Abstract
Although clinical and laboratory evidence support roles for both staphylococcal infection and environmental allergens in the pathogenesis of atopic dermatitis, human studies have largely considered these variables independently. We sought to test the hypothesis that staphylococcal superantigen influences the allergen-specific T cell response. We first mapped a Der p 1 epitope and used HLA DRB1*1501 class II tetramer-based cell sorted populations to show that specific CD4+ T cells were able to recognize the peptide presented by HLA DR-matched keratinocytes. We observed that staphylococcal enterotoxin B (SEB) enhanced the IL-4 Der p 1-specific T cell response. This response was mediated by two synergistic mechanisms: first, SEB-induced IFN-γ promoted class II and intercellular adhesion molecule-1 expression by presenting keratinocytes; and second, SEB-induced IL-4 directly amplified allergen-specific CD4+ T cell production of many cytokines. We propose that handling of staphylococcal infection is a critical step in the amplification of the allergen-specific T cell response, linking two common disease associations and with implications for the prevention and treatment of atopic disease.
Keywords: atopy, T cells, keratinocytes, Staphylococcus
There is convincing evidence that allergic reactivity to environmental challenge has a role in the pathogenesis of atopic dermatitis (AD). For example, the histology of AD shares many features with classic allergic contact delayed-type hypersensitivity, including spongiosis and a dominant T cell inflammatory infiltrate that is distributed predominantly in a dermal, and to a lesser extent epidermal pattern. Approximately 80% of individuals with AD have circulating specific IgE recognizing one or more of house dust mite, cat, dog, grass, and other ubiquitous environmental allergens (1). In many studies, allergen-specific CD4+ and CD8+ T cells have been documented to be present in the peripheral blood and lesional skin of affected individuals and to produce diverse cytokines but with a frequent T helper 2 (Th2) dominance (2, 3). House dust mite extract application to the skin of individuals with AD can reproduce eczematous changes (4, 5), and allergen avoidance has been shown by some to be partially effective (6, 7). Keratinocytes have been shown to up-regulate HLA class II and intercellular adhesion molecule-1 (ICAM-1) in AD and other inflammatory dermatoses (8, 9) and also in response to injection of IFN-γ into normal skin (10). However, in isolation, allergic reactivity to an environmental challenge does not explain all of the features of AD. Twenty percent of affected individuals do not generate IgE responses to such ubiquitous proteins, and it is unclear why disease incidence changes during age and between different geographical regions. The risk of disease is clearly influenced by genetic susceptibility factors, some of which affect the immune response, e.g., polymorphisms of the FcεR1 and IL-4R genes (11–13). However, it is becoming increasingly clear from the genetic studies that skin-specific genes, determining variables such as barrier function and innate immunity, might be primary events (14, 15) with Th2-biased immune responses perhaps being secondary.
There is also strong evidence supporting a role for Staphylococcus aureus infection in the pathogenesis of AD. Most affected individuals have S. aureus identifiable on skin swabs, often at very high density and more often present on lesional than nonlesional skin (16). Furthermore, S. aureus present on the skin of atopics is significantly more likely to express superantigen, and indeed the levels correlate with disease severity (17–19). Keratinocytes have been shown to up-regulate HLA class II and ICAM-1 after the application of staphylococcal superantigen to the skin (20). Lower levels of IgG2 antibody to S. aureus superantigen have been associated with more severe disease (21). In contrast, the histology of AD shares few features with active S. aureus infection, and staphylococcal infection of the skin of nonatopics does not usually produce an eczematous reaction. S. aureus can be isolated from the nonlesional skin of atopics, and short-term antibiotic treatment has not been convincingly shown to modify chronic AD. Therefore, although superantigen-producing S. aureus is an important associated feature of clinical AD, its presence does not explain all aspects of the disease.
We reasoned that inappropriate handling of S. aureus might influence the allergen-specific immune response. To our knowledge, there are no human data suggesting an interaction between these variables. Staphylococcal superantigen has been documented to reduce regulatory T cell activity in humans (22) and to confer T cell steroid resistance (23), but these studies have not examined the influence of superantigen on allergen-specific T cell responses. A murine study showed that combined staphylococcal enterotoxin B (SEB) and ovalbumin application to the skin increased ovalbumin-specific IgE, but again the role of specific T cells has not been addressed (24). Using house dust mite-derived Der p 1 as the model allergen, we sought to test the hypothesis that S. aureus superantigen influences the allergen-specific T cell response.
Results
Identification of Der p 1-Specific T Cells.
