Abstract
Whole-genome sequence data from samples of natural populations provide fertile grounds for analyses of intraspecific variation and tests of population genetic theory. We show that the urochordate Ciona savignyi, one of the species of ocean-dwelling broadcast spawners commonly known as sea squirts, exhibits the highest rates of single-nucleotide and structural polymorphism ever comprehensively quantified in a multicellular organism. We demonstrate that the cause for the extreme heterozygosity is a large effective population size, and, consistent with prediction by the neutral theory, we find evidence of strong purifying selection. These results constitute in-depth insight into the dynamics of highly polymorphic genomes and provide important empirical support of population genetic theory as it pertains to population size, heterozygosity, and natural selection.
Keywords: ciona, genome, heterozygosity, population size
There is increasing evidence that many animal species exhibit high levels of heterozygosity (1–4), which, according to the Neutral Theory, should be caused by elevated mutation rates or large effective population sizes (5). Large population size is, therefore, a candidate cause of elevated genetic variation. Indeed, correlations between effective population size and heterozygosity have been reported in many species, first from allozyme studies and more recently from resequencing of limited numbers of loci (6, 7). A related prediction of the Neutral Theory states that natural selection should be stronger in large populations than in smaller ones. To our knowledge, this important prediction has not been tested for lack of large-scale variation data from natural populations with large effective population sizes. We here comprehensively test the Neutral Theory's predicted relationship between population size, heterozygosity, and strength of selection. The subject of our study is the completed genome sequence of a single wild adult individual of Ciona savignyi, supplemented with limited sequence data from seven additional unrelated individuals (8).
For Ciona spp, a high rate of heterozygosity was reported in the initial publications of the C. savignyi (8) and C. intestinalis (3) genome sequences. However, genomewide quantification and characterization of variation were not conducted in either study. The assembly methodology used by the C. intestinalis project merged allelic sequences such that heterozygosity in the sequenced individual was severely underestimated, and analysis of variation in C. savignyi was confined to seven regions of the genome that totaled ≈200 kb. The results we report here, which are based on analyses of the entire genome, confirm and extend the initial estimates of heterozygosity in C. savignyi. In addition, we address a broader spectrum of variation that includes large structural polymorphism, mobile element activity, microinsertions and microdeletions, and single nucleotide polymorphism.
Our analyses of heterozygosity, population size, and selection were made possible by the unique nature of the initial genome assembly of the sequenced individual, in which allelic sequences from the sister chromosomes were forced to assemble into separate scaffolds (8). The initial assembly thus contained two copies of each genomic region, but no information as to which scaffolds were allelic. To quantify heterozygosity, we constructed a semiautomatic alignment pipeline that produced a tiling path of allelic scaffolds, and then ordered and aligned them while eliminating detectable contig-level assembly errors [see supporting information (SI) Text]. This process resulted in 374 pairwise alignments, the largest 100 of which contained 88% of the genome. The approximate size of the C. savignyi haploid genome (“haplome”) estimated from the alignments is 174 Mb.
Results
The C. savignyi Genome Exhibits Extreme Inversion, Insertion/Deletion, and SNP Heterozygosity.
To determine the rates of multiple types of polymorphism, and whether there are unusual patterns in the underlying mutational spectrum, we quantified patterns and frequencies of those types of allelic differences that could be reliably inferred from the haplome alignment (see SI Text). After elimination of sequence aligned to assembly breaks, 142.6 Mb per haplome was available for analysis of allelic differences (Table 1).
Table 1.
Polymorphism in the C. savignyi genome
| Entity | Bases, Mb | Polymorphic events, count per haplome | Per-base heterozygosity, % | Per-event heterozygosity, % |
|---|---|---|---|---|
| Length of each haplome within alignment* | 142.6 | |||
| Inversions > 1 kb/length of haplome | 2.8/142.6 | 244 | 1.96 | 0.0002 |
| Haplome-specific DNA content/length of haplome | 23.7/142.6 | 516,619 | 16.60 | 0.72 |
| 1–10 bp | 1.1/142.6 | 374,115 | 0.77 | 0.52 |
| >10 bp | 22.6/142.6 | 142,504 | 15.85 | 0.20 |
| Mobile element indels > 100 bp | 16.3/142.6 | 29,124 | 11.43 | 0.04 |
| Shared DNA content/length of haplome | 118.9/142.6 | |||
| High-quality alignment/shared DNA | 109.6/118.9 | |||
| SNPs/bases in high-quality alignment | 4.9/109.6 | 4,916,067 | 4.49 | 4.49 |
*Alignment excludes assembly breaks.
