Abstract
The high dimensionality of global transcription profiles, the expression level of 20,000 genes in a much small number of samples, presents challenges that affect the sensitivity and general applicability of analysis results. In principle, it would be better to describe the data in terms of a small number of metagenes, positive linear combinations of genes, which could reduce noise while still capturing the invariant biological features of the data. Here, we describe how to accomplish such a reduction in dimension by a metagene projection methodology, which can greatly reduce the number of features used to characterize microarray data. We show, in applications to the analysis of leukemia and lung cancer data sets, how this approach can help assess and interpret similarities and differences between independent data sets, enable cross-platform and cross-species analysis, improve clustering and class prediction, and provide a computational means to detect and remove sample contamination.
Keywords: cancer, dimension reduction, expression analysis, noise reduction, sample contamination
A major challenge in the analysis of global transcription profiles is the high level of noise and the lack of reproducibility across data sets, which results from fitting models to small numbers of samples in a high-dimensional space (i.e., thousands of genes). Ideally we would prefer to reduce the data to a small number of metagenes that better capture the essential behavior of the samples.
There are many advantages to such a metagene approach. By capturing the major, invariant biological features and reducing noise, metagenes provide descriptions of data sets that allow them to be more easily combined and compared. This is especially important when we are considering cross-platform or cross-species data. Ultimately, this can result in more sensitive clustering and classification. In addition, interpretation of the metagenes, which characterize a subtype or subset of samples, can give us insight into underlying mechanisms and processes of a disease.
Here, we describe a general methodology, metagene projection, that creates a low-dimensional representation of a training (model) data set using nonnegative metagene factors into which an independently obtained new (test) set of samples or data can be projected and analyzed. The metagene factors are a small number of gene combinations that distinguish expression patterns of subclasses in a data set. We obtain the factors by the application of nonnegative matrix factorization (NMF) (1, 2) used to extract facial features from images. We showed (3) how NMF can extract metagenes that provide stable, robust clustering of expression data. Moreover, by using gene set enrichment analysis (GSEA) to annotate the metagene factors themselves, we can gain insight into the underlying biology of both the training and test data sets.
Importantly, we illustrate the utility of metagene projection by its application to leukemia and lung cancer data sets. We show how the projection of new data sets into the space of metagene factors reduces noise and emphasizes relevant biological correlations and thus (i) enables cross-platform analysis by removing technological noise from data, (ii) enables cross-species analysis and the assessment of disease models, (iii) improves the accuracy of classification and prediction methods in the mapping of diseases types, and (iv) detects contamination in tumor samples.
Results
Overview of Method.
We consider a gene expression data set consisting of a collection of NM model samples, which we use to characterize a domain of biological (transcriptional) states of interest. The model data are represented as an nM × NM matrix, M, whose rows contain the expression levels of the nM genes in the NM samples.
Using NMF, we find a small number, k, of metagenes, positive linear combinations of the NM genes, which can be used to distinguish the transcription profiles of the subtypes contained in the model data set. Mathematically, this corresponds to finding an approximate factoring, M ≈ WM × HM, where both factors have only positive entries. WM is an nM × k matrix that defines the metagene decomposition model and whose columns specify how much each of the nM genes contributes to each of the k metagenes. HM is a k × NM matrix whose entries represent the expression levels of the k metagenes for each of the NM samples. This model selection is done in an unsupervised fashion by using either a knowledge-based or data-driven model selection approach. One can set k equal to the number of known phenotypes in the model set. Alternatively, optimal values of k can be determined based on projection stability by using consensus clustering techniques as described (3).
From the factoring of M, we are able to construct a mapping that allows us to project a data set into the space of the metagenes derived above. Mathematically, this can be accomplished by using the Moore–Penrose generalized pseudoinverse (4) of WM, so that, ĤM = (WM)−1 × M, where ĤM ≈ HM. For simplicity in notation we refer to the projected matrix as HM. After elimination of outlier samples and model refinement, we can apply the final resulting pseudoinverse to a new individual sample or entire data set and analyze that data in the context of the metagenes, which characterized the model data.
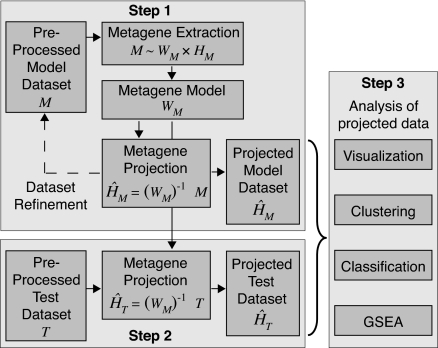
We summarize the three major steps in the metagene projection method below (Fig. 1). More detail can be found in Methods. The software is freely available from The Broad Institute web site as both R-code and a module in the GenePattern software package.
Fig. 1.
Schematic of the metagene projection methodology.
Step 1. Metagene Factor Extraction and Refinement of the Model Data Set.
