptor or receptors that transmit Fgf10 signals during bronchial tree morphogenesis also remained unknown. Peters et al. (10) reported defective branching lung morphogenesis in a dominant negative transgenic model, where truncated Fgfr2 cDNA was expressed under an alveolar mucin promoter. Dominant negative mutations are a result of heterodimerization between wild-type and truncated receptor monomers. Because heterodimers can form between different FGFR isotypes (11–13), dominant negative FGFR mutations have no isotype specificity. Therefore, the results of Peters et al. (10), although demonstrating the involvement of FGF receptors in lung morphogenesis, failed to define the specific isotype. Another indication for the role of FGFRs in lung development is the defective alveogenesis of Fgfr3–Fgfr4 double homozygotes (14).
Despite definitive information on the role of Fgf10 in limb outgrowth and lung development, the nature of the Fgfr locus, or loci, that must also contribute to these processes remained to be determined. To investigate the exact role of Fgfr2 in these processes, it was necessary to rescue the trophectoderm defects that were responsible for early lethality in previous gene-targeting experiments (9, 15). For this, a new targeting experiment coupled with tetraploid fusion chimeras was performed (16).
Materials and Methods
Gene Targeting.
Gene targeting was performed as described previously (15), except that the osdupdel vector (gift of Oliver Smithies, University of North Carolina, Chapel Hill) was used and the selection was performed at elevated G418 concentration to create homozygosity at the mutant locus (17). MF1 (albino) noninbred mice were used as embryo donors and as recipients.
Tetraploid fusion and the aggregation of ES cells were done according to Nagy et al. (16).
Histochemistry.
Bone and cartilage preparations (18) and whole-mount (19) and histological in situ hybridization (20) were performed as described. Microphotos were taken either on a Zeiss SV11 stereo microscope or on a Zeiss Axiomat research microscope.
Results
Rescue of the Trophectoderm Defect.
In a recent gene-targeting experiment we disrupted the IIIc exon (exon 9) of Fgfr2 at an EcoRV site and deleted the transmembrane exon (exon 10), as well as exon 11 and 12, which encode its first enzymatic domain. The 3′ breakpoint was a ClaI site of exon 12. This presumptive null mutation was lethal shortly after implantation, because of trophectoderm defects (15). Further interpretation of this phenotype became possible by a detailed study of early Fgfr2 expression, which revealed exclusive trophectoderm specificity from the early blastocyst to the egg-cylinder stage (21). Because the present study aimed at gaining insights into later Fgfr2 functions that were predicted by its localization during organogenesis (22, 20), the lethal trophectoderm defect had to be overcome.
Trophectoderm defects can be rescued by aggregating homozygous mutant ES cells with tetraploid normal embryos, which preferentially repopulate the extra embryonic cell lineages (16). To this end an Fgfr2−/− ES cell line was selected at increased G418 concentration (17). In this mutation, R2Δ2, the IIIc exon of Fgfr2, was disrupted and the transmembrane exon was deleted (Fig. 1). R2Δ2 differs from our previous mutation (15) in that the first kinase domain was not altered.
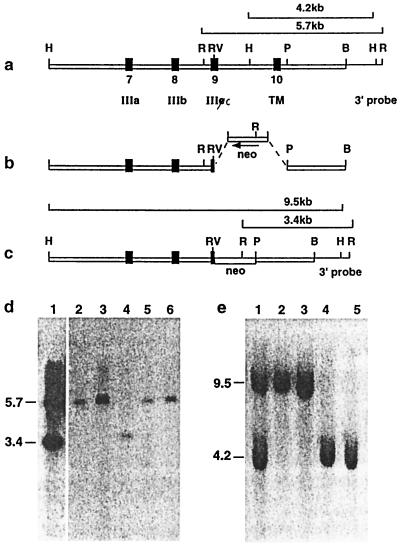
Figure 1.
