Abstract
Programmed ribosomal frameshifting is often used by viral pathogens including HIV. Slippery sequences present in some mRNAs cause the ribosome to shift reading frame. The resulting protein is thus encoded by one reading frame upstream from the slippery sequence and by another reading frame downstream from the slippery sequence. Although the mechanism is not well understood, frameshifting is known to be stimulated by an mRNA structure such as a pseudoknot. Here, we show that the efficiency of frameshifting relates to the mechanical strength of the pseudoknot. Two pseudoknots derived from the Infectious Bronchitis Virus were used, differing by one base pair in the first stem. In Escherichia coli, these two pseudoknots caused frameshifting frequencies that differed by a factor of two. We used optical tweezers to unfold the pseudoknots. The pseudoknot giving rise to the highest degree of frameshifting required a nearly 2-fold larger unfolding force than the other. The observed energy difference cannot be accounted for by any existing model. We propose that the degree of ribosomal frameshifting is related to the mechanical strength of RNA pseudoknots. Our observations support the “9 Å model” that predicts some physical barrier is needed to force the ribosome into the −1 frame. Also, our findings support the recent observation made by cryoelectron microscopy that mechanical interaction between a ribosome and a pseudoknot causes a deformation of the A-site tRNA. The result has implications for the understanding of genetic regulation, reading frame maintenance, tRNA movement, and unwinding of mRNA secondary structures by ribosomes.
Keywords: macromolecular mechanics, optical tweezers, protein synthesis, single molecules, translation
When an mRNA sequence is translated into protein by the ribosome, the nucleotide sequence is read in codons of three nucleotides and hence the mRNA in principle has three reading frames. In the vast majority of mRNAs, only one reading frame, defined by the initiation codon, is exploited and translated into protein. The elongation phase of protein synthesis is a precise process and intrinsic mechanisms exist in the ribosome to enhance translational fidelity (1, 2). The frequency of frameshift errors has been estimated to <3 × 10−5 (3, 4). However, many examples of naturally occurring and highly efficient programmed frameshift sites have been described (5, 6). There is considerable interest in how ribosomal frameshift occurs, as this may provide insight into mechanisms behind reading frame maintenance, tRNA movement, and unwinding of mRNA secondary structures by ribosomes. Typically, a −1 frameshift site comprises two elements, a slippery sequence, X XXY YYZ, where the frameshifting occurs, and additionally, a stimulatory RNA element positioned downstream in the mRNA (7). Frameshifting is thought to happen by dual tRNA slippage. In the original zero reading frame, the P-site tRNA and the A-site tRNA pair to codons XXY and YYZ, respectively, whereas after the shift to the −1 frame they pair to XXX and YYY. At the new position, the tRNAs remain paired to mRNA at the two most upstream XX and YY nucleotides in each codon.
Examples of stimulatory elements include downstream self pairing mRNA sequences called mRNA pseudoknots (Fig. 1). The requirement for these elements to function is a placement at a proper distance from the slippery sequence. In many viral frameshift sites, the stimulatory element is a pseudoknot positioned 6–9 nt downstream of the slippery sequence.
Fig. 1.
Pseudoknots and frameshift sites. (a) Sequences of frameshift sites encoded in plasmids pTH400, pTH401, and pTH421. The slippery sequence is underlined, and the stop codon UAA is marked by a black “stop sign.” pTH400 and pTH401 encode the pseudoknots PK400 and PK401. The nucleotides that can form double-stranded stems are underlined by arrows. Stem1 is indicated by black arrows, and stem2 is indicated by gray arrows. The three nucleotides present only in PK401 are in bold. (b) Schematic drawing of an mRNA where the ribosome is positioned with the slippery sequence in its A and P sites. The secondary structure of PK401 is indicated with coaxially stacked stems and single-stranded loops. Differences from PK400 are indicated by arrows. (c) A schematic drawing of a pseudoknot.
The mechanism of frameshift stimulation by pseudoknots is not well understood. Involvement of protein factors binding to the mRNA seems unlikely because, in a competition experiment, addition of excess RNA pseudoknots did not affect frameshift efficiencies (8). Furthermore, that many pseudoknot-stimulated programmed frameshifts function in heterologous organisms from different kingdoms of life makes it unlikely that the function requires transacting factors. It has been suggested that the stimulatory structure pauses the ribosome while the slippery sequence is positioned in the decoding site of the ribosome, thus increasing the chance of tRNA slippage (9). However, the data from measurements of ribosomal pausing with pseudoknots, mutated pseudoknots, and related stem-loops support the view that pausing alone cannot mediate frameshifting and that additional events are required (10, 11). The programmed frameshift in infectious bronchitis virus (IBV) has been investigated, and it was found that pseudoknots, but not similar stem-loop structures, stimulate efficient frameshifting (10).