We start with standard data preprocessing: thresholding and eliminating genes that do not vary sufficiently across the model set and rank normalizing to minimize platform idiosyncrasies. We apply NMF to factor the resulting expression matrix and derive the Moore–Penrose pseudoinverse of WM. Next, we project the model data set into metagene space and, by using a support vector machine (SVM) (5) classification step, trim outliers from the model set (model data set refinement). Finally, we refactor the expression matrix M of the refined model set, M ≈ WM× HM, and define a refined pseudoinverse or projection map. We use this refinement of the projection map in the analysis of new test data sets.
Step 2. Metagene Factor Projection of the Test Data Set.
We threshold the expression values as in step 1 and then match the genes in each test set to the corresponding genes in the model set. We then rank normalize the test samples to yield the corresponding columns in the test data expression matrix, T. Finally, we apply the pseudoinverse (WM)−1 to both M and T to obtain HM and HT, their projections into metagene space.
Step 3. Analysis of Model and Test Data Set Projection Results.
In our experience, the use of metagenes, instead of genes, as features for analysis, increases the signal-to-noise ratio and yields more robust, accurate results. Now that both the model and test data are represented in the lower dimensional metagene space, there are a variety of analyses we can apply. These include the following:
Visualization.
Model and test samples can be characterized and compared by using heat maps of the H matrices.
Clustering model and test projections.
The projection can provide a sample's class assignment by identifying the metagene with maximum expression. Alternatively, we can cluster the columns of HM and HT.
Classification of test samples.
We can use the projected data to build a multiclass predictor and assess any data set of test samples. Below, we use a one-versus-all SVM classifier (6, 7) to predict phenotypes by using the k metagenes as the input features. This method provides a predicted class and a predictive confidence by using a modified Brier score (see Methods for details).
GSEA-based metagene interpretation.
To gain biological insight into the different metagene factors, we use a variation of our GSEA methodology (8). Using the expression profile of a metagene, i.e., the corresponding row of the HM matrix, as a template, we sort the genes according to the correlation of their expression profile from the M matrix with the metagene template. We can then evaluate the “enrichment” of gene sets representing a pathway or other biological process at the top of that ranked list by using GSEA. For each metagene, one obtains a list of “enriched” gene sets and their statistical significance [see supporting information (SI) Text].
Examples.
Here, we describe three applications of the metagene projection method to highlight its utility in three cross-platform analyses, to validate disease models, to improve classification of cross-platform data sets, to assess the similarities and differences of subtypes across data sets, and to detect contamination. We start with a simple example. We then describe two more innovative results.
Cross-Platform Clustering of Leukemia Data.
We analyzed two leukemia data sets from different microarray platforms to test the method and demonstrate its power to enable cross-platform classification and to improve sensitivity in clustering. Often clustering of cross-platform data reveals the platform or originating lab as the strongest differentiating signal in the data. Importantly, we establish that the method was able to cluster the cross-platform data correctly and that these results are because of the metagene representation rather than the rank normalization step.
We considered two data sets containing samples representing three leukemia subclasses: B and T cell acute lymphoblastic leukemia (ALL-B, ALL-T) and acute myeloid leukemia (AML). The model data set consisted of 30 samples (10 ALL-B, 10 ALL-T, 10 AML) (from refs. 9 and 10). The test data set contained the 38 samples (19 ALL-B, 8 ALL-T, 11 AML) from ref. 11. The two data sets came from different laboratories and were acquired on different microarray technologies, Affymetrix U-133 for the model set and Affymetrix HU6800 for the test set (Affymetrix, Santa Clara, CA).
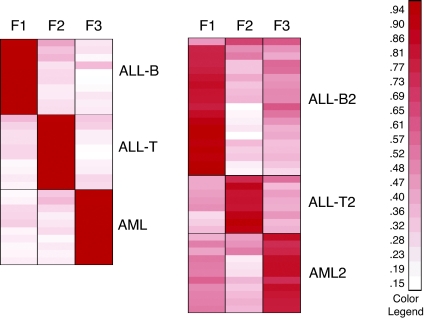
We applied the metagene projection methodology as described above. In particular, we noted that the model data set is very consistent, and no model refinement was necessary. Because the number of subtypes was known, we used k = 3 metagene factors. Fig. 2 shows the resulting heat maps for the projected model and test sets. Clearly, the metagenes are associated with the biological phenotypes (F1 ≈ ALL-B, F2 ≈ ALL-T, F3 ≈ AML) in both.
Fig. 2.
Heat maps of metagene projection of leukemia samples. These heat maps of the HM and HT matrices show the metagene expression levels for each sample. Each factor clearly corresponds to same leukemia subtype in both model (Left) and test (Right) sets.