The R2Δ2 mutation of Fgfr2. (a) Genomic fragment including exons 7–10 (solid boxes). Elements of the construct are boxed, whereas the 3′ BamHI-EcoRI probe is a single line. Diagnostic HindIII and EcoRI restriction enzyme fragments recognized by the 3′ probe are shown in a and c. (b) The disruption of exon 9 (IIIc) and deletion of exon 10 (transmembrane). Arrow shows the transcriptional orientation of the neo cassette. (c) The mutant allele. The positions of the diagnostic recombinant HindIII and EcoRI fragments are shown. (d) Southern blot ES cell clones. EcoRI digest: lane 4, homozygous homologous recombinant; lane 1, heterozygous homologous recombinants; lanes 2, 3, 5, and 6, wild-type ES cell clones. (e) Southern blot of ES cell clones. HindIII digest: lanes 2 and 3, homozygous homologous recombinants; lane 1, heterozygous homologous recombinant; lanes 4 and 5, wild type. IIIa, IIIb, and IIIc, exons encoding variants of the third Ig-like loop of the ligand-binding domain; TM, transmembrane domain of Fgfr2; B, BamHI; H, HindIII; P, PstI; R, EcoRI; RV, EcoRV.
Abrogation of Limb Outgrowth and Lung Development in R2Δ2−/− ↔ MF1 Fusion Chimeras.
Homozygous mutant ES cells were aggregated with wild-type tetraploid embryos. R2Δ2−/− ↔ MF1 fusion chimeras grew near to term [embryonic day (E)18.5], and their only external defect was the complete absence of limbs (Fig. 2 a and b). Survival throughout embryogenesis allowed us to conclude that this defect was not retardation, but an absolute lack of limb development (Fig. 2a). Lack of limb development first was observed in the fusion chimeras at E9.5. The only external indication of limb outgrowth was a transient mesenchymal swelling of the lateral mesoderm that could be detected by histology between E10 and E11.5 in both limb fields (Fig. 2 b, e, and f). The swellings were very indistinct, if visible at all, by external inspection. This slight hypertrophy disappeared between E11.5 and E12.5, and later no other external structure could be associated with limb outgrowth.
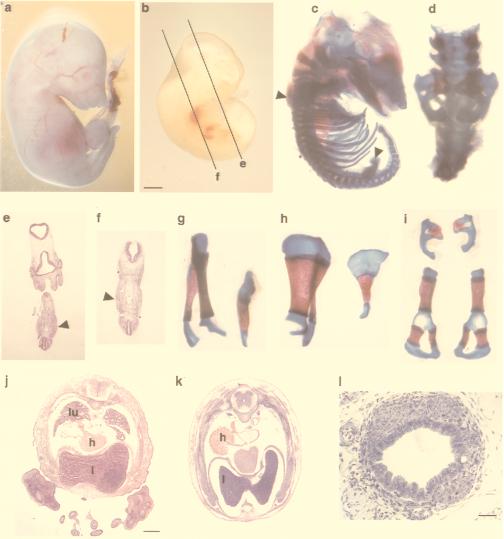
Figure 2.
Both limb and lung development are abrogated in Fgfr2−/− tetraploid fusion chimeras. (a) 18.5 dpc. (b) 11.5 dpc. Fgfr2−/− ↔ MF1 chimeras show the absence of limbs. (c, d, and g–i) Bone and cartilage preparations. Arrowheads in c show the site of the mutant scapula and pelvis. (d) Higher magnification of the pelvis in situ. Lateral (g) and dorsal (h) views of normal (Left) and mutant (Right) scapula are shown. (i) Pelvic bones (Upper, mutant; Lower, wild type). e and f show transient mesenchymal hypertrophy in the histological sections, as marked in b. (e) Hindlimb area at 11.5 dpc. (f) Forelimb area at 11.5 dpc. Sites of the hypertrophy are indicated by arrowheads. (j and k) Absence of lung development in a 14.5-dpc Fgfr2−/− ↔ MF1 chimera. (j) Wild type. (k) Mutant. (l) Cross-section of the mutant trachea displays normal histology. lu, lung; h, heart; l, liver. [Bars = 1.2 mm (b, e, and f), 1 mm (j and k), and 120 μm (l).]
Bone and cartilage preparations of E16.5 and E18.5 chimeras, however, revealed rudimentary internal structures representing the anterior and posterior limb girdles (Fig. 2 c and d). It is significant that both the early transient mesenchymal swellings and the rudimentary scapula and pelvis were located at the normal sites of fore- and hindlimb outgrowth. We assume, therefore, that the mechanism that defines the site of limb outgrowth is upstream to Fgfr2.