As shown in Fig. 1c, a pseudoknot can be viewed as a stem-loop where nucleotides in the loop form a second stem with downstream mRNA. This may lock or decrease the rotational freedom of the first stem, and hence induce supercoiling while the ribosome unfolds the first stem. Experimental data support a role for torsional restraint in positioning the ribosome to pause with the slippery sequence in the A- and P-site when unfolding pseudoknots (12). This model makes it clear that an optimal spacing of 6–9 nt between slippery sequence and pseudoknot is crucial and positions the pseudoknot close to the entrance of the mRNA tunnel of the ribosome.
Recently, the “9 Å model” was suggested for the mechanism of frameshift stimulation (13). Structural studies have revealed a 9-Å movement by the anti-codon loop of the aminoacyl-tRNA between the state of initial binding and the fully accommodated position (reviewed in ref. 14). It is expected that the codon::anti-codon bound mRNA is pulled a similar distance further into the ribosome (13). The authors suggested that a downstream mRNA pseudoknot would provide resistance to this movement by becoming wedged into the entrance of the ribosomal mRNA tunnel. These two opposing forces result in the creation of a local region of tension in the mRNA between the A-site codon and the mRNA pseudoknot. The tension can be relieved by one of two mechanisms: unwinding of the pseudoknot, allowing the ribosome to move forward, or slippage of the proximal region of the mRNA backwards by one base. Even if it slips backwards one base, then still, afterward, it will have to unwind the pseudoknot to move forward.
In this model, the stability of pseudoknots should play an important role in stimulation of frameshift. Of course, one crucial question is how “stability” is defined. A correlation has not been found between the frequency of frameshifting and the difference in Gibbs free energy between folded and unfolded pseudoknots measured from UV optical melting profiles (15). When the pseudoknot is opened by a ribosome, the action might not be thermodynamically reversible i.e., the work performed by the ribosome might be larger than ΔG, and some fraction of the work might be dissipated irreversibly.
In an attempt to simulate the action of a ribosome, we mechanically unfold the pseudoknot using optical tweezers. By applying a load on the structure, it is forced to unfold. This type of experiment is similar to previous studies on RNA hairpin folding (16, 17). The pseudoknots are unfolded at a nonzero force-loading rate and, hence, in general, do not unfold or refold through an equilibrium process. Here, we first determine the degree of frameshift stimulation effected by two pseudoknots, then focus on the mechanical unfolding and refolding events for the two different pseudoknots and for a control RNA. From the unfolding mechanics, the energetics of the process are considered. Finally, we show how the rates of frameshifting for the two investigated pseudoknots correlate with the mechanical stability of the pseudoknots.
Results
Description of Frameshift Sites.
In this work, we investigated an artificial site of programmed ribosomal frameshift resembling that of IBV, which have been studied extensively by Brierley and coworkers (10, 18–20). As a typical natural −1 frameshift site, our site includes a slippery sequence and a stimulatory element, which in this case is a 3′ pseudoknot (Fig. 1). The slippery sequence we used was UUUAAAG rather than UUUAAAC found in IBV, because XXXAAAG is found to be a more efficient slippery sequence in Escherichia coli (7), the organism we used for the measurements of in vivo frame shift efficiencies.
The choice of model pseudoknot was inspired by the work of Napthine et al. (20). They measured frameshift efficiencies in rabbit reticulocyte extracts of a series of IBV-derived pseudoknots with different lengths of stem1 sequences. Remarkably, frameshift efficiencies decreased 7-fold when the length of stem1 was reduced from the wild-type length of 11 bp to 10 bp. However, when the Napthine et al. (20) performed a structure probing analysis, both RNAs formed pseudoknots and appeared indistinguishable in conformation. Also, the predicted ΔG for stem1 of the wild-type IBV and IBV-derived pseudoknots with 11 bp and 10 bp stem1 did not correlate with the differences in frameshifting efficiency (20). In this work, we chose to compare two IBV-like pseudoknots with 11 bp and 10 bp in their stem1 (see Fig. 1 and Materials and Methods). For those two pseudoknots, we examined the frameshift efficiency in vivo and the mechanical stability in single molecule experiments. Rather than using the exact same structures as Napthine et al., the pseudoknots in this work have longer loop2 sequences, as in the wild-type IBV pseudoknot. Apart from making our experiments closer to the wild-type situation, this also increased the difference in length for folded and unfolded pseudoknots for easier spatial detection.
Pseudoknot Stimulated Frameshifts in E. coli.