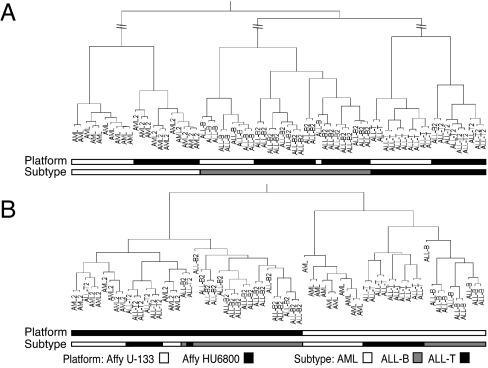
Postprojection clustering of the model samples demonstrates reduction of noise and greater emphasis of the biologically invariant signal in the data. The clusters corresponding to each phenotype have higher intracluster correlation and greater intercluster distance than obtained with the original data (SI Fig. 6). More importantly, clustering of the merged set of projected model and test samples produces very clear results with the major three clusters consisting of each leukemia subtype independent of the data set of origin (Fig. 3A and SI Fig. 7A).
Fig. 3.
Hierarchical clustering of the leukemia model and test samples. (A) Clustering of the merged test and model data sets after metagene projection, i.e., columns of the merged HM and HT matrices. (B) Clustering of merged model and test sets normalized but without projection. For clarity, some dendrogram vertical lines have been truncated in A; for full dendrograms see SI Fig. 7.
We next sought to confirm that this consistency of subtype clusters across the data sets was due to the metagene projection and not just the result of preprocessing and rank normalization. To this end, we performed two additional clusterings: one merging the model and test samples after rank normalization and clustering in the space of all filtered genes without using metagene projection (Fig. 3B and SI Fig. 7B) and another clustering the merged and rank normalized data in the space of the top-500 marker genes of each of the three subtypes in the model set, 1,500 genes in total (SI Fig. 8). This last procedure is often used for cross-platform analysis. In both alternative clusterings, not using metagene projection, the samples first split according to their data set of origin before the biological subclassification appears.
Leukemias: Improving Cross-Platform Classification and Interpretation of Subtypes.
We sought to ascertain whether metagene projection would be an effective procedure for unsupervised feature extraction (12) and dimension reduction to enable more robust and accurate classifiers. To this end, we considered 10 subclasses of leukemia (5 subtypes of ALL and 5 subtypes of AML) as represented in a model set of 170 samples from refs. 9 and 10. The test set consisted of 297 samples (13–20), obtained from eight independent published data sets. The model set samples were all acquired on the same platform in the same laboratory, whereas the test set came from multiple labs and three different microarray platforms (see SI Table 1).
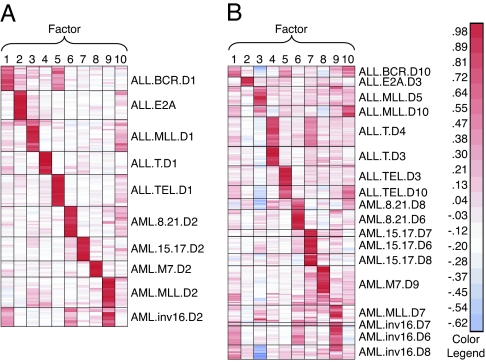
We set the number of metagene factors to the number of known phenotypes in the model set, k = 10. Metagene projection, followed by model refinement, resulted in elimination of eight outlier samples from the model set [2 of 21 AML t (8, 21); 4 of 23 AML MLL; 2 of 14 AML inv (16)] (for more detail see Methods). Fig. 4 shows the metagene expression matrices for both the model and test data sets after projection. Strikingly, we found that each leukemia subtype was characterized by essentially one metagene.
Fig. 4.
Leukemia subclasses metagene projection. Heat maps of the model (A) and test (B) sets after metagene projection show consistent representation of subtype structure across technology platform and laboratory group. SI Text contains a detailed description of the different leukemia subtypes shown here.
Next, we sought to determine whether we could build a classifier using the metagene projections that would accurately predict the subtype of the cross-platform samples in the test set. We noted that the data-driven model selection technique described in our previous work (3) indicated that k = 13 was the best choice (SI Fig 9). Thus, we evaluated SVM classifiers using both the 10- and 13-metagene models and compared them with SVM and K-nearest neighbor (K-NN) classifiers using all genes in common between the model and test data sets. SI Fig. 10 shows the comparative performance of the 10- and 13-metagene SVMs with the all-gene classifiers.
Our metagene-based classifier outperformed the classifiers based on all-genes or markers selected in all-gene space. The 13-metagene classifier attained the “best” performance, with a correct call accuracy of 88% and fewer errors than the 10-metagene model. The 10-metagene, all-gene SVM, and K-NN classifiers' correct call accuracies were 86%, 82%, and 72% respectively. We note that the SVM classifier using all common genes made fewer “confident” calls but made correspondingly fewer errors. We used 0.3 as the confidence threshold for all of the SVM multiclass predictors. Increasing this threshold will reduce both the number of correct calls and the number of errors. (SI Tables 2 and 3 contain details).