Histological investigation of 14.5- to 18.5-day-old chimeras revealed an additional Fgfr2 loss of function phenotype. In serial sections no lungs were detectable in Fgfr2−/− chimeras. Comparing transversal trunk sections of E14.5 mutant embryos revealed a shift in the position of the heart (Fig. 2k), as compared with the wild type (Fig. 2j). This feature was not constant in our material and could be associated with the mobility of the heart in the mutant’s more spacious thoracic cavity. The respiratory defect was restricted to the bronchial tree, because the mutant displayed the normal histological features of the trachea (Fig. 2l), which is similar to the targeted loss-of-function mutation of Fgf10 (6, 7). Taken together, the R2Δ2 mutation in the inner cell mass lineage caused an abrogation of limb outgrowth and lung morphogenesis that were manifest and complete until the end of gestation.
Both the limb and lung phenotypes of our R2Δ2−/− ↔ MF1 chimeras were similar to those reported for targeted Fgf10 mutations (6, 7). Thus, mutations of the ligand and the receptor abrogated both limb and lung development. The two phenotypes were similar also in their details. The transient mesenchymal swellings and the abnormalities of the anterior and posterior girdles, as well as those of the bronchial tree, were similar in the Fgf10 and in Fgfr2 mutations. A close relationship between these complex loss-of-function phenotypes indicated that Fgfr2 and Fgf10 may cooperate in limb outgrowth and lung development and suggests that Fgfr2 is the receptor that transmits Fgf10 signals in the morphogenesis of both organs.
Fgfr2 Is Required for the Development of Certain Components of the Complex Limb Girdles.
Bone and cartilage preparations of E16.5 and E18.5 mutant chimeras revealed an intact clavicula and a small and deformed scapula, with part of the blade of the scapula and the coracoid process missing (Fig. 2 c, g, and h). The posterior girdle, that is, the pelvic bones, were even more deformed, and we could identify them only by their position (Fig. 2 d and i). The girdles connect the limbs to the axial skeleton and allow them to lift and propel the body, which was a crucial step in adaptation to terrestrial life. They evolved from multiple bones during the terrestrial adaptation of vertebrates and underwent drastic reshaping and fusion. Thus, the present mammalian scapula and pelvis result from multiple fusions of numerous elements (23). Their rudimentary state in our mutant and, as it also appears from the figures provided by Min et al. and Sekine et al. (6, 7), in the Fgf10 mutations indicates that Fgfr2 and Fgf10 are required for the development of some, but not all, components of the limb girdles. It also follows from these findings as well as from the Fgf-induced de novo limb outgrowth in the chicken embryo (1–3) that the external limbs develop together with their respective limb girdles and, thus, the external limbs and certain girdle bones form one developmental unit regulated by FGF signals.
Changes of Gene Expression in the Limb Fields.
Abrogation of limb outgrowth by our mutation suggested that Fgfr2 is among the earliest mediators of limb development. To investigate the effects of our mutation, in situ hybridization was performed. Because the mutant did not have limb buds, normal controls had to be chosen at very early stages of limb outgrowth. For this, we used 9.25- to 9.5-days postcoitum (dpc) embryos. In agreement with the data of Xu et al. (9), Fgf10 expression was severely down-regulated and Fgf8 expression was virtually absent in the mutant. There was no difference between wild type and mutant in the expression of Hox-A7. Dlx2, en1, and shh also were studied, but their level of expression in the wild type was too low to appreciate the significance of their virtual absence in the mutant (not shown). Msx1 and Lhx2 expression, however, was significant in the wild-type limb field, whereas in the mutant the expression of both was inhibited (Fig. 3). Weak residual Msx1 expression was visible in the forelimb area of the 9.25-dpc mutant, without being detectable in the hindlimb field, whereas in the umbilical area the torn amnion and visceral endoderm were strongly labeled (Fig. 3 A and B). Lhx2 was strongly expressed in the incipient normal forelimb bud, whereas in the later-developing hindlimb area only weak signals were visible. In mutant limb fields, however, no Lhx2 transcripts could be detected (Fig. 3 C and D). In other areas, such as the central nervous system, branchial arches, and heart, both genes were expressed normally. Lhx2 was suggested to be responsible for limb outgrowth, as shown by abrogation of chicken limb development caused by a dominant negative Lhx2 construct that was introduced by retroviral transfer (24). Our data suggest that Lhx2 and Msx1 expression in the early limb bud depend on signaling through Fgfr2.