At least 1.96% of the bases in the genome are in inversions of size 1 kb or more (Table 1). This figure is ≈10-fold higher than in humans (9), but comparable to the inversion rate seen between human and baboon, as estimated from alignments of the Encode regions. The size distribution is expectedly skewed, with a few large events accounting for the majority of inverted sequence and small inversions accounting for the majority of events (Fig. 1A).
Fig. 1.
Characterization of inversions and indels. (A) Length distribution of inversions. Bars indicate the number of inversions in each size category as labeled on the x axis. Inversions <1 kb are not included. (B and C) Length distribution of medium and large indel polymorphisms. The red line represents indels that are aligned to repetitive sequence. Nonrepetitive indels are shown in blue. Lines are smoothed to eliminate noise. The peak centered at 891 bp contains 924 indels of length 863–913. The peak centered at 1,086 bp contains 479 indels of length 1,076–1,100. (D) Length distribution of microindels (indels <11 bp) plotted as a fraction of total indels <10 bp in length. Blue squares record microindels observed between the C. savignyi haplomes. Green triangles record microindels between the mouse and rat genomes (10).
An enormous amount of DNA, 23.7 Mb per haplome or 16.6%, is haplome-specific due to polymorphic insertions or deletions (indels; Table 1). There are 1,033,225 indel events between the two haplomes that have an N50 size of 574 bases. The indel size distribution exhibits a highly nonrandom pattern of peaks at lengths suggestive of specific mobile element classes (Fig. 1 B and C). If these indels are indeed indicative of ongoing mobile element activity, the majority of long indels should be aligned to mobile elements. This is, in fact, the case for 76% of all indel events >100 bp (Table 1 and Fig. 1 B and C). We estimate that mobile element activity accounts for 78% of haplome-specific DNA, but for a minority of indel events.
The vast majority of indel events are of size 1–10 bp (microindels; Table 1). The relative size distribution of microindels is extraordinarily similar to that inferred from a comparison of the rat and mouse genomes (10) and to that seen within the human genome (11), with 43% being single-base indels and having an exponential fall-off of frequency with increasing size (Fig. 1D). The remarkable congruence of the size distributions underscores the conservation of the molecular processes that generate microindels in animal genomes. The genomewide average SNP heterozygosity is 4.5% (Table 1), a figure that is in agreement with the previous study of 220 kb of finished sequence from the sequenced individual (8). This extreme degree of SNP heterozygosity is precisely confirmed in finished BAC sequence from seven unrelated individuals, underscoring that the sequenced individual is not an outlier that happens to be unusually polymorphic. Nucleotide diversity in 4-fold degenerate (4D) sites is an unprecedented 8.0% (SI Table 2). Assuming that 4D sites reflect the neutral diversity in the population, the level of single-nucleotide polymorphism in C. savignyi is approximately two percentage points higher than the neutral divergence between human and Old World monkeys (12). The mutational spectrum of SNPs is not unusual, as the transition/transversion rate ratio of 2.45 (SI Table 3) is similar in other species that, like Ciona, do not exhibit a substantial bias against CpG dinucleotides (1). In summary, the C. savignyi genome bears extremely high rates of several types of polymorphism that exhibit standard characteristics.
Because of their high rates, SNPs and microindel polymorphisms lent themselves to high-resolution analysis of the distribution of heterozygosity within the C. savignyi genome (Fig. 2). First, the distributions of heterozygosity measured in nonoverlapping windows of 1 kb (SNPs) and 5 kb (microindels) are overdispersed compared with those from a randomized alignment (SI Fig. 5). Second, we observe a correlation between the frequency of SNPs and microindels across the genome (R2 = 0.25 in 1-kb windows), which is in contrast to comparisons between distinct species, in which a correlation between SNPs and microindels is not observed (10). Both phenomena could be caused by structural or functional heterogeneity across the genome or heterogeneity in the time to coalescence. We observe no correlation between SNP or microindel frequency and repeat density, gene density, or GC content, and therefore propose that these phenomena are a reflection of the time to coalescence between the two alleles in a given region, as has been observed for SNPs in humans (13).
Fig. 2.