Closer examination of the confusion matrices for the 10- and 13-metagene classifiers revealed that two thirds of the errors resulted from placing ALL-BCR, AML-t (8, 21), AML-M7, and AML-MLL samples into the AML-inv16 class. We believe this results from shared metagene signals, which can be seen in the heat map in Fig. 4B. A GSEA analysis of the metagene factors, described below, uncovered a biological interpretation for some of the errors. This also led us to explore the extent to which cross talk between the AML and ALL data in the model might be affecting our ability to predict the classes in the test set. Interestingly, we found that building 10-metagene, five-class classifiers for just the ALL [respectively AML] subtypes improved accuracy substantially to 97% (130 samples) with 1.5% no calls (2 samples) and 1.5% errors (2 samples) [92% (150 samples) with 3% no calls (4 samples) and 5% errors (9 samples)]. The all-gene SVM and 9-NN predictors also improved accuracy, but the metagene-based classifier continues to make more correct calls and fewer no-calls (SI Fig. 10).
These are remarkably good multiclass, cross-platform classification results. It was difficult to make direct comparisons with other approaches in the literature, because the specific data sets or data preparation were not always available. However, the metagene-based approach appears to outperform other leukemia cross-platform classification approaches: 93–96% accuracy on ALL subtypes and 68–78% on AML subtypes (21); ≈40% accuracy on AML subtypes (22).
Finally, we applied GSEA analysis to help interpret the metagenes characterizing the leukemia subtypes. Interestingly, many of the results agreed with the current understanding of these subclasses, and others posed new hypotheses. We present them as an illustration of the power of the metagene projection method to provide biological insights. The top two gene sets enriched in F4 (i.e., high in ALL T Cell) are (i) a set of E2F1 targets known to be activated in T Cell ALL (23) and (ii) a set of genes down-regulated by ET-743 treatment, which is known to induce apoptosis in acute T cell leukemia Jurkat cells (24). Metagene F9, high in AML-MLL, shows enrichment for chromosome band 11q13, which is known to be frequently coamplified with MLL in AML patients (25).
F6 is highly expressed in t (8, 21) and also up-regulated in inv (16) subclasses of AML. The mechanism of leukemogenesis in AML in both these subtypes is disruption of the core binding factor (CBF) transcriptional complex, comprised of the RUNX1 and CBFB proteins. In t (8, 21), RUNX1 is fused to the CBFA2T1 gene, and inv (16) causes a CBFB-MYH11 fusion gene. Both fusion genes disrupt the CBF complex, which is required for normal hematopoietic differentiation. Patients with t (8, 21) and inv (16) also have similar clinical features: both subclasses are associated with a relatively good prognosis and particular benefit from consolidation chemotherapy with high-dose cytarabine. F6 therefore identifies patients harboring distinct cytogenetic abnormalities with a common molecular mechanism and clinical phenotype. Intriguingly, F9 also shows strong correlation with both AML.MLL and AML-inv (16). This leads us to speculate some common program of these two AML subtypes.
In this example, we have shown that metagene projection is an effective approach to building multiclass classification models across different platforms and sources of data that are accurate, robust, and interpretable.
Lung Cancer: Cross-Platform Comparison, Contamination Detection, and Interpretation of Cell Line Models.
We next investigated whether metagene projection would enable us to evaluate consistency in a collection of cross-platform data sets, validate cell lines as good models for different tumor types, and, importantly, provide a method to computationally extract some of the expression signal of normal tissue contamination from tumor samples.
For our model set, we used a subset of data set A from ref. 26, BOS, consisting of 30 lung adenocarcinomas, 20 squamous tumors, and 17 normal lung samples. Our test set derived from seven independent data sets (refs. 27–32 and one unpublished set, see SI Table 4). Note that these data sets were acquired on four different microarray platforms by six different laboratories.
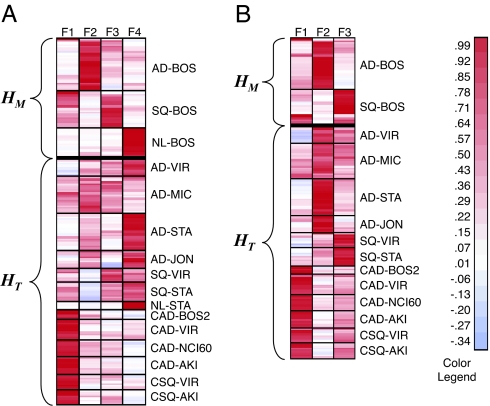
We first built a four-metagene model from the BOS model set as described above. Although the model set included three major subtypes, the data-driven NMF model selection procedure indicated that four factors was the smallest optimal solution greater than the number of known phenotypes (SI Fig. 11). After SVM model refinement, one outlier adenocarcinoma sample was removed from the model set, and the metagene factors recalculated. Fig. 5A contains the HM matrix of metagene expression levels. From the HM matrix, we can see that metagenes F2, F3, and F4 characterize the adenocarcinoma, squamous, and normal samples respectively, whereas the F1 metagene picks up an additional signature in a subset of the adenocarcinoma and squamous samples. Next, we projected all of the test data sets into metagene space (HT in Fig. 5A) and found an unexpected result. The normal test samples NL-STA continued to be characterized by F4. However, although the adenocarcinoma and squamous samples still showed F2 and F3 metagene signatures, respectively, they also showed significant expression in the F4 “normal” metagene. This led us to speculate that these samples might have varying degrees of contamination by stroma or normal tissue, which we might be able to extract computationally.