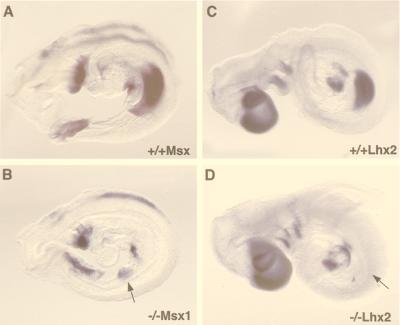
Figure 3.
Effect of the Fgfr2 mutation on the expression of Msx1 and Lhx2 in mouse embryos (9.25-dpc embryos). (A and B) Msx1 expression. (C and D) Lhx2 expression. A shows Msx1 expression in the first branchial arch and heart, the torn visceral endoderm covering the umbilical area, and the incipient forelimb bud of the wild type. Transcripts are detectable at all of these sites in the mutant (B) except the forelimb bud area, where only weak signals are seen. Lhx2 in the wild type (C) is expressed in the forebrain and facial area, the branchial arches, the heart, and the forelimb bud as well as weakly in the area of the prospective hindlimb bud. The mutant (D) is distinguished by the complete absence of Lhx2 transcripts in both limb fields. Arrows in B and D indicate the probable site of the forelimb field in the mutant.
Interactions Between the Splicing Alternatives of Fgfr2 and Their Ligands.
Previous studies indicated that the two splicing alternatives of Fgfr2, Fgfr2-IIIb and Fgfr2-IIIc, may be active in epithelial–mesenchymal interactions. This was first suggested by the preferential expression of Fgfr-IIIb in epithelial tissues and by Fgfr2-IIIc in mesenchymal tissues (20). Mesenchymal distribution of Fgf10 and its effect on ectopic limb outgrowth suggested that epithelial FGF isotypes, such as Fgf4 and Fgf8, that are localized in the apical ectodermal ridge may establish an epithelial–mesenchymal interaction with Fgf10 in the progress-zone mesenchyme (3). Deng’s laboratory, based on reported ligand-binding specificity of Fgfr2 and its splice variants (25), connected the limb-outgrowth defect of their Fgfr2 mutation with interactions between the splicing alternatives of Fgfr2 and their various ligands, and they proposed that Fgf10 in the limb mesenchyme interacts with Fgfr2-IIIb in the surface ectoderm, whereas Fgf8 in the epithelial apical ectodermal ridge interacts with Fgfr2-IIIc in the progress-zone mesenchyme (9). We reinvestigated the expression of the splicing alternatives of Fgfr2 (20).
The primary aim of the present in situ hybridization experiments was to investigate whether interaction loops also may exist in lung morphogenesis. We found here, too, that Fgfr2-IIIb is expressed in an epithelium in the bronchial epithelium of the developing lung (Fig. 4h), whereas transcripts of the mesenchymal Fgfr2-IIIc alternative were localized mainly to the mesenchyme (Fig. 4i). We argued that because Fgf10, which is chemotactic for developing epithelial lung buds (26), is expressed in the lung mesenchyme (27), a mesenchyme-to-epithelium interaction between Fgf10 and Fgfr2-IIIb could take place. This mesenchyme-to-epithelium interaction then would represent the first link of the interaction loop of lung development (Table 1). It was less clear whether, as the second link of interactions in lung development, signaling could originate from an epithelial Fgf isotype toward the mesenchymal Fgfr2-IIIc receptor. An epithelial Fgf isotype indeed does exist in the lung, because we have detected the expression of Fgf9 transcripts in the pleura of the developing lung (Fig. 4 j and k). Because Fgf9 is a ligand of Fgfr2-IIIc (25), an Fgf9-to-FGFR2-IIIc interaction could represent the second link. Therefore, a similar interaction loop may be active in lung and limb development (Table 1). These as well the published (9, 15) Fgfr2 mutations affect the entire locus, including transcriptional alternatives; hence, comprehensive analysis of their ligand–receptor interactions will require exon-specific gene targeting.