SNP and μ-indel heterozygosity are variable across the C. savignyi genome. Map of heterozygosity of SNPs (top bar of each pair) and μ-indels of size 1 (bottom bar) across the 99 longest allelic regions, encompassing 153 Mb (an estimated 88% of the genome). Region ID is on the left. Each colored square represents a nonoverlapping window of 50 kb, with the color range scaled to cover 3 SDs on either side of the mean heterozygosity, as measured in 50-kb windows (SNP mean = 4.6%; μ-indel mean = 0.27%). Gray represents no estimate because of missing sequence for one of the two alleles (see SI Text). Boxed charts show heterozygosity measured in 5-kb windows (smoothed to decrease noise) across the indicated 500-kb regions.
C. savignyi's Extreme Heterozygosity Is Driven by a Large Effective Population Size.
We next sought to identify the underlying cause of the elevated heterozygosity in C. savignyi. Population heterozygosity, θ, is a function of effective population size, Ne, and mutation rate, μ: θ = 4 Neμ (5). Hence, extreme heterozygosity in C. savignyi could be caused by either a large population size or elevated mutation rates. Invoking roughly equally elevated mutation rates for multiple distinct types of polymorphism does not seem parsimonious, suggesting that a large population size may be the better explanation. We therefore estimated effective population size by using recombination rate estimates, relying on the relationship Ne = ρ/4c (ρ is the population recombination parameter, representing the frequency of recombination events among sampled individuals since their most recent common ancestor; c is the per-site, per-generation recombination rate) (5). We directly obtained a range of values for c by typing five large genomic regions in a genetic cross. The average value of c was of 5 × 10−8, but all five values were used in our analyses to capture variation in recombination rates across the genome (see SI Text). ρ was obtained with the aforementioned seven BAC sequences, which constitute a third allele, as follows: Using alignments of the BACs to the sequenced individual's allelic regions, we analyzed the lengths of SNP-based “haplotype blocks,” which are defined as runs of SNPs in which two chromosomes share one allele and the third has another (Fig. 3A). A comparison between the observed block lengths and those calculated from an alignment in which the order of positions is randomized shows an excess of long blocks (Fig. 3A), which is caused by linkage disequilibrium (14). We then generated hypothetical block length distributions by simulation, using a range of values for Ne (Fig. 3B). We find that the C. savignyi haplotype block length distribution is most similar to simulated distributions generated at an Ne of 1.5 million individuals (Fig. 3C and SI Fig. 6).
Fig. 3.
Estimation of effective population size by comparison between observed and simulated haplotype block structures. (A) Frequency distribution of haplotype blocks; length is measured in number of concordant SNPs. The red line is the distribution observed from the three-way alignment of the two haplotypes of the sequenced individual and ≈200 kb of finished BAC sequence from unrelated individuals. The blue line is the distribution observed when the order of alignment columns is randomized. (B) Distribution of the proportion of total SNPs contained at each haplotype block length from 1 to 10. Colored bars show the distributions from population recombination simulations for a range of discrete population sizes (Ne). The red line is the observed distribution as in A. (C) Similarity of observed distribution of haplotype block lengths to each simulated distribution. Each data point represents the mean similarity of the observed haplotype block distribution to 1,000 independent simulated distributions at the specified Ne. Black standard error bars are barely visible. The arrow indicates the point of maximum similarity, at an Ne of 1.5 million individuals.
A large population size does not preclude the possibility that an elevated mutation rate also contributes to the extreme heterozygosity. We calculate the point mutation rate μ to be 1.3 × 10−8 per generation per base by solving θ = 4 Neμ using our estimates of Ne = 1.5 × 106 and θ4D = 0.08. Alternatively, we calculate μ to be 7.6 × 10−9 per generation per base by setting θ to the genomewide SNP heterozygosity (0.045). Both estimates of μ are well within the range previously reported for vertebrate and invertebrate species (15). We conclude that the extreme heterozygosity in C. savignyi is caused by a large effective population size and not an elevated mutation rate.
Evidence for Strong Purifying Selection in C. savignyi.
The neutral theory (5) predicts highly efficient natural selection as a consequence of a large effective population size, as the efficacy of selection is determined by the selection coefficient, s, of a mutation, and the effective population size. Mutations are strongly selected against only if they reduce fitness by s ≫ 1/4 Ne (16). Hence, as effective population size increases, purifying selection should be able to remove alleles with smaller selection coefficients. Because of the large population size, the variation present in C. savignyi should therefore reflect stronger purifying selection than that seen in organisms with smaller effective population sizes. Testing whether these important predictions of the neutral theory hold, we performed analyses to quantify the strength of purifying selection in the C. savignyi population.