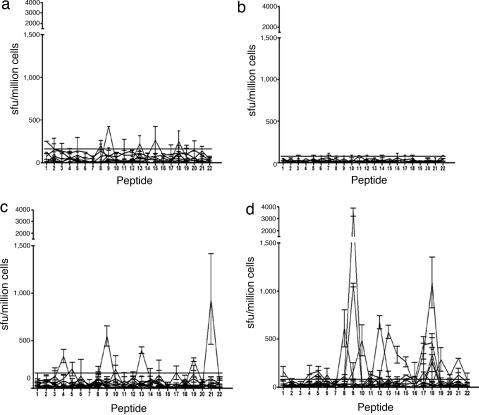
To test our hypothesis, we first had to characterize the immunogenic regions of a common allergen. To determine the breadth of responses to potential epitopes within the Der p 1 protein, cells derived from patients and controls were tested by ELISpot to overlapping peptides (20 aa in length) from the mature protein. To examine the cytokine polarization of antigen-specific T cells, we compared the IFN-γ and IL-4 responses to these peptides by using cultured ELISpot. Although some (40%) nonatopic individuals did demonstrate positive responses to Der p 1 peptides (Fig. 1 a and b), they were predominantly mediated through production of IFN-γ. Atopics demonstrated significantly more frequent (P = 0.0006) and stronger (P < 0.0001) IL-4 responses compared with nonatopic controls (Fig. 1 c and d). Eighty-five percent of atopics responded to at least one Der p 1-derived peptide through production of IL-4. Responses to individual peptides frequently showed both IFN-γ and IL-4 responses but in certain individuals were preferentially IL-4 or IFN-γ. Antigen-specific responses were recorded for 20 of 22 overlapping peptides, correlating to multiple potential T cell epitopes arising throughout the mature protein. CD8+ T cell depletion confirmed that these responses were largely mediated by CD4+ T cells.
Fig. 1.
Cultured IFN-γ and IL-4 ELISpot responses to Der p 1 peptides by nonatopics (a and b) and individuals with AD (c and d). Cultured ELISpot data from nonatopics (NA) (n = 10) and individuals with AD individuals (n = 13). Responses to overlapping peptides of Der p 1 as defined by ELISpot IFN-γ (a and c) and IL-4 (b and d) cytokine secretion measured as spot- forming units (sfu) per million cells are shown. The horizontal bar represents the cutoff for a positive response (mean + 3 × SD of control wells of the population). Data are presented as mean ± SD of sfu.
Fine Mapping and Restriction of Peptide 18 Response.
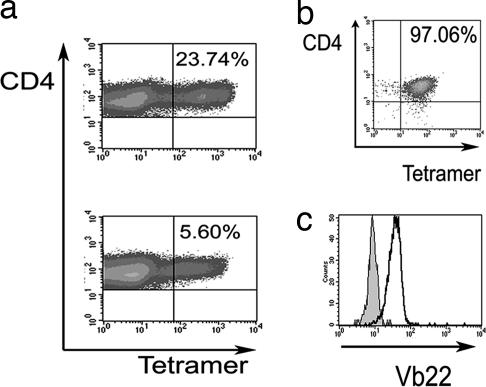
All individuals expressing DRB1*1501 showed T cell responses to peptide 18, which were present both ex vivo and after in vitro culture [supporting information (SI) Fig. 5]. Target cells were sensitized for recognition by pulsing with peptide and by incubation with recombinant Der p 1 protein, indicating functional processing and presentation. An HLA DR-blocking antibody showed partial inhibition of the response to ≈60% (data not shown). Fibroblasts transfected to express HLA DRB1*1501 (kindly given by Lars Fugger, Medical Research Council Human Immunology Unit, Oxford, U.K.) were able to present both peptide and recombinant protein to the T cell line (SI Fig. 5). Serial truncation of the 20 aa peptide showed the optimal epitope to be AVNIVGYSNAQGVDY (SI Fig. 5). We proceeded to use HLA DRB1*1501 tetrameric complexes based on the Der p 1 peptide to facilitate the derivation of antigen-specific CD4+ T cells. We were able to generate large numbers of tetramer-binding cells after short-term incubation of peripheral blood mononuclear cells (PBMCs) from atopics in the presence of specific peptide or recombinant Der p 1 protein (Fig. 2a). Fig. 2b shows the tetramer staining (97.06% positive) of one tetramer-sorted cell population derived from a single tetramer-positive cell, which was shown to express Vβ22 (Fig. 2c). Overall, these data confirm that Der p 1 is a common immunogen in individuals with AD and that it is possible to map optimal CD4+ T cell epitopes. Having identified a dominant epitope within Der p 1, we were able to proceed to test our hypothesis that superantigen influences allergen-specific immunity.
Fig. 2.
HLA DRB1*1501 tetramer staining of T cells. (a) Flow cytometric analysis of a short-term T cell line grown with peptide 18.10 (Upper) or recombinant Der p 1 (Lower). (b) Tetramer-sorted clonal population. (c) The histogram demonstrates a control Vβ 22-negative cell population (filled) vs. CD4+ tetramer-positive cells used in b (open).
SEB Enhances the IL-4 Der p 1-Specific T Cell Response.