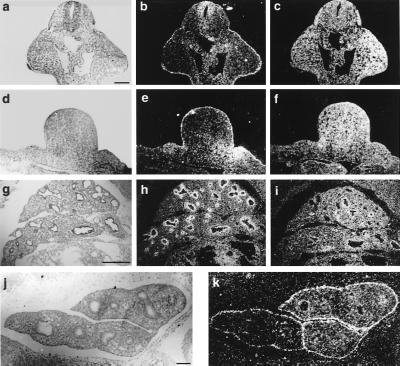
Figure 4.
Localized transcription of Fgfr2-IIIb, IIIc, and Fgf9. (a, d, g, and j) Bright-field illumination. (b, c, e, f, h, i, and k) Dark-field illumination. (a–c) Coronal section of forelimb buds. (d–f) Transversal section of left hindlimb bud, 10.5 dpc. (g–i) Lobes of right lung, 11.5 dpc. (j and h) Lung Fgfr2-IIIb is expressed in the surface ectoderm of the fore- and hindlimbs (b and e), as well as in the bronchial epithelium (h). Note increased Fgfr2-IIIb expression in the posterior area of the hindlimb bud (e). Fgfr2-IIIc is expressed in the mesenchyme of limb buds (c and f) and lung (i). Fgf9 is expressed in the surface ectoderm or pleura of the developing lung (k). [Bars = 150 μm (a–f), 200 μm (g–i), and 180 μm (j and k).]
Table 1.
Epithelial–mesenchymal circuits in limb outgrowth and lung morphogenesis
| Site | Transcriptional localization
|
||
|---|---|---|---|
| Epithelium | Mesenchyme | ||
| Limb outgrowth | Fgfr2-IIIb | ← | Fgf10 |
| Fgf8 and 4 | → | Fgfr2-IIIc | |
| Lung morphogenesis | Fgfr2-IIIb | ← | Fgf10 |
| Fgf9 | → | Fgfr2-IIIc | |
Discussion
Rescuing the trophectoderm defect in our Fgfr2 mutation led to phenotypes in limb and lung. This is strong evidence for the trophectoderm specificity of the early defects displayed by both targeted Fgfr2 mutations (9, 15). Although limb and lung defects were the only manifestations of Fgfr2 loss of function in the embryonic cell lineages, Fgfr2 is expressed in the development of multiple organs, in addition to limbs and the lung (20, 22). Moreover, its dominant mutations lead to abnormal congenital bone development in man (28). It is therefore likely that this receptor is involved in additional processes of organogenesis besides lung and limb development. Nevertheless, Fgfr2 is unlikely to be their only mediator, because they were not affected by the targeted recessive loss-of-function mutations (9, 15). Thus, although Fgfr2, together with other Fgfr isotypes, may contribute to multiple events of morphogenesis, its specific roles in certain aspects of limb outgrowth and in the morphogenesis of the bronchial tree are absolutely required and may not normally be replaced by other Fgfr isotypes. This is not to debate the involvement of other Fgfrs in limb and lung development, because Fgfr1−/− ES cell chimeras displayed neural tube and limb defects (8), targeted disruption of Fgfr3 caused limb bone overgrowth (29, 30), and Fgfr3–Fgfr4 double mutants displayed alveolar defects (14).
It seems significant to us that all three unique functions of Fgfr2 are connected to events in the evolution of higher vertebrates. Its trophectoderm-specific early functions take place in the extraembryonic tissues, and, thus, they are amniote characteristics (15, 21). Its involvement in limb and limb-girdle development points to a role of Fgfr2 in the adaptation of semiaquatic forms to terrestrial life and so does its importance for lung morphogenesis. It is therefore possible that FGFR2, which differs in its size from all other FGF receptors (31), arose as a specific adaptation. Alternatively, its regulation may have been modified at an advanced stage of vertebrate evolution.
Previous data suggest that Fgfr2 in the trophectoderm of the early embryo interacts with Fgf4, which is expressed in the inner cell mass (15). The present findings indicate that in inner cell- mass-derived embryonic lineages, Fgfr2, with Fgf10 as its ligand, mediates the outgrowth of the single epithelial bud of the limb as well as the complex bronchial tree. The epithelial–mesenchymal FGF-FGFR circuits of the mammalian limb and lung (Table 1) include highly conserved interactions. An FGFR homologue is responsible for morphogenic cell migration in the insect trachea (32, 33). Moreover, epithelial buds expressing the receptor grow toward their ligand in adjacent cell layers (34), both in the tracheal organ of Drosophila and in the mammalian lung (27). Hence, epithelial–mesenchymal interactions between FGF and FGFR may be a general paradigm for FGF-mediated developmental signaling.