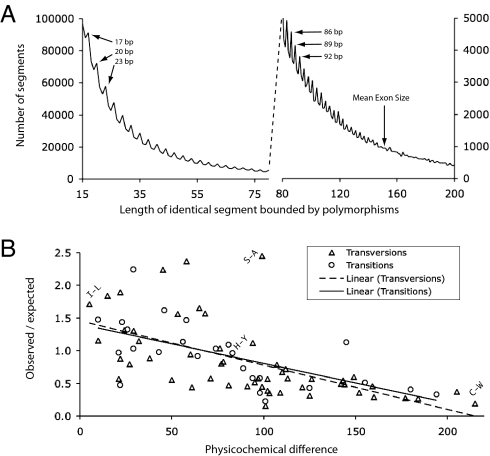
We find a tantalizing footprint of efficient purifying selection in the length distribution of segments of perfect identity between the haplomes (Fig. 4A). In the absence of selection, mutations would be distributed such that the frequency of identical segments decays exponentially as a function of length. By contrast, the C. savignyi genome exhibits a “sawtooth” pattern with a periodicity of three, wherein there are more segments of length 3N + 2 than of length 3N or 3N + 1, even though there is the expected general trend of shorter segments being more common than longer ones. As third position changes in codons that bound N nonpolymorphic codons yield an identical segment of length 3N + 2, the sawtooth pattern is suggestive of purifying selection in coding regions. The pattern is lost just before 150 bp, the mean length of exons in C. intestinalis (3).
Fig. 4.
Purifying selection in the C. savignyi genome. (A) Length distribution of segments of 100% identity in the C. savignyi genome. Peaks occur at lengths 3N + 2, presumably because of the excess of segments bounded by third-position polymorphisms in degenerate codons. The excess of segments of length 3N + 2 is lost near the mean exon length of 150 bp. (B) Nonsynonymous coding polymorphisms in the C. savignyi genome are biased against physiochemically different amino acids. The y axis records the ratio of the observed frequency of polymorphic instances of a pair of amino acids to the frequency expected with no selection. The x axis records the physiochemical difference (20) between amino acid pairs. High values indicate greater difference. Each data point represents one amino acid pair, and all pairs that can be substituted via a single nucleotide change are included (SI Table 7). Four representative points are labeled with single-letter amino acid abbreviations. The dashed line represents the regression for amino acid differences caused by transversions (R2 = 0.36); the solid line indicates those caused by transitions (R2 = 0.40).
To quantify rigorously the strength of purifying selection in coding regions we performed two analyses. First, from the available 207,310 polymorphic codons (SI Table 4 and SI Table 5) we calculated the ratio of the rate of nonsynonymous substitutions (pA) to that of synonymous substitutions (pS) to be 0.07. By contrast, the pA/pS ratio within humans, which have a much smaller effective population size, is higher at 0.23 (17); intermediate pA/pS values of 0.14 and 0.15 have been reported for zebrafish (ref. 18; no published population size estimate) and Drosophila melanogaster (19), for which effective population size estimates range from 104 to 106, with an intermediate value likely being realistic. Thus, as predicted by the neutral theory, C. savignyi, the species with the largest effective population size, exhibits a signature of the most efficient purifying selection among all species for which pertinent data are available.
Second, we examined the physicochemical characteristics of the amino acid changes that are caused by 30,895 nonsynonymous SNPs (SI Table 6). For each of the possible changes, we calculated the ratio of observed-to-expected frequency and found this ratio to be strongly anticorrelated with physicochemical distance (20) between the encoded amino acids (R2 = 0.37; Fig. 4B and SI Table 7). This result shows that SNPs that generate more dissimilar amino acids are, on average, subject to stronger purifying selection than those that generate similar amino acid variants. To our knowledge, this type of analysis has not been done before, and so we lack a comparison with other species. Nonetheless, the fact that more than one-third of the variance in heterozygosity of nonsynonymous SNPs can be explained by Grantham's values (20), which combine two physicochemical properties and the atomic composition of the amino acids, is astounding. The remarkably low value of pA/pS and the strong anticorrelation between physicochemical distance and nonsynonymous SNP frequency underscores the effectiveness of purifying selection in the C. savignyi population.