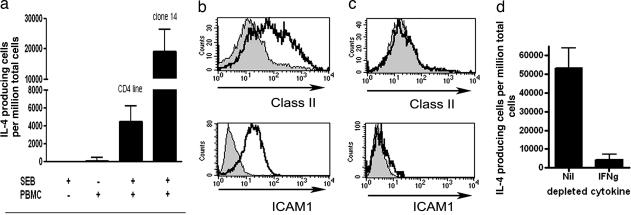
Using DRB1*1501-expressing keratinocytes to present the cognate antigen to Der p 1-specific T cells, we showed that untreated keratinocytes present peptide relatively poorly to antigen-specific CD4+ T cells (Fig. 3a). However, if the keratinocytes were first incubated overnight with the supernatant from SEB-stimulated PBMCs, there was a dramatic increase in the ability of the keratinocytes to present antigen to allergen-specific CD4+ T cells (Fig. 3a). Thus, it is possible that a factor(s) present within the supernatant of SEB-stimulated PBMCs enhanced the capacity of keratinocytes to present antigen to CD4+ T cells. HLA-mismatched DR15-negative PBMCs were used to allow exclusion of any carryover of other cells with the potential capacity to present antigen to the DR15-restricted T cells (Fig. 3a). No enhancement occurred with mismatched PBMCs alone or from SEB alone, thus excluding any potential direct effect of SEB on the allergen-specific CD4+ T cells (Fig. 3a). The enhancement occurred when we used PBMCs derived from either atopic or nonatopic individuals and therefore were not dependent on differential SEB reactivity of atopic PBMCs. The keratinocytes did not express CD80/86 and were not able to prime new T cell responses. Overall, these findings suggested that within the peptide-specific T cell line, few of the cells were SEB-reactive and that the enhancement was caused by soluble factor(s) derived from SEB stimulation of the PBMCs. However, to confirm that this theory was indeed the case, we generated a number of Der p 1-specific CD4+ T cell clones by using tetramer-guided cell sorting to obtain a purified clonal population that we could be confident was SEB-nonreactive. Of the several clones generated, we observed that one clone (clone 14) expressed Vβ 7 [ImMunoGeneTics database entry BV4-1 (refs. 25 and 26)] which is SEB-nonreactive in humans (27). We then proceeded to use ELISpot to investigate whether clone 14 was able to recognize peptide-pulsed keratinocytes after the latter had been incubated with the supernatant from SEB-stimulated PBMCs, and we showed that this theory was the case (Fig. 3a). Overall, these findings documented that factor(s) produced from SEB interaction with PBMCs promoted the antigen-presenting capacity of keratinocytes. Using ELISpot, we showed that SEB stimulation of PBMCs could induce production of both IFN-γ and IL-4 (mean 2,577 ± 765 and 808 ± 186 cytokine-producing cells per million PBMCs respectively). We therefore hypothesized that IFN-γ or IL-4 may have contributed to the enhanced presentation effect, and we proceeded to address this question and the underlying mechanisms.
Fig. 3.
SEB enhances the IL-4 Der p 1-specific T cell response. (a) HLA DRB1*1501 keratinocytes (HaCaT cells) were incubated overnight with SEB alone or with unstimulated PBMCs or SEB-stimulated PBMCs. The keratinocytes were subsequently washed, pulsed with peptide, and then incubated with a peptide-specific T cell line or clone 14 in an IL-4 ELISpot assay. Only when keratinocytes were incubated with the supernatant from SEB-stimulated PBMCs were they able to present antigen to the peptide-specific CD4+ T cell line and clone. Responses are demonstrated as sfu by using IL-4 ELISpot with mean ± SD. (b) Flow cytometric analysis of HLA class II and ICAM-1 expression of keratinocytes incubated overnight with supernatant from PBMCs alone (gray area) or PBMCs and SEB (black line). (c) As b but with IFN-γ-depleted supernatant. (d) IL-4 ELISpot assay using the undepleted (Nil) or IFN-γ-depleted (IFNg) supernatant-incubated keratinocytes to present peptide 18.10 to specific T cell line. Data are presented as mean ± SD.
SEB-Induced IFN-γ Enhances Class II and ICAM-1 Expression by Keratinocytes, Resulting in Increased Antigen Presentation to T Cells.
Incubation of the keratinocytes with the supernatant derived from SEB and PBMCs resulted in significant enhancement of the class II and ICAM-1 expression by the keratinocytes (Fig. 3). Removal of IFN-γ from the supernatant inhibited up-regulation of class II and ICAM-1 and resulted in loss of the enhanced presentation (Fig. 3). Furthermore, the increase in class II and ICAM-1 expression could be replicated by the addition of IFN-γ (300 units/ml) overnight to both HaCat cultured keratinocytes and primary keratinocytes and resulted in significantly (P < 0.01) increased presentation to T cells (SI Fig. 6). These data show that SEB-induced IFN-γ promotes antigen presentation by keratinocytes, which associates with induction of class II and ICAM-1 expression. Thus, SEB not only directly stimulates the SEB-reactive T cells within a given population but also has the capacity to enhance epithelial antigen presentation to all CD4+ T cells regardless of whether they carry SEB-reactive TCR.
SEB-Induced IL-4 Further Amplifies Th2 Cytokine Production.
We reasoned that IL-4 produced by both SEB-reactive and allergen-reactive T cells within a mixed-T cell population may promote further amplification of Th2 cytokine production, and we set out to test this possibility.