Three plasmid constructs were made for measurements of frameshift efficiencies. All three encode the slippery sequence and a 6-nt spacer (Fig. 1). This sequence is followed by either a pseudoknot with 11 bp in stem1, a pseudoknot with 10 bp in stem1, or no pseudoknot as a control. The shortest pseudoknot is henceforth named “PK400,” the longer pseudoknot is named “PK401,” and the control is named “PK421.” DNA oligonucleotides encoding these elements were inserted in the end of an orf that originates from bacteriophage T7 gene10 (see Materials and Methods). Translation of the gene10 orf and the slippery sequence without frameshift will lead to termination at a UAA stop codon in the spacer between slippery sequence and the pseudoknot. The result is the release of a 28-kDa termination product. If the ribosomes shift to read the −1 frame at the slippery sequence, the UAA stop codon is out of frame, and translation continues through the pseudoknot and into a lacZ orf. The −1 frame shift allows continuous translation to the end of the lacZ orf and results in a 147-kDa frameshift product. The relative amounts of termination and frame shift products were used to estimate the frameshift frequencies. The proteins were metabolically labeled by addition of [35S]methionine in pulse–chase experiments with cultures of E. coli expressing the individual constructs (see Materials and Methods). After harvesting the cultures, the proteins were separated by gel electrophoresis and visualized by autoradiography (Fig. 2). Prominent bands corresponding to the termination and frameshift products were identified by comparison of proteins harvested from IPTG-induced and uninduced cultures. IPTG is a specific inducer of transcription from the ptac promoter driving the expression of the plasmid encoded hybrid T7 gene10::lacZ gene fusions. The presence of the frameshift products in cultures of PK400 and PK401 indicates efficient ribosomal frameshift with the pseudoknots encoded in the construct. However, for the control culture, containing the gene fusion without a pseudoknot, only the shorter termination product could be detected thus confirming the dependency of frameshifting at slippery sequences on the presence of stimulatory pseudoknots. The frameshift frequencies were quantified by measuring the relative amounts of frame shifted and terminated proteins in the individual lanes (see Materials and Methods). We found that the PK401 construct, encoding the pseudoknot with an 11-bp stem1, as in the wild-type IBV pseudoknot, gave the highest frequency of frameshift, namely 14 ± 1.5% (mean ± SEM). The pseudoknot with the shorter 10-bp stem1 yielded 5.9 ± 0.40% frameshift, a >2-fold reduction caused by a difference by a single base pair in stem1. The PK421 control construct, which does not contain a pseudoknot, yielded a frameshift frequency below our detection limit of ≈0.5% frameshift.
Fig. 2.
Frameshift assay. Autoradiogram of SDS-polyacrylamide gel showing proteins metabolically labeled by [35S]methionine. Expression from plasmids of genes encoding frameshift sites are induced by the presence of IPTG. For the constructs encoding pseudoknots, PK400 or PK401, induction leads to synthesis of two proteins (see arrows): one protein from ribosomes terminated at the zero frame stop codon, which follows the slippery sequence, and another protein from ribosomes which frameshifted to the −1 frame. Only the termination product is detected for the construct with no pseudoknot, PK421. Percent frameshift is given above each lane.
Mechanical Unfolding and Refolding of Pseudoknots.
The mechanical unfolding of pseudoknots was performed with an optical trap by exerting a stretching force on individual mRNA molecules containing the structure-forming sequences.
Tethers of RNA pseudoknots PK400 or PK401 with DNA handles were formed (see Materials and Methods) with one end of the tether specifically attached by a biotin–streptavidin bond to a bead held by an optical trap while the other end of the tether was specifically attached by a digoxygenin–antidigoxygenin bond to another bead held by a micropipette (Fig. 3). The optical trap and the detection system has been described (21, 22) and were used to measure the forces acting on the bead in the trap. The optical tweezers exert a harmonic force, Ftrap, on the trapped particle, Ftrap = ktrap x, where x is the deviation from the equilibrium position and ktrap is denoted the trap stiffness. For the positions visited by the bead in the trap, ktrap can be considered constant and found by proper calibration (22, 23). Hence, with x measured Ftrap is also known. During a stretching or relaxing experiment, the trap was stationary while the micropipette was moved by a piezo electric stage. Several cycles of stretching and relaxing were performed for each molecule.
Fig. 3.
Mechanical stretching setup. Schematic drawing of the experiment. (a) The RNA with complementary DNA handles is attached to beads with biotin-streptavidin and digoxygenin-antidigoxygenin bonds. The RNA/DNA heteroduplexes are 426 and 415 bp, respectively, and leave the middle region of the RNA free to form tertiary structures. The possible nucleotide sequences of the middle region are listed in Fig. 1a. (b) One bead is placed in the force measuring laser trap, whereas the other bead is attached to a micropipette. The micropipette was moved with respect to the laser trap to stretch and relax the molecule. The image is drawn to scale.