Fig. 5.
Metagene projection of the lung cancer data set. Heat maps showing projection of model and test data sets into four-metagene space F1-F4 (A) and three-metagene space F1-F3 after numerical removal of normal component F4 and reconstruction of model (B). AD, adenocarcinoma; SQ, squamous; C, cell lines; NL, normal lung.
To remove the normal signature, we set the F4 metagene factor coefficient in the HM matrix to zero and multiplied it by the original WM to yield a matrix M̃ that reproduces the original data but without the contribution of F4. We then excluded the normal tissue samples from the model data set because they only had residual values, factored the resulting data matrix to extract the three remaining metagene factors, and projected all of the samples as was done before. The resulting expression profiles of the metagenes in the model and test sets are seen in Fig. 5B. Eliminating the contribution of the F4 metagene, we find the dominant signatures in the adenocarcinoma and squamous samples are F2 and F3, respectively, as in the model set, and F1 retains its role as the signature of the cell lines. Thus, we were able to numerically “modulate” a specific metagene to computationally reduce contamination in the tumor samples.
The most striking feature of the metagene projection of the test samples is that the adenocarcinoma and squamous cell lines do not project with the corresponding tumor classes. This has been reported in the literature (27). Using the GSEA approach we described above, we can gain some biological insight into the metagene, F1, which characterizes the cell lines. SI Table 5 shows the top-20 gene sets enriched in F1.
Metagene F1 is enriched in gene sets associated with rapamycin response (mTOR activation), protein production (genes down-regulated by amino acid starvation), lack of differentiation, the mitochondria, oxidative phosphorylation, and BRCA1 signaling. We have observed some of these gene sets before as part of a group of gene sets enriched in poor-outcome lung adenocarcinoma patients in three different data sets (8). This leads us to speculate that F1 represents transcriptional programs associated with hyperactivation of AKT/mTOR, an associated mTOR-mediated increase of protein production and high proliferation, and a lack of differentiation.
In this example, we have shown the power of the metagene projection to define a common space of transcriptional variation in which we can analyze and assess multiple data sets across different technology platforms and laboratories. Despite the diversity of platforms, sample sources, and different experimental conditions, most test samples project with their biological counterparts. Moreover, we have shown that metagene projection provides a method for computationally reducing sample contamination, which enables more coherent projection of tumor samples. Finally, the combination of metagene projection and GSEA analysis allows us to gain insights into more robust, invariant biological features of different phenotypes and tumor subtypes.
Discussion
Traditional approaches to microarray analysis focus on identifying marker genes, which are correlated with a phenotype of interest, and on using them to build classifiers for samples whose phenotype may be unknown or to gain some insight into the underlying biology of a cellular state. These strategies often fail when classifiers are applied to data from other laboratories or derived on different technology platforms or when used to try to assess the validity of a disease model.
Lower-dimensional projections and decompositions of DNA microarray data, such as principal component analysis, singular value decomposition, and NMF, have been used to analyze transcriptional states (3, 33–37). Primarily, these approaches were applied in the context of a single data set for clustering or visualization.
We introduced a metagene projection method to assess the validity of a Snf5 knockout mouse as a murine model for Snf5-deficient human rhabdoid tumors (38), and found that the murine Snf5 model samples were closely related to the human rhabdoid samples (from both model and test sets) and distinct from the controls. The model and test sets were obtained on different microarray platforms in addition to being cross-species. This approach combined our previous work, using NMF to identify a small number of gene combinations (metagenes) whose profiles best represent the most distinguishing features of the expression patterns of the subclasses in a data set, with our previously published gene expression data set derived from a collection of human pediatric brain tumors (rhabdoid, medulloblastoma, glioma, and normal cerebellum) (33). A corresponding projection map, the Moore-Penrose generalized pseudo-inverse of one of the factor matrices, allowed us to analyze new data in the context of the space of metagenes arising from the original data set.
This article presents a refinement of that method, which is more sensitive, robust, and broadly applicable to cross-platform and cross-species analysis and classification (see SI Text). In addition, we have shown how the projection can be used to highlight the biologically invariant aspects and commonalities of the subclasses, assess the similarities and differences between suitable chosen sets of model and test samples, and, surprisingly, to computationally remove contaminating signals from tumor data.
The method, as presented here, has a number of advantages over other approaches. Metagene projection, together with NMF, reduces dimensionality and summarizes the salient features of a data set with coherent patterns shared by multiple genes and samples. In contrast to approaches using principal component analysis or singular value decomposition, it yields a sparser representation of the original model data set optimized for the number of factors specified. NMF factors are nonnegative and more localized and therefore easier to interpret and analyze. We note here that Alter and Golub (39) applied the pseudoinverse to genomic data by using the singular value decomposition.