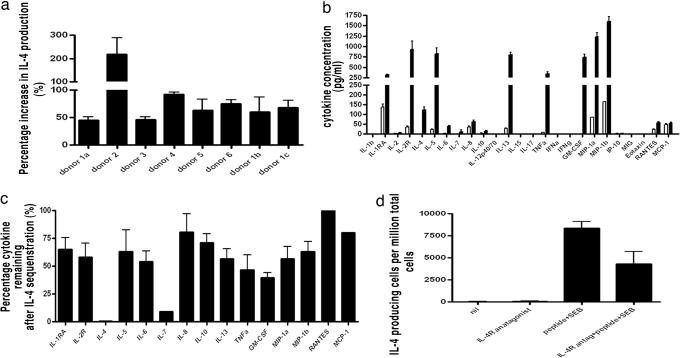
We used p18-specific polyclonal T cell lines derived from six individuals and found that combined SEB and peptide stimulation led to significantly enhanced production of IL-4 compared with the sum of each stimulant alone (Fig. 4a). This finding suggested that within a mixed-T cell population, including both SEB-reactive and peptide-reactive T cells, combined stimulation leads to synergistic production of IL-4. We used two approaches to investigate whether IL-4 itself was central to this Th2 amplification. We first undertook analysis of the supernatant of the peptide-stimulated polyclonal T cell line and showed broad cytokine production including both Th1 and Th2 cytokines (Fig. 4b). We repeated the experiment in conditions that would sequester IL-4 (IL-4low) from the supernatant as it was produced. In IL-4low conditions, the production of many cytokines was significantly reduced (Fig. 4c). To confirm these findings, we tested whether IL-4 receptor inhibition would affect IL-4 production because Th2 cells are known to express the IL-4 receptor (28). In conditions of a blocking IL-4 receptor antibody, T cells produced significantly less IL-4 when stimulated with combined SEB and peptide (Fig. 4d). We showed no influence of IL-4 depletion on cell growth during the overnight assay and confirmed that there was no difference in annexin-V expression (SI Fig. 7). Overall, these data showed that IL-4 produced by SEB-reactive and peptide-reactive T cells promotes the further production of IL-4 and many other cytokines. Thus again, SEB not only directly stimulates the SEB-reactive T cells within a given population but also has the capacity to enhance IL-4 production indirectly by all Th2 cells within a given population regardless of whether they carry SEB-reactive TCR or not.
Fig. 4.
SEB enhances IL-4 responses of antigen-specific T cells by IL-4 mediated activation. (a) IL-4 ELISpot assay. Polyclonal peptide-specific T cell lines from six separate donors show significantly (P < 0.01) enhanced IL-4 production when stimulated with combined peptide + SEB compared with the sum of the isolated peptide and SEB responses. Three different lines were tested from donor 1. (b) Cytokine bead assay. Cytokine production by peptide-stimulated (filled) antigen-specific T cell line from donor 1 compared with irrelevant stimulus (open). The line produces both Th1 and Th2 cytokines in response to peptide. (c) Percentage reduction of cytokine production in a low IL-4 environment. Of the cytokines showing responses in b, the majority were significantly (P < 0.01) reduced in conditions that sequestered IL-4. (d) IL-4 ELISpot assay. Blocking IL-4R signaling results in significantly (P < 0.01) reduced IL-4 production after combined stimulation of peptide-specific T cell line with SEB and peptide. Data are presented as the mean ± SD of sfu.
Overall, SEB acts beyond just the stimulation of the SEB-reactive T cells within a mixed population of cells and will provide a far greater adjuvant effect through both enhancing epithelial presentation of antigen and by providing amplification to existing Th2 cells at an inflammatory site.
Discussion
Having first identified a CD4+ T cell epitope within Der p 1, we have shown that S. aureus enterotoxin B superantigen is a potent adjuvant for human allergen-specific Th2 responses through production of IL-4 and IFN-γ. We have documented that Der p 1 epitope-specific T cell production of Th2 cytokines is enhanced by SEB-induced IFN-γ and IL-4. The IFN-γ promotes class II and ICAM-1 expression by epithelial cells, which facilitates presentation of antigen to CD4+ T cells. In addition, the SEB-induced IL-4 further amplifies the production of many cytokines by allergen-specific CD4+ T cells. Both of these pathways combine to produce dramatic enhancement of the allergen-specific Th2 response. Overall, our data show that superantigens potentially have a much broader influence than thought previously and are able to amplify T cell responses far beyond those just involving SEB-reactive TCR. When such amplification includes promoting an inflammatory Th2 environment to harmless allergens, this possibility may offer a survival advantage to S. aureus as discussed below. IFN-γ is known to promote Th1 and to inhibit Th2 responses. However the current data show that through facilitating increased epithelial presentation of antigen to Th2 cells, IFN-γ is ultimately also able to promote increased IL-4 production and to contribute to disease. Although AD is a complex disease and is likely to have heterogeneous mechanisms differing between patient groups, we believe that in many individuals such an interaction between S. aureus and allergen reactivity may play an important role in disease pathogenesis, particularly in the determination of disease progression and severity.
A wealth of evidence has highlighted the importance of S. aureus and allergens in atopic disease pathogenesis, but there are surprisingly few studies that have investigated potential interactions and synergy between these variables. The cutaneous presence of S. aureus correlates with active AD, and use of antibiotics during acute flares of disease is beneficial. Intravenous Ig, which contains anti-staphylococcal antibody, is beneficial in individuals with severe AD (29). However, the use of antibiotics in combination with allergen avoidance (or immunotherapy) has not been assessed for prevention or treatment of chronic disease. Differential handling of S. aureus may influence the likelihood of subsequent development of a disease-relevant allergen-specific immune response. Recent data suggest that cutaneous expression of antimicrobial peptides/proteins may influence disease, and indeed altered defensin expression in lesional AD skin has been documented (30, 31). Furthermore, AD has been linked to a region of chromosome 1 that contains numerous genes thought to encode antimicrobial proteins (32). At any point, the vast majority of skin-homing T cells are believed to be resident in the skin (33) and thus are likely to represent a diverse pool of cells with the potential TCR capacity to bind to SEB. Many different cutaneous stimuli may have the capacity to induce IFN-γ, and indeed localized exacerbation of AD with local traumatic events such as scratching is well documented. However, the additional potent induction of IL-4 by SEB would be consistent with a mechanism in which S. aureus further facilitates Th2 amplification and chronic disease. In addition, the selective stimulation of an IL-4 allergen-specific T cell response may have several survival advantages for S. aureus. IL-4 is known to promote reduction in cutaneous defensins (34), increase expression of potential S. aureus adhesion molecules (35, 36), reduce potentially damaging Th1 cytokine responses and phagocytic infiltration (37, 38), reduce skin-homing receptor expression by pathogen-specific T cells, and reduce superantigen-induced lethality of reactive T cells (39). By diverting cellular immunity to a Th2 response to ubiquitous environmental allergens, S. aureus promotes an inflammatory surface site conducive to survival and efficient contact spread to neighboring hosts without typically causing host lethality.
