Life is complicated. Things would be much simpler for myosin V if it had to contend only with the in vitro environments created to facilitate elucidation of its mechanochemical properties: isolated actin filament tracks with simple geometries, minimal physical barriers to movement, and no need to interface with microtubule motors that normally cooperate with it (1). But in the cell, myosin V must deal with actin filament networks composed of randomly oriented and/or branched filaments, numerous physical barriers to movement of its vesicular cargo, and the necessity to work in a coordinated fashion with plus-end-directed microtubule motors in a partnership that couples long-range vesicle movement on microtubules with short-range actin-based movement by myosin V in the cell periphery (2, 3). In the paper by Ali et al. in a recent issue of PNAS (4), the authors have begun to tackle this complexity by designing in vitro motility assays in which myosin V encounters either intersecting/branched actin filaments or actin–microtubule intersections. The results are both informative and surprising.
In the first set of experiments, the authors visualized by total internal reflection fluoresence (TIRF) microscopy the movement of a quantum dot-labeled heavy meromyosin (HMM)-like fragment of mouse myosin Va [a dimeric fragment containing the myosin's motor domains but lacking its C-terminal cargo-binding globular tail domains (GTD)] on actin filaments applied to a coverslip by a two-step application, such that the second set of filaments lay at various angles across the top of the first set of filaments. What they observed is that the myosin is perfectly capable of either stepping over an intersecting filament or switching seamlessly to movement on the intersecting filament. Moreover, the myosin can switch filaments that intersect at angles as acute as 150°, suggesting that it is quite flexible, a property that bodes well for its movement on isotropic actin meshworks in vivo. The authors provide evidence that the distance between the trailing head of the myosin and the intersecting actin filament predicts fairly well the three possible outcomes of the encounter: stepping over the filament, stepping onto the filament, or terminating movement; and they integrate these results with current knowledge regarding the myosin's step size. Finally, they show that the myosin is adept at handling branched actin filament arrays generated by the Arp2/3 complex. Specifically, myosin V has no problem switching from the “mother” filament to the “daughter” filament jutting out from the side of the mother filament at the typical ≈70° angle produced by Arp2/3. Overall, these experiments are exciting because they provide the first glimpse of how this important vesicle motor might deal effectively with the complex actin filament geometries found inside living cells.
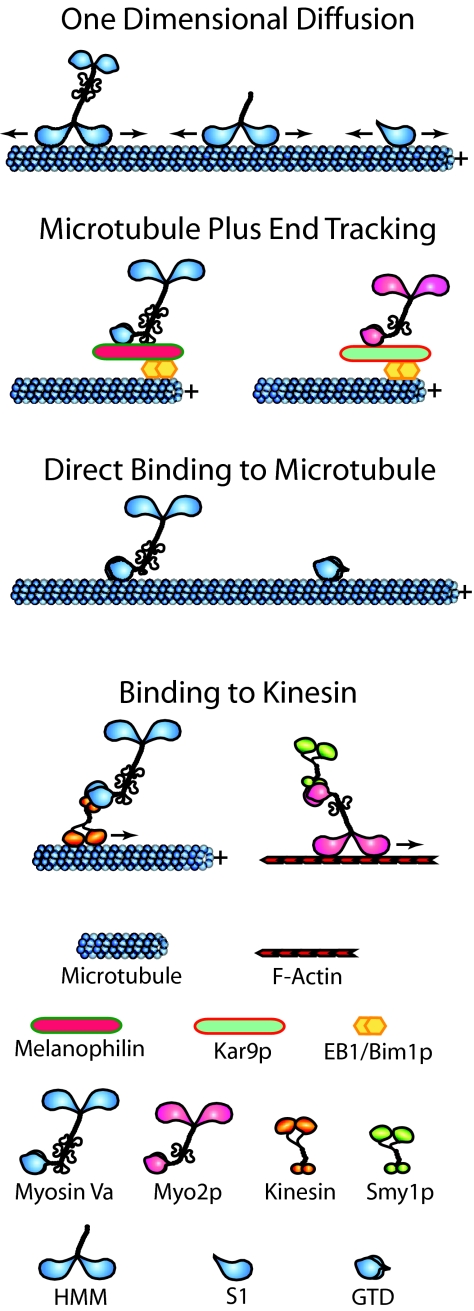
The real surprise the authors provide (4), however, came from imaging the movement of myosin V that was added to coverslips where microtubules had been applied along with actin filaments. Amazingly, the myosin was seen to associate with microtubules directly or by stepping off an intersecting actin filament and to then to undergo rapid one-dimensional diffusion along the microtubule lattice (Fig. 1). Some of the properties of myosin V's one-dimensional diffusion are very similar to those of mitotic centromer-associated kinesin (MCAK), a kinesin-13 family member that uses this mechanism to find the plus and minus ends of the microtubule, where it then catalyzes microtubule depolymerization (5). For example, in both cases, the distribution of displacements in successive images is a Gaussian centered at zero displacement, consistent with a one-dimensional diffusive search, and the calculated diffusion constants for myosin V and MCAK are both ≈0.3 μm2/sec. Ali et al. (4) report a maximal instantaneous velocity of ≈3 μm/sec for myosin V's diffusion on the microtubule, although this number is somewhat misleading because it invites comparison to the speed of real motor protein movement. The actual maximum distance traveled by the myosin per second during its rapid diffusion on the microtubule is probably much greater than 3 μm; MCAK takes 12,000 steps per second when diffusing, which is ≈100-fold faster than kinesin-1 steps directionally on the microtubule (5). As with MCAK, the typical distance scanned on the microtubule by myosin V HMM during each diffusive event is quite small (≈2 μm per event for HMM vs. ≈1 μm for MCAK) because diffusion, although very fast, occurs with equal frequency in both directions. One striking difference, however, is in the average lifetime of a diffusive movement. For MCAK, it is <1 sec, whereas for myosin V, typical events appear to persist for tens of seconds. Myosin V's ability to diffuse along the surface of a microtubule appears to be based on an electrostatic interaction because the frequency of events drops rapidly with rising ionic strength, and myosin V exhibited minimal diffusion on microtubules assembled using tubulin lacking its acidic E hook (which also blocks the diffusion of MCAK). The authors (4) also show that a subfragment 1 (S1)-like fragment of myosin Va (a monomeric fragment spanning the myosin's motor domain) diffuses on the microtubule as effectively as the HMM-like fragment. The authors conclude by suggesting that this form of myosin V motility could serve a number of purposes in vivo. For example, it could promote the physical interaction of myosin V with microtubule motors like kinesin, and it could help myosin V locate organelles undergoing microtubule-dependent movement.