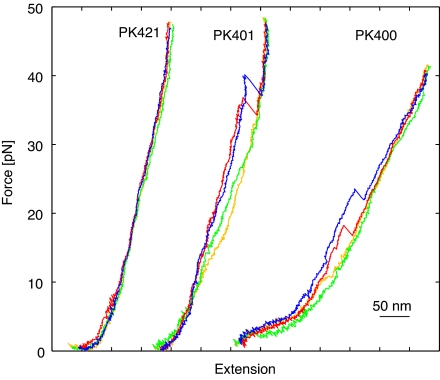
Fig. 4 shows typical force-extension curves from stretching and relaxing cycles of the two pseudoknots PK400 and PK401 as well as of the PK421 control. However, the stretching–relaxing curves can have quite different appearances, and more curves are shown in supporting information (SI). For PK400, the rip-data presented stems from 32 stretching curves and for PK401 100 rip events were analyzed. For both PK400 and PK401, a sudden elongation of the molecules is observed in the stretching curve at 31 ± 1.9 pN and 39 ± 1.5 pN (mean ± SEM), respectively, corresponding to an unfolding of the pseudoknot. The distributions of unfolding forces are shown in Fig. 5a. Henceforth, this sudden elongation will be termed a rip. A Student's t test showed that the rupture force of PK400 is significantly lower than the rupture force of PK401. At the loading rate used (≈10 pN/s), the variation in unfolding forces was relatively broadly distributed with standard deviations of 10 and 15 pN for PK400 and PK401, respectively. We used only two criteria to determine whether the structure being stretched was, indeed, an RNA tether containing a pseudoknot; however, other criteria were also tested (see Materials and Methods). The final numbers of the average unfolding forces depended slightly on the chosen criteria, but regardless of the criteria used, the unfolding force of PK400 was always significantly lower than the unfolding force of PK401.
Fig. 4.
Stretch and relax curves of single molecules. Force and change in extension were measured in several cycles of stretching and relaxing of a single molecule. Here, data from two cycles are shown for pseudoknot containing RNA, PK400 and PK401, and for the no-pseudoknot control PK421. Red, first stretch; yellow, first relax; blue, second stretch; green, second relax.
Fig. 5.
Histograms of unfolding forces and rip lengths. The distributions of unfolding forces (a) and rip lengths (b) are shown in histograms. The values were estimated from individual stretching curves of PK400 and PK401 RNAs. Also, Gaussian curves are drawn by using the calculated mean and standard deviation from the data sets.
Most often, only one rip is observed in the stretching and relaxing curves, thus indicating that the pseudoknots seemed to unfold in a single step. Only in 4 of the 32 analyzed stretching traces originating from PK400 a close inspection of the force versus time traces shows an intermediate state (SI Fig. 6). For these traces, it is noteworthy that, consistently in all data sets, the first unfolding part is longer (12 nm ± 2.4 nm) than the second (9 nm ± 2.8) (mean ± SD), although the difference is not statistically significant. None of the 100 analyzed traces from unfolding of PK401 showed an intermediate state.
Qualitatively, the relaxation curves differ substantially. For both pseudoknots, we grouped the relax traces into five categories, depending on how the relax curve fitted with the corresponding stretching curve: (i) One clear re-folding transition, (ii) one smaller refolding event plus a gradual refolding (SI Fig. 7a), (iii) two clear refolding events (SI Fig. 7b), (iv) only gradual and slow refolding (SI Fig. 7c), and (v) none of the others. The distribution of relaxation curves into these categories is given in Table 1.
Table 1.
Nature and distribution of refolding traces
| Type of knot, no. of traces | One clear step, % | One smaller step plus gradual refolding, % | Two clear refolding steps, % | Only gradual refolding, % | None of the other categories, % |
|---|---|---|---|---|---|
| PK400, n = 22 | 5.7 | 26 | 8.0 | 39 | 22 |
| PK401, n = 73 | 25 | 20 | 18 | 28 | 8.9 |
Clearly, the category iii in Table 1 shows that in a substantial fraction of the refolding traces, there are two distinguishable events. For PK401, these two distinguishable events happen at 9 ± 4 pN and 19 ± 4 pN (mean ± SD), respectively. This might originate from a two step refolding of the pseudoknot, folding, e.g., stem 1 first and then stem 2. The refolding data from PK400 was too sparse to allow for a similar analysis of the forces.
Rip Lengths.
Fig. 5b shows the distribution of riplengths during unfolding of the pseudoknots. The mean rip lengths were 17 ± 1.2 nm for the PK400 and 21 ± 1.0 nm (mean ± SEM) for the PK401 pseudoknots. These values are also given in Table 2 in the Rexp column. A Student's t test shows that these numbers are significantly different.
Table 2.
Experimental and calculated thermodynamical parameters for mechanical unfolding of the two RNA pesudoknots
| Pseudoknot | ΔGtheory0, kJ/mol | Wstr,theory, kJ/mol | ΔGtotal,theory, kJ/mol | Rtheory, nm | Rexp, nm | Wtotal, kJ/mol |
|---|---|---|---|---|---|---|
| PK400 | 151 | 111 | 262 | 27.8 | 17 ± 1.2 | 317 ± 48 |
| PK401 | 165 | 127 | 292 | 30.1 | 21 ± 1.0 | 501 ± 36 |
ΔGtheory0 is the standard Gibbs free energy of unfolding the pseudoknot at 37°C in 1 M Na+ obtained from http://bibiserv.techfak.uni-bielefeld.de/pknotsrg/. Wstr,theory is the calculated work it takes to stretch the RNA tether from zero to the unfolding force using Eq. 1. ΔGtotal,theory = ΔGtheory0 + Wstr,theory. Rtheory is the calculated change in length of the RNA tether during the unfolding process, and Rexp is the similar value experimentally measured. Wtotal is the area underneath the F − x curve during unfolding.