A large effective population size should also result in highly efficient positive selection, provided that potentially advantageous variants segregate in the population. To address this possibility, we calculated pA/pS ratios across 28,489 high-confidence exons with known frame, where a pA/pS of >1 might indicate positive selection. This type of analysis is the only one we could perform, given that two alleles do not lend themselves to detection of selective sweeps or other signatures of positive selection. Only 17, or 0.06%, of analyzed exons had a pA/pS of >1, and similarity searches and manual examination of the relevant gene models allowed no conclusions as to a shared biological function. By contrast, >78% of exons had a pA/pS of <0.1. Notwithstanding our limited power to detect positive selection, this result further underscores the prevalence of purifying selection in the C. savignyi population.
Discussion
Our analysis of C. savignyi highlights a paradox of the neutral theory. Because of the sheer amount of polymorphism, populations with large sizes and high heterozygosity carry a large genetic burden despite the more efficient purifying selection. This idea is underscored by the fact that geneticists working with C. savignyi now exploit selfing of these hermaphroditic animals to recover deleterious recessive phenotypes at a high rate (21–23). Although the extent of heterozygosity in C. savignyi is extreme by any metric, and in fact surpasses the amount of divergence seen between many species, we do not expect it to remain an outlier. A representative view of the genomic variation in natural populations has been obscured by the preponderance of inbred laboratory and agricultural strains in sequencing projects. As whole-genome sequencing continues to sample wild populations, a more balanced view of the variation segregating within them will emerge.
The strength of purifying selection acting on the extreme genomic variation in C. savignyi also underscores the apparent robustness of the cellular and developmental machinery of a species that contains such genomic variation, considering that a sufficient fraction of individuals in each generation must be viable and have the potential to reproduce. First, the “average” cellular machinery engaged in recombination, replication, and gene expression that is carried in the population must be able to function reliably upon the vast diversity of genomes; second, the population diversity in functional elements (regulatory elements, exons, etc.) that engage in physical interactions must not be so high that it results in too high a chance of deleterious synthetic (genetic) interactions. Given its wealth of easily ascertained polymorphism, C. savignyi provides an excellent natural laboratory for further exploration of the dynamics of variation in natural populations and the functional consequences of such variation.
Methods
Alignment and Haplome Assembly.
The alignment pipeline applied to the initial assembly is described briefly in SI Text. The total length of the reconstructed haplomes is 323,246,196 bp plus 12,758,832 N contig break placeholders. The haplome assembly consists of 374 pairs of allelic sequence, arbitrarily labeled A and B; LAGAN (24) alignments of all pairs are available at http://mendel.stanford.edu/SidowLab/Ciona.html.
Inversion Identification.
Nineteen of the inversions, which cover ≈1.2% of bases in the genome, were identified in our alignment pipeline, manually examined, and reoriented in the final alignment. These regions were excluded from subsequent automated inversion calling, as were 12 allelic sequence pairs that contained large palindromic low complexity regions. Additional smaller inversions were called automatically with SLAGAN (25). A subset of inversion breakpoints was experimentally verified in a previous study (8). We estimated a 2.1% human–baboon inversion rate from 28 Mb of alignments of all ENCODE (26) target regions; 113 inversions were found, which contained a total of 588 kb.
Indel and SNP Identification.
Indels were parsed directly from the haplome alignment by counting the number and size of alignment gaps in regions that did not contain or border assembly breaks. SNPs were identified only in aligned positions that passed a strict alignment quality filter (SI Text). Nucleotide diversity in 4D sites was calculated from alanine, glycine, proline, threonine, serine (TCN codons only), and valine codons by dividing the number of codons with a synonymous substitution by the total number of identical or synonymous instances of that amino acid.
Repeat Identification.
Repeats were identified with RepeatMasker (http://repeatmasker.org) using a de novo repeat library constructed by the RECON (27) program and hand-curated to remove multicopy genes, tRNA, and rRNA elements (SI Text). The library is available at http://mendel.stanford.edu/SidowLab/Ciona.html.
Estimation of Per-Site Recombination Rate.