There is complementary work of Huang (40) and Bild (41), which is conceptually similar to ours in the sense of combining dimensionality reduction and classification models, but has distinct objectives. Their main goal is to provide an exquisitely specific predictor of pathway activation, which has been experimentally characterized by the overexpression of a single gene. In contrast, our goal is to model global transcriptional states, rather than specific pathways, and to use them to describe an entire range of biological behavior, e.g., different morphologies, lineages, etc. Thus, the specific methodologies and techniques we use are also quite different.
Classifiers built in metagene, rather than all-gene, space are more robust, reproducible, and generalizable across platforms and laboratories because the projection can reduce noise and technology-based variation more than simple normalization. In particular, we found this approach to be very sensitive in the complex, cross-platform, multiclass setting of the leukemia data sets. Others have studied cross-platform classification in lung cancer (42, 43). However, they use the test data explicitly to choose similarly correlated genes as features, rather than relying solely on the model set.
Most importantly, metagene models built on previously acquired or published data sets enable the use of prior knowledge to help characterize and analyze new data. This is seen in our work validating a mouse model for human rhabdoid tumors (38). We also used this approach to analyze samples from malaria-infected patients using signatures derived from publicly available yeast data (P.T., D.S., J.P.M., unpublished work). Thus, we see that this metagene projection method not only decreases noise by reducing the dimensionality of microarray data, but can also provide a powerful knowledge-based approach to the cross-platform, cross-species analysis of microarray data.
Methods
Data Set Preprocessing and Normalization.
For Affy Hu6800 and U133 microarrays, we threshold at 20 and 100,000 units. Gene filtering excludes genes with <5-fold and 500 units of maximum difference for the first leukemia example, 8-fold/800 for the second leukemia example, and 3-fold/300 for the lung. We rank the genes according to their expression levels and replace the value by 10,000 × (rank(gene) − 1)/(number of genes − 1).
Metagene Factor Extraction.
We use NMF with 2,000 iterations and stopping criterion as described (3).
Metagene Model Selection.
We select k based either on the known number of phenotypes or by using the values determined by projection stability described (3). Optimal solutions are peaks in the cophenetic coefficient as a function of k.
Data Set Refinement.
We train a SVM on HM to predict each class, and we remove samples that are errors (known phenotypes) or no calls (discovered classes). In our experience, the number of outliers is quite small compared with the size of the classes if the number of metagenes is chosen as described above.
Calculating the Pseudoinverse of WM.
We use “ginv” from R's MASS package.
Metagene Projection of Model and Test Set Samples.
To project the model set, we use the pseudoinverse of WM. For each data set in the test set, we match the genes to the corresponding rows of WM (i.e., genes in the model set). We calculate the pseudoinverse for that set of rows and apply it to obtain the corresponding columns of HT for that specific data set. This procedure adapts the projection to the particular test data set and, by tolerating unmatched genes between model and test set, supports the projection of data sets from different platforms. If too many unmatched genes result in weak amplitudes in HT, we rescale the columns of HT so the sum of the squares of their row-entries is equal to one. This postnormalization is optional.
Clustering.
We use “hclust” (complete linkage) from R's STATS package.
Classification and Prediction Confidence.
We use the “svm” function from R's e1071 package (one vs. all, radial function kernel). The predicted class is the one with the highest probability, and a predictive confidence 1 ≥ Cp ≥ 0 is computed by using a modification of the Brier skill score (44):
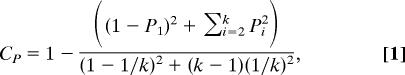 |
where P1 > P2 > …> Pk. is the sorted list of k output probabilities for a given sample. Cp < 0.3 is a no call. The K-NN classifier in the leukemia example used 50 marker genes and nine nearest neighbors. For the SVM using all genes we use a “linear” kernel.
Supplementary Material
Acknowledgments
We thank J. P. Brunet, T. Golub, E. Lander, and M. Meyerson for helpful conversations and for reviewing this manuscript.
Abbreviations
- GSEA
gene set enrichment analysis
- NMF
nonnegative matrix factorization
- SVM
support vector machine.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0701068104/DC1.