Because the standard worm like chain (WLC) model (24) only holds for the forces below 10 pN, the extensible worm like chain (EWLC) model inspired by Odijk (25) was used in this analysis. According to the EWLC model, the force-extension relation can be written as (26)
where kB, T, K, LP, x, and F are Boltzmann constant, absolute temperature, stretch modulus, persistent length, end to end distance, and stretching force, respectively. A theoretical estimation of rip length was calculated from Eq. 1, setting the stretch modulus K to 1 nN (26), the persistence length LP to 1 nm (27) and the contour length of single-stranded RNA L to 0.59 nm/nt (27). The original spatial extension of the pseudoknots was calculated by approximating their 3D structure as helixes of 16 and 17 bp with a rise of 0.28 nm/bp as in a canonical RNA A-helix (28). The pseudoknots and RNA/DNA duplex handles are much stiffer than the rest of the structure and were treated as nonelastic. Based on these approximations, the predicted rip lengths, Rtheory, are 27.8 and 30.1 nm for pseudoknots PK400 and PK401, respectively. Thus, the observed rip length is substantially shorter than the predicted rip length.
Discussion
In this work, two pseudoknots, differing only by 3 nt, have been compared by single-molecule stretching experiments and a ribosomal frameshift assay in living cells. The pseudoknots efficiently stimulated frameshift in vivo with a 2-fold difference in frequency. It seems reasonable that frameshifting efficiency does not depend on the pseudoknot only, but also on the primary sequence and the context, e.g., the slippery sequence, the spacing, and the translation system including ribosomes and tRNAs. Thus, a 7- to 8-fold difference was observed when a set of pseudoknots, resembling PK400 and PK401, was tested by in vitro translation in a mammal (rabbit) reticulocyte lysate (20).
In an attempt to mimic the action of a ribosome, RNA tethers including one of the two pseudoknots were mechanically unfolded using optical tweezers. The unfolding happened most often in a single step and the unfolding forces were found to be significantly different for the two pseudoknots. The pseudoknot giving rise to the highest degree of frameshifting (PK401) also demanded for the largest unfolding force. Hence, the unfolding force appears to correlate with the degree of frameshifting that the pseudoknot gives rise to. The “looser” pseudoknot, PK400, was seen occasionally (in 12.5% of the cases) to unfold through an intermediate step. The unfolding distances of the two steps were almost equal and in accordance with, for example, first the unfolding of stem2 and thereafter stem1. For PK401, the mechanically “stronger” pseudoknot, no similar intermediates were observed in the rip characterizing the unfolding. More often, the refolding traces, of both PK400 and PK401, showed intermediate steps.
The control molecule with no pseudoknots encoded was found to yield stretch and relax curves without rips but otherwise identical to the pseudoknot containing RNAs (Fig. 4). Seen from a polymer physics point of view, the force-extension behavior of the control tether is nontrivial, as it consists of two DNA–RNA hybrid handles with a small section in the middle consisting of ssRNA (27 nt). The contour lengths of the DNA–RNA handles are only a few times their persistence lengths, and hence, it does not make sense to fit worm-like chain or similar theories to this part of the data.
Thermodynamics.
The total work of unfolding the pseudoknot was found from the experimental data as the area under force-extension curve at the unfolding region, the obtained values are given as Wtotal in Table 2. During the process, the pseudoknot is both unfolded and somewhat stretched by the applied force. Wtotal includes both the irreversible and reversible parts of the work of the unfolding process as well as the work of stretching the single-stranded RNA segment at that particular force. This experimentally obtained total work can be compared with the theoretically predicted estimates of the free-energy cost of unfolding a pseudoknot plus stretching it; theoretical estimates of the change in free energy of unfolding the structures, ΔG°theory, were calculated by the pknotsRG algorithm (ref. 29 and http://bibiserv.techfak.uni-bielefeld.de/pknotsrg/), which works at 37°C and 1 M Na+ (results are shown in Table 2). The experimental conditions were somewhat different, namely, 20°C, 10 mM Tris·HCl (pH 7.5), 250 mM NaCl, and 10 mM Mg2Cl. A theoretical estimate of the work of stretching the unfolded RNA tether, Wstr,theory, from zero to it's extension at the unfolding force was calculated by integration of Eq. 1 with respect to extension (27) (see Table 2 for results). The ΔGtotal,theory column of Table 2 shows the total energy, ΔG°total,theory = ΔG°theory + Wstr,theory, which on theoretical grounds is expected to go into the unfolding and stretching process. Comparing ΔG°total,theory to the experimentally obtained Wtotal, it is clear that the latter is much larger, thus implying that a significant amount of the performed work is dissipated irreversibly under the unfolding/stretching process where the loading rate was ≈10 pN/s. It is striking that the measured Wtotal (Table 2) is the only parameter in Table 2 that yields a difference between the two pseudoknots that is comparable to the 2-fold difference between the in vivo frame shifting effects of the same pseudoknots.