The hygiene hypothesis predicts that reduced infections in early childhood modify risk of atopic disease and remains controversial (40). The current data suggest that certain infections can promote allergic inflammation and raise the possibility that the influence of infections or antibiotics may in some cases be complicated by other intermediate variables, such as staphylococcal carriage. It also offers potential explanations for differences in incidence between geographical regions, in migrants, and during childhood. Changes in colonizing bacteria or acquisition of immunity to such populations (21) might modify the risk of developing allergen-specific immunity. We believe that the proposed mechanism may generically apply to other hypersensitivities, particularly when S. aureus has been implicated (e.g., hay fever), but it is likely that in different settings, there will be other relevant adjuvants, such as respiratory syncytial virus infection in asthmatics. The mechanism would also fit with other models of infection-induced flares in autoimmune disease. Furthermore, because we now have the relevant scientific tools to study the CD4+ T cell response in detail, it will also be important to investigate changes in the first months of life to define the earliest events in atopic disease development.
The study data have implications for the management of many individuals with AD. Combined allergen avoidance (or immunotherapy) with prolonged anti-staphylococcal treatment may offer therapeutic benefit. New treatments and/or vaccination with less likelihood for development of resistance are on the horizon and would be important for this undertaking (41). Understanding the components of successful immunotherapy will be critically important as we move further toward more targeted therapeutics. Another interesting strategy may incorporate inhibition of superantigen-mediated proinflammatory signaling such as SOCS3 inhibition of STAT1 phosphorylation (42).
Overall, we have shown that staphylococcal-derived superantigen is a potent adjuvant for allergen-specific Th2 responses. In the setting of AD, this finding links two of the most common disease associations and may explain chronic disease exacerbation seen with recurrent staphylococcal infections. It also potentially provides a mechanism that draws together existing diverse genetic, immunological, and clinical data and has implications for the pathogenesis and management of atopic disease.
Materials and Methods
Subjects.
Thirteen moderate to severely symptomatic AD patients were recruited through the department of dermatology, Churchill Hospital, Oxford, U.K., under ethical approval from the Oxfordshire Clinical Research Committee. All AD patients fulfilled the diagnostic criteria for AD as defined by the U.K. AD diagnostic criteria, which are specific and sensitive for AD (43, 44). Ten nonatopic controls without AD were also recruited. The mean house dust mite IgE (d1, ImmunoCAP, assay range 0–6; Amersham Pharmacia, Piscataway, NJ) for each group was: AD, 4.25 (±1.76, SD); nonatopic, 0.0 (±0.0, SD). All individuals with disease had detectable house dust mite-specific IgE. Donors were tissue typed by PCR–SSP phototyping (45).
T Cell Culture.
PBMCs were separated from heparinized whole blood on Lymphoprep (Axis-Shield PoC, Oslo, Norway), washed, and resuspended in RPMI medium 1640 (Invitrogen, Carlsbad, CA) supplemented with 2 mM l-glutamine/50 units/ml penicillin/50 μg/ml streptomycin/10% (vol/vol) human serum (R10H). Using the known sequence for Der p 1 (Swiss-Prot locus DERP1_DERPT, accession no. P08176), overlapping peptides (by 10 aa) spanning the whole length of the mature protein were constructed in house or at a separate source (Sigma–Aldrich, St. Louis, MO). Peptides were added each at a final concentration of 4 μM. The cells were incubated at 37°C with 5% CO2. At days 3 and 7, IL-2 was added to a final concentration of 100 units/ml. At day 10, the cells were removed from the plate, washed twice in sterile PBS, and returned overnight to unused wells in R10H. At day 11, the cells were then used in ELISpot analyses as below at 4 × 104 per well.
ELISpot Analyses.
ELISpot analyses were performed as described previously (46). Ex vivo PBMCs were tested at 2 × 105 per well; for T cell lines, input cell numbers were 4 × 104 per well; and highly enriched tetramer sorted T cells were tested at 2–2.5 × 103 per well. Peptide was added at a final concentration of 8 μM. Recombinant Der p 1 (Indoor Biotechnologies, Charlottesville, VA) was added at a final concentration of 10 μg/ml. SEB (confirmed endotoxin-free by mass spectrometry; Sigma) was added to a final concentration between 0.75 and 5 μg/ml (as indicated). The plates were incubated overnight at 37°C and 5% CO2 and were developed with streptavidin-alkaline phosphatase (Mabtech AB, Cincinnati, OH) and alkaline phosphatase conjugate substrate kit (Bio-Rad, Hercules, CA). As a control, all experiments included culture medium alone and or an irrelevant peptide. Peptide-specific reactivity was calculated by subtracting the counts from control wells, and the results were expressed as sfu per million lymphocytes plated. Positive responses were recorded as those responses greater than the mean of all the background samples plus 3 × the SD of the population.