Acknowledgments
We thank Mrs. Marina Gerstenstein (Lautenberg Institute, Toronto) for teaching us tetraploid fusion, Dr. Avi Orr-Urtreger (Institute of Genetics, Rabin Medical Center, Tel Aviv) for help with in situ hybridization, and Professor Benny Shilo of our department for comments on the manuscript. This work was supported by a Research Center Grant of the Israel Science Fund and by an Infrastructure Grant of the Ministry of Science.
Abbreviations
- FGF
fibroblast growth factor
- ES
embryonic stem
- E
embryonic day
- dpc
days postcoitum
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Cohn M J, Izpisua-Belmonte J C, Abud H, Heath J K, Tickle C. Cell. 1995;80:739–746. doi: 10.1016/0092-8674(95)90352-6. [DOI] [PubMed] [Google Scholar]
- 2.Crossley P H, Minowada G, MacArthur C A, Martin G R. Cell. 1996;84:127–136. doi: 10.1016/s0092-8674(00)80999-x. [DOI] [PubMed] [Google Scholar]
- 3.Ohuchi H, Nakagawa T, Yamamoto A, Araga A, Ohata T, Ishimaru Y, Yoshioka H, Kuwana T, Nohno T, Yamasaki M, et al. Development (Cambridge, UK) 1997;124:2235–2244. doi: 10.1242/dev.124.11.2235. [DOI] [PubMed] [Google Scholar]
- 4.Abud H, Skinner J A, McDonald F J, Bedford M T, Lonai P, Heath J K. Dev Genet. 1996;181:51–65. doi: 10.1002/(SICI)1520-6408(1996)19:1<51::AID-DVG6>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 5.Martin G R. Genes Dev. 1998;12:1571–1586. doi: 10.1101/gad.12.11.1571. [DOI] [PubMed] [Google Scholar]
- 6.Min H, Danilenko D M, Scully S A, Bolon B, Ring B D, Tarpley J E, DeRose M, Simonet W S. Genes Dev. 1998;12:3156–3161. doi: 10.1101/gad.12.20.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sekine T, Ohuchi H, Fujiwara M, Yamasaki M, Yoshizawa T, Sato T, Yagishita N, Matsui D, Koga Y, Itoh N, et al. Nat Genet. 1999;21:138–141. doi: 10.1038/5096. [DOI] [PubMed] [Google Scholar]
- 8.Deng C, Bedford M, Li C, Xu X, Yang X, Dunmore J, Leder P. Dev Biol. 1997;185:42–54. doi: 10.1006/dbio.1997.8553. [DOI] [PubMed] [Google Scholar]
- 9.Xu X, Weinstein M, Li C, Naski M, Cohen R I, Ornitz D M, Leder P, Deng C. Development (Cambridge, UK) 1998;125:753–765. doi: 10.1242/dev.125.4.753. [DOI] [PubMed] [Google Scholar]
- 10.Peters K, Werner S, Liao X, Wert S, Whitsett J, Williams L. EMBO J. 1994;13:3296–3301. doi: 10.1002/j.1460-2075.1994.tb06631.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bellot F, Crumley G, Kaplow J M, Schlessinger J, Jaye M, Dionne C A. EMBO J. 1991;10:2849–2854. doi: 10.1002/j.1460-2075.1991.tb07834.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ueno H, Gunn M, Dell K, Tseng A J, Williams L. J Biol Chem. 1992;267:1470–1476. [PubMed] [Google Scholar]
- 13.Celli G, LaRochelle W J, Mackem S, Merlino G. EMBO J. 1998;17:1942–1655. doi: 10.1093/emboj/17.6.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weinstein, M., Xu, X., Ohyama, K. & Deng, C.-X. (1998) 125, 3615–3623. [DOI] [PubMed]