An estimate for the per-generation per-site recombination rate was obtained by typing 92 meioses of an outbred cross in five distinct regions of the C. savignyi genome totaling 4.6 Mb, or 2.6% of the genome. Markers were spaced approximately every 200 kb across the five regions, which ranged in length from 572 kb to 1.1 Mb. The average physical distance per map unit was ≈250 kb/cM and ranged from 130 to 550 kb/cM across the five regions.
Calculating the Population Recombination Parameter ρ.
Most available methods of calculating ρ were written for typical SNP data sets that are comprised of many alleles at discontinuous, short loci. All such methods we tried failed to produce an estimate of ρ from our data, which was comprised of only three alleles but contained complete sequence over a large region. We therefore estimated ρ by comparing the length distribution of observed and simulated haplotype blocks of concordant SNPs. The MS program (28) was used to simulate 1,000 independent replicates of three sequences with θ = 0.045 at a succession of values of ρ corresponding to values of Ne ranging from 100,000 to 20 million. Similarity between the observed and simulated distributions was calculated as the sum of the absolute value of the difference in frequency at each block length. Block lengths of >20 were condensed into one category. Further details are available in SI Text.
Identification of Coding Variants.
Coding regions and exons were identified by homology to the C. intestinalis v2.0 gene set (http://genome.jgi-psf.org/Cioin2 and SI Text). Counts of all aligned codons and amino acids are provided in SI Table 5 and SI Table 6. pA/pS was estimated from a concatenation of all annotated codons by using CODEML (29) with the F3 × 4 codon frequency model; pA = 0.0059 and pS = 0.0825. Physicochemical distance between amino acids was measured with the Grantham matrix (20), a composite metric of volume, polarity, and atomic composition. Expected frequencies of polymorphic amino acid pairs were normalized to account for codon frequency and the difference in the rate of transitions and transversions. A total of 4,678 amino acid polymorphisms that are the result of more than one nucleotide change were not included in this analysis, but are recorded in SI Table 6. For detection of potential positive selection on exons, 28,489 exons that exhibited more than three silent changes between the two alleles (from a total of 52,372 exons identified by homology to C. intestinalis) were analyzed for their pA/pS ratios.
Supplementary Material
Acknowledgments
We thank Zhirong Bao and Sean Eddy for generating the C. savignyi repeat library; George Asimenos (Stanford University) for providing human–baboon alignments of Encode target regions; Jade Vinson for prepublication access to the Arachne assembly; William Smith and Di Jiang (University of California, Santa Barbara, CA) for providing crossed individuals; and Mehdi Yahyanejad for extensive discussions and advice regarding estimation of the population recombination parameter. This work was supported by grants from the National Institutes of Health/National Institute of General Medical Sciences and National Institutes of Health/National Research Initiative Competitive Grants Program, a National Science Foundation graduate fellowship (to M.B.), a Stanford Graduate Fellowship (to K.S.S.), and the Stanford Genome Training Program (National Institutes of Health/National Human Genome Research Institute) (M.M.H. and K.S.S.).
Abbreviation
- 4D
4-fold degenerate.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0700890104/DC1.
References
- 1.Moriyama EN, Powell JR. Mol Biol Evol. 1996;13:261–277. doi: 10.1093/oxfordjournals.molbev.a025563. [DOI] [PubMed] [Google Scholar]
- 2.Holt RA, Subramanian GM, Halpern A, Sutton GG, Charlab R, Nusskern DR, Wincker P, Clark AG, Ribeiro JM, Wides R, et al. Science. 2002;298:129–149. doi: 10.1126/science.1076181. [DOI] [PubMed] [Google Scholar]