DRB1*1501 Keratinocyte Presentation to CD4+ Cells (47).
HaCaT keratinocytes (gift from N. Fusenig, German Cancer Research Center, Heidelberg, Germany) were grown in DMEM/10% FCS at 37°C with 5% CO2. HLA DR expression was confirmed with a pan anti-HLA DR FITC-conjugated antibody source in flow cytometry. As a control, immortalized non-DRB1*1501-expressing keratinocytes (gift from E. O'Toole, Department of Dermatology, Royal Hospitals NHS Trust, London, U.K.) were used, and they were grown in the same manner. The recombinant protein (75 μg/ml) and irrelevant and peptide 18 (both 40 μM) were pulsed for 1 h. Before incubation with the T cell line, HaCaTs were washed three additional times to remove any excess protein or peptide.
IFN-γ Depletion of Supernatant from SEB-Stimulated PBMCs.
R10H was run through washed protein G–Sepharose 4 Fast Flow (Amersham Biosciences) beads in an Econo-Column (Bio-Rad) (R10col). The supernatant was then incubated for 60 min at room temperature with mouse IgG1 anti-cytokine Ab (anti-IFN-γ at 30 μg/ml; Mabtech AB). Both the anti-IFN-γ-treated supernatant and the control were passed through the protein G column, which binds mouse IgG1, thus depleting IFN-γ. Depletion was checked by ELISA.
Flow Cytometry.
Four-color flow cytometric analysis was performed by using a FACScalibur (Becton Dickinson, San Jose, CA) with CellQuest software (Becton Dickinson). DRB1*150101 iTAg (Beckman Coulter, San Diego, CA) tetramer staining was carried out at 37°C, 5% CO2 for 1 h before cell surface antibody staining. Directly conjugated Abs were added according to the manufacturers' instructions, and the samples were incubated for 20 min at 4°C. Samples were fixed in 2% formaldehyde.
Cytokine and Chemokine Analyses.
T cells derived from a line grown against peptide 18 were incubated on a flat-bottomed 96-well plate. Input cell numbers were 4 × 104 per well. Peptide 18 or irrelevant peptide was added at a final concentration of 8 μM. SEB was added at a final concentration of 1 μg/ml. The plates were incubated at 37°C and 5% CO2. After 16 h, the plates were centrifuged, and 50 μl of supernatant from cell stimulation wells were analyzed with a Human Antibody Bead 25-plex kit for the Luminex system (Biosource International, Camarillo, CA), according to the manufacturer's instructions. Plate-bound anti-IL-4 antibody (Mabtech AB) was used to deplete IL-4 and confirmed with measurement of cytokine levels. Unstimulated (irrelevant) controls were used to correct for the effect on cytokine production by changes in cell number between IL-4normal and IL-4low experiments. A single correction factor was used for all cytokines so that the overall pattern of cytokine production remained unaltered.
Statistical Analysis.
Statistical analyses were calculated by using Prism software (GraphPad, San Diego, CA). A two-tailed Mann–Whitney test was used to detect a statistical difference between strength of response in AD and nonatopics. A two-tailed Fisher's exact test was used to test the number of individuals demonstrating a positive cytokine response in each group.
Supplementary Material
Acknowledgments
We thank all of the patients and controls as well as staff at the Department of Dermatology, Churchill Hospital, Oxford, and at the Weatherall Institute of Molecular Medicine, Oxford, for their valuable contributions. We very much appreciate the suggestions and critical review from Prof. Andrew McMichael, Prof. Lars Fugger and Prof. F. Y. Liew. This work was supported by the Wellcome Trust, the Medical Research Council, and the Barrie Trust.
Abbreviations
- AD
atopic dermatitis
- ICAM-1
intercellular adhesion molecule-1
- PBMCs
peripheral blood mononuclear cells
- SEB
staphylococcal enterotoxin B
- sfu
spot-forming units
- Th2
T helper 2.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0700733104/DC1.