Models for Pseudoknot-Stimulated Frameshifting.
The fact that the difference in unfolding forces correlates positively with frameshift frequencies for the two pseudoknots investigated is consistent with the 9 Å model of frameshift stimulation. This model builds on the possible resistance of the pseudoknot to unfold at its entrance into the mRNA channel of the ribosome. The pseudoknot is thought to block the translation pathway and force the ribosome to slip one base downstream at the slippery sequence to adapt to the physical obstacle. This step normally causes a 9-Å allosteric-induced movement in the ribosome upon GTP hydrolysis at the incoming ternary complex in the translational cycle (13). Recently, this model has found support from structural cryo-electromicroscopic observations that interaction between a ribosome and a pseudoknot causes a deformation of the A-site tRNA (30).
Indeed, the work we found necessary to unfold our pseudoknots by stretching (see Table 2) was much greater than the energy that can be obtained by hydrolysis of GTP into GDP + Pi (≈35 kJ/mol). Thus, we expect that unfolding the pseudoknot can be an energetic barrier to the movement of the ribosome, and that this is the reason why the frequency of frameshifting positively correlates to the mechanical strength of the tertiary structure. In comparison to a simple hairpin structure, the pseudoknot has lost its rotational freedom in the stem1 helix due to the “locking” via base pairing in stem2. This locking may cause a need for more base pairs to be broken simultaneously, probably in stem2, before the structure resolves. This process may involve ribosome associated RNA helicases (30, 31) at least in some organisms.
Opposed to the suggested pausing at pseudoknots, hairpin structures have been shown not to slow down ribosome movement in vivo (32) and unable to assist frameshifting in IBV (13). Nevertheless, the downstream stimulatory structure in the E. coli dnaX frameshift is a hairpin, and a correlation between frameshift efficiency and the predicted strength (ΔG°) of different mutant hairpins have been found (34). Presently, theoretical estimates of hairpin stability might be more precise than those of the more complex pseudoknots. Indeed, for a hairpin, ΔG° values from reversible mechanical unfolding matched theoretical estimates (16). Regardless of the extent and role of pausing in frameshifting, the results of Larsen et al. (34) support the finding that downstream mRNA structure strength correlates with the ability to stimulate frameshift on a slippery sequence.
Naptine et al. (20) investigated the effect on frameshifting on shortening the length of stem1 of a pseudoknot, and they found the frameshifting efficiency to be closely related to the length of this stem, if shorter than 11 bp, essentially no frameshifting was observed. Among other effects investigated (e.g., slippery sequence-pseudoknot spacing distance), they found the length of stem1 to be most important. This finding is in accordance with our observations, where we postulate that the mechanical stability of the pseudoknot, which is also determined by the length of, for example, stem1, is crucial for the degree of frameshifting.
In another class of frameshift-stimulating pseudoknots, the stem1 is as short as 5–7 bp. The function of these pseudoknots in frameshifting seems to depend on an extra loop2–stem1 interaction that facilitates a special kinked tertiary structure (19). To our knowledge, it remains to be determined whether this extra loop2–stem1 interaction increases the physical strength of this particular class of pseudoknots.
In conclusion, we find that two pseudoknots stemming from the frameshifting site in IBV and differing only in 3 of 68 nucleotides give rise to a factor of 2 difference in frameshifting frequencies. Unfolding the structure by optical tweezers shows that the two structures unfold at forces that are different; the pseudoknot giving rise to the lower degree of frameshifting is easier to unfold than the pseudoknot giving rise to the higher degree of frameshifting. This finding leads us to propose that the frameshifting efficiency of a given pseudoknot is correlated to its mechanical strength. In the future, this postulate should be supported with similar experiments on a variety of pseudoknots. Our observations and postulates are in accordance with the 9 Å model (13) and with the mechanical explanation of pseudoknot function suggested by recent cryoelectronmicroscopy observations (30).
Materials and Methods
Plasmids and Frameshift assay.
Plasmids encoding the sequences shown in Fig. 2 were made by inserting synthetic DNA oligomeres, containing the sequences, between unique HindIII and ApaI restriction endonuclease sites in pOFX302 (33) and verified by DNA sequencing.
The frequencies of ribosomal frameshifting were estimated in protein extracts from cells labeled in pulse labeling experiments. See SI Text.