- 15.Arman E, Haffner-Krausz R, Chen Y, Heath J K, Lonai P. Proc Natl Acad Sci USA. 1998;95:5082–5087. doi: 10.1073/pnas.95.9.5082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nagy A, Rossant J, Abramow-Newerly W, Roder J C. Proc Natl Acad Sci USA. 1993;90:8424–8428. doi: 10.1073/pnas.90.18.8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mortensen R M, Conner D A, Chao S, Geisterfer-Lowrance A A T, Seidman J G. Mol Cell Biol. 1992;12:2391–2395. doi: 10.1128/mcb.12.5.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaufman M H. The Atlas of Mouse Development. London: Academic; 1992. [Google Scholar]
- 19.Conlon R A, Herrmann B G. Methods Enzymol. 1992;225:361–372. doi: 10.1016/0076-6879(93)25026-x. [DOI] [PubMed] [Google Scholar]
- 20.Orr-Urtreger A, Bedford M T, Burakova T, Arman E, Zimmer Y, Yayon A, Givol D, Lonai P. Dev Biol. 1993;158:475–486. doi: 10.1006/dbio.1993.1205. [DOI] [PubMed] [Google Scholar]
- 21.Haffner-Krausz R, Gorivodsky M, Chen Y, Lonai P. Mech Dev. 1999;85:167–172. doi: 10.1016/s0925-4773(99)00082-9. [DOI] [PubMed] [Google Scholar]
- 22.Orr-Urtreger A, Givol D, Yayon A, Yarden Y, Lonai P. Development (Cambridge, UK) 1991;113:1419–1434. doi: 10.1242/dev.113.4.1419. [DOI] [PubMed] [Google Scholar]
- 23.Jarvik E. Basic Structure and Evolution of Vertebrates. London: Academic; 1980. [Google Scholar]
- 24.Rodriguez-Esteban C, Schwabe J W R, De La Pena J, Rincon-Limas D E, Magallon J, Botas J, Izpisua Belmonte J C. Development (Cambridge, UK) 1998;125:3925–3934. doi: 10.1242/dev.125.20.3925. [DOI] [PubMed] [Google Scholar]
- 25.Ornitz D M, Xu J, Colvin J S, McEwen D G, MacArthur C A, Coulier F, Gao G, Goldfarb M. J Biol Chem. 1996;271:15292–15297. doi: 10.1074/jbc.271.25.15292. [DOI] [PubMed] [Google Scholar]
- 26.Park W Y, Miranda B, Lebeche D, Hashimoto G, Cardoso W V. Dev Biol. 1998;201:125–134. doi: 10.1006/dbio.1998.8994. [DOI] [PubMed] [Google Scholar]
- 27.Bellusci S, Grindley J, Emoto H, Itoh N, Hogan B L M. Development (Cambridge, UK) 1997;124:4867–4878. doi: 10.1242/dev.124.23.4867. [DOI] [PubMed] [Google Scholar]
- 28.Wilkie A O M, Morriss-Kay G M, Jones E Y, Heath J K. Curr Biol. 1995;5:500–507. doi: 10.1016/s0960-9822(95)00102-3. [DOI] [PubMed] [Google Scholar]
- 29.Colvin J S, Bohne B A, Harding G W, McEwen D G, Ornitz D M. Nat Genet. 1996;12:390–397. doi: 10.1038/ng0496-390. [DOI] [PubMed] [Google Scholar]
- 30.Deng C, Wynshaw-Boris A, Zhou F, Kuo A, Leder P. Cell. 1996;84:911–921. doi: 10.1016/s0092-8674(00)81069-7. [DOI] [PubMed] [Google Scholar]
- 31.Twigg S R F, Oldridge M, Heath J K, Wilkie A O M. Hum Mol Gen. 1998;7:685–691. doi: 10.1093/hmg/7.4.685. [DOI] [PubMed] [Google Scholar]
- 32.Klambt C, Glazer L, Shilo B-Z. Genes Dev. 1992;6:1668–1678. doi: 10.1101/gad.6.9.1668. [DOI] [PubMed] [Google Scholar]
- 33.Reichman-Fried M, Dickson B, Hafen E, Shilo B-Z. Genes Dev. 1994;8:428–439. doi: 10.1101/gad.8.4.428. [DOI] [PubMed] [Google Scholar]
- 34.Sutherland D, Samakovlis C, Krasnow M. Cell. 1996;87:1091–1101. doi: 10.1016/s0092-8674(00)81803-6. [DOI] [PubMed] [Google Scholar]