References
- 1.Juhlin L, Johansson GO, Bennich H, Hogman C, Thyresson N. Arch Dermatol. 1969;100:12–16. doi: 10.1001/archderm.100.1.12. [DOI] [PubMed] [Google Scholar]
- 2.Wierenga EA, Snoek M, de Groot C, Chretien I, Bos JD, Jansen HM, Kapsenberg ML. J Immunol. 1990;144:4651–4656. [PubMed] [Google Scholar]
- 3.van der Heijden FL, Wierenga EA, Bos JD, Kapsenberg ML. J Invest Dermatol. 1991;97:389–394. doi: 10.1111/1523-1747.ep12480966. [DOI] [PubMed] [Google Scholar]
- 4.Darsow U, Ring J. Clin Exp Dermatol. 2000;25:544–551. doi: 10.1046/j.1365-2230.2000.00695.x. [DOI] [PubMed] [Google Scholar]
- 5.Ring J, Darsow U, Behrendt H. J Am Acad Dermatol. 2001;45:S49–S52. doi: 10.1067/mjd.2001.117015.24. [DOI] [PubMed] [Google Scholar]
- 6.Friedmann PS. Clin Exp Dermatol. 1999;24:433–437. doi: 10.1046/j.1365-2230.1999.00525.x. [DOI] [PubMed] [Google Scholar]
- 7.Clark RA, Adinoff AD. J Am Acad Dermatol. 1989;21:863–869. doi: 10.1016/s0190-9622(89)70269-3. [DOI] [PubMed] [Google Scholar]
- 8.Singer KH, Tuck DT, Sampson HA, Hall RP. J Invest Dermatol. 1989;92:746–750. doi: 10.1111/1523-1747.ep12722441. [DOI] [PubMed] [Google Scholar]
- 9.Bieber T, Dannenberg B, Ring J, Braun-Falco O. Clin Exp Dermatol. 1989;14:35–39. doi: 10.1111/j.1365-2230.1989.tb00880.x. [DOI] [PubMed] [Google Scholar]
- 10.Barker JN, Allen MH, MacDonald DM. Br J Dermatol. 1990;122:451–458. doi: 10.1111/j.1365-2133.1990.tb14721.x. [DOI] [PubMed] [Google Scholar]
- 11.Cox HE, Moffatt MF, Faux JA, Walley AJ, Coleman R, Trembath RC, Cookson WO, Harper JI. Br J Dermatol. 1998;138:182–187. doi: 10.1046/j.1365-2133.1998.02108.x. [DOI] [PubMed] [Google Scholar]
- 12.Novak N, Kruse S, Kraft S, Geiger E, Kluken H, Fimmers R, Deichmann KA, Bieber T. J Invest Dermatol. 2002;119:870–875. doi: 10.1046/j.1523-1747.2002.00191.x. [DOI] [PubMed] [Google Scholar]
- 13.Song Z, Casolaro V, Chen R, Georas SN, Monos D, Ono SJ. J Immunol. 1996;156:424–429. [PubMed] [Google Scholar]
- 14.Cookson W. Nat Rev Immunol. 2004;4:978–988. doi: 10.1038/nri1500. [DOI] [PubMed] [Google Scholar]
- 15.Palmer CN, Irvine AD, Terron-Kwiatkowski A, Zhao Y, Liao H, Lee SP, Goudie DR, Sandilands A, Campbell LE, Smith FJ, et al. Nat Genet. 2006;38:441–446. doi: 10.1038/ng1767. [DOI] [PubMed] [Google Scholar]
- 16.Guzik TJ, Bzowska M, Kasprowicz A, Czerniawska-Mysik G, Wojcik K, Szmyd D, Adamek-Guzik T, Pryjma J. Clin Exp Allergy. 2005;35:448–455. doi: 10.1111/j.1365-2222.2005.02210.x. [DOI] [PubMed] [Google Scholar]
- 17.Zollner TM, Munk ME, Keller T, Nuber V, Boehncke WH, Kaufmann SH, Duijvestijn AM, Sterry W, Kaufmann R. Immunol Lett. 1996;49:111–116. doi: 10.1016/0165-2478(95)02491-3. [DOI] [PubMed] [Google Scholar]
- 18.Zollner TM, Wichelhaus TA, Hartung A, Von Mallinckrodt C, Wagner TO, Brade V, Kaufmann R. Clin Exp Allergy. 2000;30:994–1000. doi: 10.1046/j.1365-2222.2000.00848.x. [DOI] [PubMed] [Google Scholar]
- 19.Bunikowski R, Mielke ME, Skarabis H, Worm M, Anagnostopoulos I, Kolde G, Wahn U, Renz H. J Allergy Clin Immunol. 2000;105:814–819. doi: 10.1067/mai.2000.105528. [DOI] [PubMed] [Google Scholar]
- 20.Morishita Y, Tada J, Sato A, Toi Y, Kanzaki H, Akiyama H, Arata J. Clin Exp Allergy. 1999;29:1110–1117. doi: 10.1046/j.1365-2222.1999.00593.x. [DOI] [PubMed] [Google Scholar]
- 21.Mrabet-Dahbi S, Breuer K, Klotz M, Herz U, Heeg K, Werfel T, Renz H. Clin Exp Allergy. 2005;35:274–281. doi: 10.1111/j.1365-2222.2005.02192.x. [DOI] [PubMed] [Google Scholar]
- 22.Ou LS, Goleva E, Hall C, Leung DY. J Allergy Clin Immunol. 2004;113:756–763. doi: 10.1016/j.jaci.2004.01.772. [DOI] [PubMed] [Google Scholar]
- 23.Li LB, Goleva E, Hall CF, Ou LS, Leung DY. J Allergy Clin Immunol. 2004;114:1059–1069. doi: 10.1016/j.jaci.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 24.Savinko T, Lauerma A, Lehtimaki S, Gombert M, Majuri ML, Fyhrquist-Vanni N, Dieu-Nosjean MC, Kemeny L, Wolff H, Homey B, Alenius H. J Immunol. 2005;175:8320–8326. doi: 10.4049/jimmunol.175.12.8320. [DOI] [PubMed] [Google Scholar]