Preparation of the Samples for Single-Molecule Experiments.
RNA was synthesized in vitro by T7 RNA polymerase as recommended by the manufacturer (Promega). First, DNA templates for the RNA synthesis were produced by PCR. Plasmids pTH400, pTH401, or pTH421 served as template in the PCRs. Primers were TH412: ATAATTCGCGTCTGGCCTTC and TH414: TAATACGACTCACTATAGGGAGAGTATACCTCTCAGTTGGGTG. The 5′ end of TH414 contains the T7 promoter (underlined). Run-off RNA synthesis are expected to produce 939-, 942-, or 876-nt RNA strands from the three templates.
Upstream and downstream handles DNA were each synthesized by assymetric PCR. The downstream handle DNA had a digoxygenin group on its 5′ nucleotide, and the upstream handle was labeled with biotin in its 3′ end enzymatically (see SI Text for details). Handles were annealed to RNA by mixing approximately equal amounts of the nucleotide species in buffer R (10 mM Tris·HCl, pH 7.5/250 mM NaCl/10 mM Mg2Cl) or in 10 mM sodiumphosphate (pH 6.0), 250 mM NaCl, and 10 mM Mg2Cl. The mixture was heated to 65°C for 8 min and allowed to cool down to room temperature for >30 min. Annealed RNA and handles were stored at −70°C until usage. To bind the handle/RNA to beads, appropriate dilutions of the handle/RNA mixture and 2.1-μm streptavidin-coated polystyrene beads (Bangs) were mixed and incubated for 1 h at room temperature with gentle mixing. The ratio of handle/RNA to 2.1-μm beads was chosen so that no more than approximately four of the five beads would later form a tether (see SI Text) to the 2.88-μm anti-digoxygenin-coated bead. This criterion insures a low likelihood of getting tethers of more than one handle/RNA molecule. After binding the RNA to the 2.1-μm beads, the mixture was diluted in a dilution of 2.88-μm anti-digoxygenin-coated beads and transferred to the sample chamber for the microscope. Experiments were performed at 22°C.
Data Analysis.
The stage signal was smoothed with a sliding window 3,000 data points wide, before the time series were averaged in 10-ms nonoverlapping windows. The force exerted on the bead in the trap and its position were calculated by using both coordinates of the quadrant photodiode, whereas the first point of the time series was defined as origin for the coordinate system. The change in tether length was calculated by subtraction of the movement of the bead in the trap from the stage movement.
Due to the short overall length of the tether, it is difficult by eye to decide whether an RNA tether is present between the two beads; if the beads are connected, this could also be due to the van der Waals attraction between them, caused by a polystyrene “hair” sticking out of one of the beads or simply some kind of dirt in the sample. To avoid “false” tethers/attractions between the beads in our data set, all data sets were subject to two filtering criteria: (i) The first part, i.e., the first 250 ms, of the force-extension curve needs to be flat (slope less than ± 50 nm/s). This filter would remove tethers that were too short. (ii) The “noise” of the first part (250 ms), of the curve should not be zero, more precisely, traces for which the standard deviation was between 3.5 and 7 nm passed this filter. These criteria made sure that the cases where the two beads were attached to each other did not pass. Other filters were also tested (e.g., only using data showing rips, only using data where we had force measurements >70 pN), but regardless of how these criteria were chosen, the rupture forces of the two different pseudoknots were consistently significantly different. Thus, this is a very robust result. The presented data represent repetitive pulls of at least eight individual molecules of each of the pseudoknots PK400, PK401, or the control PK421.
Supplementary Material
Acknowledgments
We thank Stanley Brown for suggestions and helpful discussions and John Atkins for comments on the manuscript. This work was supported by grants from the Carlsberg Foundation, BioNET sponsored by the Villum Kann Rasmussen Foundation, and the Faculty of Science, University of Copenhagen.
Abbreviation
- IBV
infectious bronchitis virus.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0608668104/DC1.