References
- 1.Lee DD, Seung HS. Nature. 1999;401:788–791. doi: 10.1038/44565. [DOI] [PubMed] [Google Scholar]
- 2.Lee DD, Seung HS. Adv Neural Info Proc Syst. 2001;13:556–562. [Google Scholar]
- 3.Brunet JP, Tamayo P, Golub TR, Mesirov JP. Proc Natl Acad Sci USA. 2004;101:4164–4169. doi: 10.1073/pnas.0308531101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ben-Israel A, Greville TNE. Generalized Inverses: Theory and Applications. New York: Springer; 2003. [Google Scholar]
- 5.Cristianini N, Shawe-Taylor J. An Introduction to Support Vector Machines and Other Kernel-Based Learning Methods. New York: Cambridge Univ Press; 2000. [Google Scholar]
- 6.Ramaswamy S, Tamayo P, Rifkin R, Mukherjee S, Yeang CH, Angelo M, Ladd C, Reich M, Latulippe E, Mesirov JP, et al. Proc Natl Acad Sci USA. 2001;98:15149–15154. doi: 10.1073/pnas.211566398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rifkin R, Mukherjee S, Tamayo P, Ramaswamy S, Yeang CH, Angelo M, Reich M, Poggio T, Lander ES, Golub TR, Mesirov J. SIAM Rev. 2003;45:706–723. [Google Scholar]
- 8.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ross ME, Zhou X, Song G, Shurtleff SA, Girtman K, Williams WK, Liu HC, Mahfouz R, Raimondi SC, Lenny N, et al. Blood. 2003;102:2951–2959. doi: 10.1182/blood-2003-01-0338. [DOI] [PubMed] [Google Scholar]
- 10.Ross ME, Mahfouz R, Onciu M, Liu HC, Zhou X, Song G, Shurtleff SA, Pounds S, Cheng C, Ma J, et al. Blood. 2004;104:3679–3687. doi: 10.1182/blood-2004-03-1154. [DOI] [PubMed] [Google Scholar]
- 11.Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri MA, et al. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 12.Guyon IM, Gunn SR, Nikravesh M, Zadeh L. Feature Extraction, Foundations and Applications. Heidelberg: Physica, Springer; 2006. [Google Scholar]
- 13.Yeoh EJ, Ross ME, Shurtleff SA, Williams WK, Patel D, Mahfouz R, Behm FG, Raimondi SC, Relling MV, Patel A, et al. Cancer Cell. 2002;1:133–143. doi: 10.1016/s1535-6108(02)00032-6. [DOI] [PubMed] [Google Scholar]
- 14.Armstrong SA, Staunton JE, Silverman LB, Pieters R, den Boer ML, Minden MD, Sallan SE, Lander ES, Golub TR, Korsmeyer SJ. Nat Genet. 2002;30:41–47. doi: 10.1038/ng765. [DOI] [PubMed] [Google Scholar]
- 15.Chiaretti S, Li X, Gentleman R, Vitale A, Vignetti M, Mandelli F, Ritz J, Foa R. Blood. 2004;103:2771–2778. doi: 10.1182/blood-2003-09-3243. [DOI] [PubMed] [Google Scholar]
- 16.Bullinger L, Dohner K, Bair E, Frohling S, Schlenk RF, Tibshirani R, Dohner H, Pollack JR. N Engl J Med. 2004;350:1605–1616. doi: 10.1056/NEJMoa031046. [DOI] [PubMed] [Google Scholar]
- 17.Valk PJ, Verhaak RG, Beijen MA, Erpelinck CA, Barjesteh van Waalwijk van Doorn-Khosrovani S, Boer JM, Beverloo HB, Moorhouse MJ, van der Spek PJ, Lowenberg B, Delwel R. N Engl J Med. 2004;350:1617–1628. doi: 10.1056/NEJMoa040465. [DOI] [PubMed] [Google Scholar]
- 18.Gutierrez NC, Lopez-Perez R, Hernandez JM, Isidro I, Gonzalez B, Delgado M, Ferminan E, Garcia JL, Vazquez L, Gonzalez M, San Miguel JF. Leukemia. 2005;19:402–409. doi: 10.1038/sj.leu.2403625. [DOI] [PubMed] [Google Scholar]
- 19.Bourquin JP, Subramanian A, Langebrake C, Reinhardt D, Bernard O, Ballerini P, Baruchel A, Cave H, Dastugue N, Hasle H, et al. Proc Natl Acad Sci USA. 2006;103:3339–3344. doi: 10.1073/pnas.0511150103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fine BM, Stanulla M, Schrappe M, Ho M, Viehmann S, Harbott J, Boxer LM. Blood. 2004;103:1043–9. doi: 10.1182/blood-2003-05-1518. [DOI] [PubMed] [Google Scholar]
- 21.Nilsson B, Andersson A, Johansson M, Fioretos T. Haematologica. 2006;91:821–4. [PubMed] [Google Scholar]
- 22.Warnat P, Eils R, Brors B. BMC Bioinformatics. 2005;6:265. doi: 10.1186/1471-2105-6-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lemasson I, Thebault S, Sardet C, Devaux C, Mesnard JM. J Biol Chem. 1998;273:23598–604. doi: 10.1074/jbc.273.36.23598. [DOI] [PubMed] [Google Scholar]