Fig. 1.
Myosin V:microtubule interactions. See text for details.
Although Ali et al.'s (4) demonstration that myosin V can undergo one-dimensional diffusion on microtubules in vitro is both exciting and very thought-provoking, it might be a bit premature to include this form of motility in Mother Nature's tool box for myosin V. The major question is whether the myosin diffuses on microtubules in vivo. One significant concern is that the frequency of myosin V's diffusion events in vitro is very close to zero at an ionic strength approaching physiological. It is somehow unsettling that the motor domain of conventional type II myosin also undergoes one-dimensional diffusion on microtubules. Is this a general property of myosin head domains or some kind of nonspecific in vitro phenomenon? In support of specificity, the authors (4) do state that the full-length myosin V molecule can also diffuse on microtubules. It will be interesting to determine whether this behavior is restricted to the myosin's extended active 11S conformation or whether its folded mechanically quiescent 14S conformation can also diffuse (6, 7). It would also be informative to know whether myosin V can switch back and forth from diffusion on the microtubule to movement on actin. In summary, although it is fair to say that published results of living cells expressing GFP-tagged myosin V do not reveal evidence of the myosin undergoing one-dimensional diffusion along microtubules, it is equally fair to say that this phenomenon could well have been missed in those studies. Experiments in living cells designed specifically to see this phenomenon are now in order.
In considering Ali et al.'s results (4), it is worth taking stock of other ways in which myosin V has been reported to interact with microtubules (summarized in Fig. 1). First, type V myosins have been shown to associate with the plus end of growing microtubules by hitchhiking on the plus-end-tracking protein EB1. This phenomenon was shown first in budding yeast, where the type V myosin Myo2p was shown to target to the plus end of astral microtubules emanating from the budward-directed spindle pole by binding to the bridging protein Kar9p, which in turn is bound to yeast's EB1 homolog Bim1p present at the microtubule tip (8). This microtubule plus-end complex plays an important role in preanaphase spindle positioning (8, 9). Subsequently, mouse myosin Va was shown to target to the plus end of growing microtubules by binding to melanophilin present at the plus end by virtue of its interaction with EB1 (10). Melanophilin's other known role is in linking myosin Va to Rab27a present on the surface of melanosomes (11–14). The plus-end complex of myosin Va–melanophilin–EB1 may play a role in focusing at the microtubule plus end the transition of melanosomes from microtubule- to actin-based movement (“track switching”) (10), although versions of melanophilin that cannot see EB1 still rescue melanosome distribution when introduced into cultured leaden (melanophilin-null) melanocytes (15). Second, purified myosin Va has been shown to bind directly and with reasonable affinity (Kd = 70 nM) to MAP-free microtubules in vitro (16). Binding is mediated by the myosin's globular tail domain, and full length myosin Va generates cross-linked gels of actin and microtubules. Moreover, in the presence of calcium and ATP, myosin Va drives the contraction of these gels, suggesting that it mechanically couples microtubules to actin filaments. Interestingly, endogenous myosin Va has been localized to microtubule-rich domains in both interphase and dividing cells (see refs. in ref. 16). Third, previous yeast two-hybrid data revealed an interaction between an internal fragment of myosin Va's globular tail domain and a fragment of conventional kinesin's stalk domain, suggesting that myosin Va might associate with microtubules indirectly by binding to kinesin and that the coordination of the activities of these two motors might involve their direct physical interaction (17). We note, however, that myosin Va:kinesin interaction has never been verified biochemically by using intact purified proteins, and there is scant evidence that they interact directly in vivo. Having said that, there is clear physical, genetic, and cell biological evidence that yeast Myo2p interacts with Smy1p, a kinesin-like molecule (18, 19). The rub here is that Smy1p does not appear to be a functioning microtubule motor; its ability to suppress a temperature-sensitive mutation in Myo2p does not require microtubules, its motor domain sequence is fairly divergent, and introduction of a mutation that should destroy its motor activity does not destroy its function. The best guess is that Myo2p:Smy1p interaction dates back to a distant time when budding yeasts were hyphal and used Smy1p to drive long-range microtubule-dependent movement in hyphal extensions. With the subsequent loss of a requirement for such long-range movement, Smy1p accumulated mutations that abrogated its function as a microtubule motor, but Myo2p:Smy1p interaction has been retained during evolution because it facilitates Myo2p's function in some way.