References
- 1.Ninio J. Biochimie. 1975;57:587–595. doi: 10.1016/s0300-9084(75)80139-8. [DOI] [PubMed] [Google Scholar]
- 2.Rodnina MV, Gromadski KB, Kothe U, Wieden HJ. FEBS Lett. 2005;579:938–942. doi: 10.1016/j.febslet.2004.11.048. [DOI] [PubMed] [Google Scholar]
- 3.Atkins JF, Elseviers D, Gorini L. Proc Natl Acad Sci USA. 1972;69:1192–1195. doi: 10.1073/pnas.69.5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kurland CG. In: Nonsense Mutations and tRNA Suppressors. Celis JE, Smith JD, editors. London: Academic; 1979. pp. 98–108. [Google Scholar]
- 5.Farabaugh PJ. Microbiol Rev. 1996;60:103–134. doi: 10.1128/mr.60.1.103-134.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gesteland RF, Atkins JF. Annu Rev Biochem. 1996;65:741–768. doi: 10.1146/annurev.bi.65.070196.003521. [DOI] [PubMed] [Google Scholar]
- 7.Weiss RB, Dunn DM, Shuh M, Atkins JF, Gesteland RF. New Biol. 1989;1:159–169. [PubMed] [Google Scholar]
- 8.ten Dam E, Brierley I, Inglis S, Pleij C. Nucleic Acids Res. 1994;22:2304–2310. doi: 10.1093/nar/22.12.2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rice NR, Stephens RM, Burny A, Gilden RV. Virology. 1985;142:357–377. doi: 10.1016/0042-6822(85)90344-7. [DOI] [PubMed] [Google Scholar]
- 10.Kontos H, Napthine S, Brierley I. Mol Cell Biol. 2001;21:8657–8670. doi: 10.1128/MCB.21.24.8657-8670.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tu C, Tzeng TH, Bruenn JA. Proc Natl Acad Sci USA. 1992;89:8636–8640. doi: 10.1073/pnas.89.18.8636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Plant EP, Dinman JD. Nucleic Acids Res. 2005;33:1825–1833. doi: 10.1093/nar/gki329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Plant EP, Jacobs KL, Harger JW, Meskauskas A, Jacobs JL, Baxter JL, Petrov AN, Dinman JD. RNA. 2003;9:168–174. doi: 10.1261/rna.2132503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Noller HF, Yusupov MM, Yusupova GZ, Baucom A, Cate JH. FEBS Lett. 2002;514:11–16. doi: 10.1016/s0014-5793(02)02327-x. [DOI] [PubMed] [Google Scholar]
- 15.Giedroc DP, Theimer CA, Nixon PL. J Mol Biol. 2000;298:167–185. doi: 10.1006/jmbi.2000.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liphardt J, Onoa B, Smith SB, Tinoco IJ, Bustamante C. Science. 2001;292:733–737. doi: 10.1126/science.1058498. [DOI] [PubMed] [Google Scholar]
- 17.Onoa B, Tinoco I., Jr Curr Opin Struct Biol. 2004;14:374–379. doi: 10.1016/j.sbi.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 18.Brierley I, Digard P, Inglis SC. Cell. 1989;57:537–547. doi: 10.1016/0092-8674(89)90124-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liphardt J, Napthine S, Kontos H, Brierley I. J Mol Biol. 1999;288:321–335. doi: 10.1006/jmbi.1999.2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Napthine S, Liphardt J, Bloys A, Routledge S, Brierley I. J Mol Biol. 1999;288:305–320. doi: 10.1006/jmbi.1999.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berg-Sorensen K, Oddershede L, Florin EL, Flyvbjerg H. J Appl Phys. 2003;93:3167–3176. [Google Scholar]
- 22.Oddershede L, Grego S, Nørrelykke SF, Berg-Sorensen K. Probe Microscopy. 2001;2:129–137. [Google Scholar]
- 23.Hansen P, Tolic-Norrelykke I, Flyvbjerg H, Berg-Sorensen K. Comput Phys Commun. 2006;174:518–520. [Google Scholar]
- 24.Marko JF, Siggia ED. Macromolecules. 1995;28:8759–8770. [Google Scholar]
- 25.Odjik T. Macromolecules. 1995;28:7016–7018. [Google Scholar]
- 26.Vanzi F, Takagi Y, Shuman H, Cooperman BS, Goldman YE. Biophys J. 2005;89:1909–1919. doi: 10.1529/biophysj.104.056283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tinoco I., Jr Annu Rev Biophys Biomol Struct. 2004;33:363–385. doi: 10.1146/annurev.biophys.33.110502.140418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holbrook SR, Kim SH. Biopolymers. 1997;44:3–21. doi: 10.1002/(SICI)1097-0282(1997)44:1<3::AID-BIP2>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 29.Reeder J, Giegerich R. BMC Bioinformatics. 2004;5:104. doi: 10.1186/1471-2105-5-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Namy O, Moran SJ, Stuart DI, Gilbert RJ, Brierley I. Nature. 2006;441:244–247. doi: 10.1038/nature04735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takyar S, Hickerson RP, Noller HF. Cell. 2005;120:49–58. doi: 10.1016/j.cell.2004.11.042. [DOI] [PubMed] [Google Scholar]
- 32.Sorensen MA, Kurland CG, Pedersen S. J Mol Biol. 1989;207:365–377. doi: 10.1016/0022-2836(89)90260-x. [DOI] [PubMed] [Google Scholar]
- 33.Rettberg CC, Prere MF, Gesteland RF, Atkins JF, Fayet O. J Mol Biol. 1999;286:1365–1378. doi: 10.1006/jmbi.1999.2546. [DOI] [PubMed] [Google Scholar]
- 34.Larsen B, Gesteland RF, Atkins JF. J Mol Biol. 1997;271:47–60. doi: 10.1006/jmbi.1997.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.