- 3.Dehal P, Satou Y, Campbell RK, Chapman J, Degnan B, De Tomaso A, Davidson B, Di Gregorio A, Gelpke M, Goodstein DM, et al. Science. 2002;298:2157–2167. doi: 10.1126/science.1080049. [DOI] [PubMed] [Google Scholar]
- 4.Sodergren E, Weinstock GM, Davidson EH, Cameron RA, Gibbs RA, Angerer RC, Angerer LM, Arnone MI, Burgess DR, Burke RD, et al. Science. 2006;314:941–952. doi: 10.1126/science.1133609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kimura M. The Neutral Theory of Molecular Evolution. Cambridge, UK: Cambridge Univ Press; 1983. [Google Scholar]
- 6.Hartl DL, Clark AG. Principles of Population Genetics. Sunderland, MA: Sinauer; 1997. [Google Scholar]
- 7.Bazin E, Glemin S, Galtier N. Science. 2006;312:570–572. doi: 10.1126/science.1122033. [DOI] [PubMed] [Google Scholar]
- 8.Vinson JP, Jaffe DB, O'Neill K, Karlsson EK, Stange-Thomann N, Anderson S, Mesirov JP, Satoh N, Satou Y, Nusbaum C, et al. Genome Res. 2005;15:1127–1135. doi: 10.1101/gr.3722605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tuzun E, Sharp AJ, Bailey JA, Kaul R, Morrison VA, Pertz LM, Haugen E, Hayden H, Albertson D, Pinkel D, et al. Nat Genet. 2005;37:727–732. doi: 10.1038/ng1562. [DOI] [PubMed] [Google Scholar]
- 10.Cooper GM, Brudno M, Stone EA, Dubchak I, Batzoglou S, Sidow A. Genome Res. 2004;14:539–548. doi: 10.1101/gr.2034704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bhangale TR, Rieder MJ, Livingston RJ, Nickerson DA. Hum Mol Genet. 2005;14:59–69. doi: 10.1093/hmg/ddi006. [DOI] [PubMed] [Google Scholar]
- 12.Elango N, Thomas JW, Yi SV. Proc Natl Acad Sci USA. 2006;103:1370–1375. doi: 10.1073/pnas.0510716103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reich DE, Schaffner SF, Daly MJ, McVean G, Mullikin JC, Higgins JM, Richter DJ, Lander ES, Altshuler D. Nat Genet. 2002;32:135–142. doi: 10.1038/ng947. [DOI] [PubMed] [Google Scholar]
- 14.Pritchard JK, Przeworski M. Am J Hum Genet. 2001;69:1–14. doi: 10.1086/321275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lynch M. Mol Biol Evol. 2006;23:450–468. doi: 10.1093/molbev/msj050. [DOI] [PubMed] [Google Scholar]
- 16.Ohta T. Nature. 1973;246:96–98. doi: 10.1038/246096a0. [DOI] [PubMed] [Google Scholar]
- 17.Chimpanzee Sequence Analysis Consortium. Nature. 2005;437:69–87. [Google Scholar]
- 18.Guryev V, Koudijs MJ, Berezikov E, Johnson SL, Plasterk RH, van Eeden FJ, Cuppen E. Genome Res. 2006;16:491–497. doi: 10.1101/gr.4791006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fay JC, Wyckoff GJ, Wu CI. Nature. 2002;415:1024–1026. doi: 10.1038/4151024a. [DOI] [PubMed] [Google Scholar]
- 20.Grantham R. Science. 1974;185:862–864. doi: 10.1126/science.185.4154.862. [DOI] [PubMed] [Google Scholar]
- 21.Hendrickson C, Christiaen L, Deschet K, Jiang D, Joly JS, Legendre L, Nakatani Y, Tresser J, Smith WC. Methods Cell Biol. 2004;74:143–170. doi: 10.1016/s0091-679x(04)74007-8. [DOI] [PubMed] [Google Scholar]
- 22.Jiang D, Munro EM, Smith WC. Curr Biol. 2005;15:79–85. doi: 10.1016/j.cub.2004.12.041. [DOI] [PubMed] [Google Scholar]
- 23.Jiang D, Tresser JW, Horie T, Tsuda M, Smith WC. J Exp Biol. 2005;208:433–438. doi: 10.1242/jeb.01420. [DOI] [PubMed] [Google Scholar]
- 24.Brudno M, Do CB, Cooper GM, Kim MF, Davydov E, Green ED, Sidow A, Batzoglou S. Genome Res. 2003;13:721–731. doi: 10.1101/gr.926603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brudno M, Malde S, Poliakov A, Do CB, Couronne O, Dubchak I, Batzoglou S. Bioinformatics. 2003;19(Suppl 1):i54–i62. doi: 10.1093/bioinformatics/btg1005. [DOI] [PubMed] [Google Scholar]
- 26.Encode Project Consortium. Science. 2004;306:636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- 27.Bao Z, Eddy SR. Genome Res. 2002;12:1269–1276. doi: 10.1101/gr.88502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hudson RR. Bioinformatics. 2002;18:337–338. doi: 10.1093/bioinformatics/18.2.337. [DOI] [PubMed] [Google Scholar]
- 29.Yang Z. Comput Appl Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.