- 25.Giudicelli V, Chaume D, Lefranc MP. Nucleic Acids Res. 2004;32:W435–W440. doi: 10.1093/nar/gkh412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lefranc MP, Giudicelli V, Kaas Q, Duprat E, Jabado-Michaloud J, Scaviner D, Ginestoux C, Clement O, Chaume D, Lefranc G. Nucleic Acids Res. 2005;33:D593–D597. doi: 10.1093/nar/gki065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Choi YW, Kotzin B, Herron L, Callahan J, Marrack P, Kappler J. Proc Natl Acad Sci USA. 1989;86:8941–8945. doi: 10.1073/pnas.86.22.8941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bosco A, McKenna KL, Devitt CJ, Firth MJ, Sly PD, Holt PG. J Immunol. 2006;176:4766–4777. doi: 10.4049/jimmunol.176.8.4766. [DOI] [PubMed] [Google Scholar]
- 29.Jolles S. Clin Exp Dermatol. 2002;27:3–7. doi: 10.1046/j.0307-6938.2001.00955.x. [DOI] [PubMed] [Google Scholar]
- 30.Ong PY, Ohtake T, Brandt C, Strickland I, Boguniewicz M, Ganz T, Gallo RL, Leung DY. N Engl J Med. 2002;347:1151–1160. doi: 10.1056/NEJMoa021481. [DOI] [PubMed] [Google Scholar]
- 31.Nomura I, Gao B, Boguniewicz M, Darst MA, Travers JB, Leung DY. J Allergy Clin Immunol. 2003;112:1195–1202. doi: 10.1016/j.jaci.2003.08.049. [DOI] [PubMed] [Google Scholar]
- 32.Sugiura H, Ebise H, Tazawa T, Tanaka K, Sugiura Y, Uehara M, Kikuchi K, Kimura T. Br J Dermatol. 2005;152:146–149. doi: 10.1111/j.1365-2133.2005.06352.x. [DOI] [PubMed] [Google Scholar]
- 33.Clark RA, Chong B, Mirchandani N, Brinster NK, Yamanaka K, Dowgiert RK, Kupper TS. J Immunol. 2006;176:4431–4439. doi: 10.4049/jimmunol.176.7.4431. [DOI] [PubMed] [Google Scholar]
- 34.Nomura I, Goleva E, Howell MD, Hamid QA, Ong PY, Hall CF, Darst MA, Gao B, Boguniewicz M, Travers JB, Leung DY. J Immunol. 2003;171:3262–3269. doi: 10.4049/jimmunol.171.6.3262. [DOI] [PubMed] [Google Scholar]
- 35.Cho SH, Strickland I, Tomkinson A, Fehringer AP, Gelfand EW, Leung DY. J Invest Dermatol. 2001;116:658–663. doi: 10.1046/j.0022-202x.2001.01331.x. [DOI] [PubMed] [Google Scholar]
- 36.Cho SH, Strickland I, Boguniewicz M, Leung DY. J Allergy Clin Immunol. 2001;108:269–274. doi: 10.1067/mai.2001.117455. [DOI] [PubMed] [Google Scholar]
- 37.Xiao T, Kagami S, Saeki H, Sugaya M, Kakinuma T, Fujita H, Yano S, Mitsui H, Torii H, Komine M, et al. J Dermatol Sci. 2003;31:111–117. doi: 10.1016/s0923-1811(02)00149-4. [DOI] [PubMed] [Google Scholar]
- 38.Xiao T, Fujita H, Saeki H, Mitsui H, Sugaya M, Tada Y, Kakinuma T, Torii H, Nakamura K, Asahina A, Tamaki K. Cytokine. 2003;23:126–132. doi: 10.1016/s1043-4666(03)00221-7. [DOI] [PubMed] [Google Scholar]
- 39.Lin YT, Wang CT, Hsu CT, Wang LF, Shau WY, Yang YH, Chiang BL. J Immunol. 2003;171:1102–1108. doi: 10.4049/jimmunol.171.2.1102. [DOI] [PubMed] [Google Scholar]
- 40.Bach JF. J Autoimmun. 2005;25(Suppl):74–80. doi: 10.1016/j.jaut.2005.09.024. [DOI] [PubMed] [Google Scholar]
- 41.Raghavan M, Linden PK. Drugs. 2004;64:1621–1642. doi: 10.2165/00003495-200464150-00002. [DOI] [PubMed] [Google Scholar]
- 42.Jo D, Liu D, Yao S, Collins RD, Hawiger J. Nat Med. 2005;11:892–898. doi: 10.1038/nm1269. [DOI] [PubMed] [Google Scholar]
- 43.Williams HC, Burney PG, Pembroke AC, Hay RJ. Br J Dermatol. 1996;135:12–17. [PubMed] [Google Scholar]
- 44.Williams HC, Burney PG, Hay RJ, Archer CB, Shipley MJ, Hunter JJ, Bingham EA, Finlay AY, Pembroke AC, Graham-Brown RA, et al. Br J Dermatol. 1994;131:383–396. doi: 10.1111/j.1365-2133.1994.tb08530.x. [DOI] [PubMed] [Google Scholar]
- 45.Bunce M, O'Neill CM, Barnardo MC, Krausa P, Browning MJ, Morris PJ, Welsh KI. Tissue Antigens. 1995;46:355–367. doi: 10.1111/j.1399-0039.1995.tb03127.x. [DOI] [PubMed] [Google Scholar]
- 46.Lalvani A, Brookes R, Hambleton S, Britton WJ, Hill AVS, McMichael AJ. J Exp Med. 1997;186:859–865. doi: 10.1084/jem.186.6.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boukamp P, Petrussevska RT, Breitkreutz D, Hornung J, Markham A, Fusenig NE. J Cell Biol. 1988;106:761–771. doi: 10.1083/jcb.106.3.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.