Although elegant single-molecule experiments and structural studies draw us ever closer to a full molecular description of myosin V's mechanochemical properties, large questions remain regarding how this motor protein moves vesicles inside cells. By striving to mimic in part the complex environment that myosin V must cope with in vivo, the in vitro experiments of Ali et al. (4) represent an important step toward resolving these questions.
Footnotes
The authors declare no conflict of interest.
See companion article on page 4332 in issue 11 of volume 104.
References
- 1.Sellers JR, Veigel C. Curr Opin Cell Biol. 2006;18:68–73. doi: 10.1016/j.ceb.2005.12.014. [DOI] [PubMed] [Google Scholar]
- 2.Wu X, Bowers B, Rao K, Wei Q, Hammer JA., III J Cell Biol. 1998;143:1899–1918. doi: 10.1083/jcb.143.7.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Langford GM. Traffic. 2003;3:859–865. doi: 10.1034/j.1600-0854.2002.31202.x. [DOI] [PubMed] [Google Scholar]
- 4.Ali MY, Krementsova EB, Kennedy GG, Mahaffy R, Pollard T, Trybus KM, Warshaw DM. Proc Natl Acad Sci USA. 2007;104:4332–4336. doi: 10.1073/pnas.0611471104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Helenius J, Brouhard G, Kalaidzidis Y, Diez S, Howard J. Nature. 2006;441:115–119. doi: 10.1038/nature04736. [DOI] [PubMed] [Google Scholar]
- 6.Thirumurugan K, Sakamoto T, Hammer JA, III, Sellers JR, Knight PJ. Nature. 2006;442:212–215. doi: 10.1038/nature04865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu J, Taylor DW, Krementsova EB, Trybus KM, Taylor KA. Nature. 2006;442:208–211. doi: 10.1038/nature04719. [DOI] [PubMed] [Google Scholar]
- 8.Yin H, Pruyne D, Huffaker TC, Bretscher A. Nature. 2000;406:1013–1015. doi: 10.1038/35023024. [DOI] [PubMed] [Google Scholar]
- 9.Beach DL, Thibodeaux J, Maddox P, Yeh E, Bloom K. Curr Biol. 2000;10:1497–1506. doi: 10.1016/s0960-9822(00)00837-x. [DOI] [PubMed] [Google Scholar]
- 10.Wu XS, Tsan GL, Hammer JA., III J Cell Biol. 2006;171:201–207. doi: 10.1083/jcb.200503028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu XS, Rao K, Zhang H, Wang F, Sellers JR, Matesic LE, Copeland NG, Jenkins NA, Hammer JA., III Nat Cell Biol. 2002;4:271–278. doi: 10.1038/ncb760. [DOI] [PubMed] [Google Scholar]
- 12.Fukuda M, Kuroda TS. J Biol Chem. 2002;277:43096–43103. doi: 10.1074/jbc.M203862200. [DOI] [PubMed] [Google Scholar]
- 13.Strom M, Hume AN, Tarafder AK, Barkagianni E, Seabra MC. J Biol Chem. 2002;277:25423–25430. doi: 10.1074/jbc.M202574200. [DOI] [PubMed] [Google Scholar]
- 14.Wu X, Sakamoto T, Zhang F, Sellers JR, Hammer JA., III FEBS Lett. 2006;580:5863–5868. doi: 10.1016/j.febslet.2006.09.047. [DOI] [PubMed] [Google Scholar]
- 15.Hume AN, Tarafder AK, Ramalho JS, Sviderskaya EV, Seabra MC. Mol Biol Cell. 2006;17:4720–4735. doi: 10.1091/mbc.E06-05-0457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cao TT, Chang W, Masters SE, Mooseker MS. Mol Biol Cell. 2004;15:151–161. doi: 10.1091/mbc.E03-07-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang JD, Brady ST, Richards BW, Stenolen D, Resau JH, Copeland NG, Jenkins NA. Nature. 1999;397:267–270. doi: 10.1038/16722. [DOI] [PubMed] [Google Scholar]
- 18.Lillie SH, Brown SS. J Cell Biol. 1998;140:873–883. doi: 10.1083/jcb.140.4.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beningo KA, Lillie SH, Brown SS. Mol Biol Cell. 2000;11:691–702. doi: 10.1091/mbc.11.2.691. [DOI] [PMC free article] [PubMed] [Google Scholar]