- 24.Gajate C, An F, Mollinedo F. J Biol Chem. 2002;277:41580–9. doi: 10.1074/jbc.M204644200. [DOI] [PubMed] [Google Scholar]
- 25.Zatkova A, Ullmann R, Rouillard JM, Lamb BJ, Kuick R, Hanash SM, Schnittger S, Schoch C, Fonatsch C, Wimmer K. Genes Chromosomes Cancer. 2004;39:263–76. doi: 10.1002/gcc.20002. [DOI] [PubMed] [Google Scholar]
- 26.Bhattacharjee A, Richards WG, Staunton J, Li C, Monti S, Vasa P, Ladd C, Beheshti J, Bueno R, Gillette M, et al. Proc Natl Acad Sci USA. 2001;98:13790–5. doi: 10.1073/pnas.191502998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Virtanen C, Ishikawa Y, Honjoh D, Kimura M, Shimane M, Miyoshi T, Nomura H, Jones MH. Proc Natl Acad Sci USA. 2002;99:12357–62. doi: 10.1073/pnas.192240599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Staunton JE, Slonim DK, Coller HA, Tamayo P, Angelo MJ, Park J, Scherf U, Lee JK, Reinhold WO, Weinstein JN, et al. Proc Natl Acad Sci USA. 2001;98:10787–92. doi: 10.1073/pnas.191368598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gemma A, Li C, Sugiyama Y, Matsuda K, Seike Y, Kosaihira S, Minegishi Y, Noro R, Nara M, Seike M, et al. BMC Cancer. 2006;6:174. doi: 10.1186/1471-2407-6-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beer DG, Kardia SL, Huang CC, Giordano TJ, Levin AM, Misek DE, Lin L, Chen G, Gharib TG, Thomas DG, et al. Nat Med. 2002;8:816–824. doi: 10.1038/nm733. [DOI] [PubMed] [Google Scholar]
- 31.Garber ME, Troyanskaya OG, Schluens K, Petersen S, Thaesler Z, Pacyna-Gengelbach M, van de Rijn M, Rosen GD, Perou CM, Whyte RI, et al. Proc Natl Acad Sci USA. 2001;98:13784–13789. doi: 10.1073/pnas.241500798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jones MH, Virtanen C, Honjoh D, Miyoshi T, Satoh Y, Okumura S, Nakagawa K, Nomura H, Ishikawa Y. Lancet. 2004;363:775–781. doi: 10.1016/S0140-6736(04)15693-6. [DOI] [PubMed] [Google Scholar]
- 33.Pomeroy SL, Tamayo P, Gaasenbeek M, Sturla LM, Angelo M, McLaughlin ME, Kim JY, Goumnerova LC, Black PM, Lau C, et al. Nature. 2002;415:436–442. doi: 10.1038/415436a. [DOI] [PubMed] [Google Scholar]
- 34.Kim PM, Tidor B. Genome Res. 2003;13:1706–1718. doi: 10.1101/gr.903503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alter O, Brown PO, Botstein D. Proc Natl Acad Sci USA. 2000;97:10101–10106. doi: 10.1073/pnas.97.18.10101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moloshok TD, Klevecz RR, Grant JD, Manion FJ, Speier WFT, Ochs MF. Bioinformatics. 2002;18:566–575. doi: 10.1093/bioinformatics/18.4.566. [DOI] [PubMed] [Google Scholar]
- 37.Dueck D, Morris QD, Frey BJ. Bioinformatics. 2005;21(Suppl 1):i144–i151. doi: 10.1093/bioinformatics/bti1041. [DOI] [PubMed] [Google Scholar]
- 38.Isakoff MS, Sansam CG, Tamayo P, Subramanian A, Evans JA, Fillmore CM, Wang X, Biegel JA, Pomeroy SL, Mesirov JP, Roberts CW. Proc Natl Acad Sci USA. 2005;102:17745–17750. doi: 10.1073/pnas.0509014102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alter O, Golub GH. Proc Natl Acad Sci USA. 2006;103:11828–11833. doi: 10.1073/pnas.0604756103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang E, Ishida S, Pittman J, Dressman H, Bild A, Kloos M, D'Amico M, Pestell RG, West M, Nevins JR. Nat Genet. 2003;34:226–230. doi: 10.1038/ng1167. [DOI] [PubMed] [Google Scholar]
- 41.Bild AH, Yao G, Chang JT, Wang Q, Potti A, Chasse D, Joshi MB, Harpole D, Lancaster JM, Berchuck A, et al. Nature. 2006;439:353–357. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 42.Parmigiani G, Garrett-Mayer ES, Anbazhagan R, Gabrielson E. Clin Cancer Res. 2004;10:2922–2927. doi: 10.1158/1078-0432.ccr-03-0490. [DOI] [PubMed] [Google Scholar]
- 43.Hayes DN, Monti S, Parmigiani G, Gilks CB, Naoki K, Bhattacharjee A, Socinski MA, Perou C, Meyerson M. J Clin Oncol. 2006;24:5079–5090. doi: 10.1200/JCO.2005.05.1748. [DOI] [PubMed] [Google Scholar]
- 44.Brier GW. Monthly Weather Rev. 1950;78:1–3. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.