Abstract
Glycosylation of macrolide antibiotics confers host cell immunity from endogenous and exogenous agents. The Streptomyces antibioticus glycosyltransferases, OleI and OleD, glycosylate and inactivate oleandomycin and diverse macrolides including erythromycin, respectively. The structure of these enzyme–ligand complexes, in tandem with kinetic analysis of site-directed variants, provide insight into the interaction of macrolides with their synthetic apparatus. Erythromycin binds to OleD and the 23S RNA of its target ribosome in the same conformation and, although the antibiotic contains a large number of polar groups, its interaction with these macromolecules is primarily through hydrophobic contacts. Erythromycin and oleandomycin, when bound to OleD and OleI, respectively, adopt different conformations, reflecting a subtle effect on sugar positioning by virtue of a single change in the macrolide backbone. The data reported here provide structural insight into the mechanism of resistance to both endogenous and exogenous antibiotics, and will provide a platform for the future redesign of these catalysts for antibiotic remodelling.
Keywords: enzymology, glycobiology, erythromycin, streptomyces, glycoside
Microorganisms, primarily Streptomyces, produce an array of clinically significant bioactive molecules that include antibiotics and antitumor and immunosuppressant agents. Polyketides represent the most extensive group of bioactive molecules, which include macrolide polyketide antibiotics that target Gram-positive bacteria. Because resistance to these antimicrobial agents is not yet widespread among pathogens; they have been viewed as the “last line of defense against bacterial pathogens” (World Health Organization, January 30, 1998) (1). Macrolide antibiotics comprise a macrocyclic backbone which following synthesis is decorated with diverse glycans that play a key role in the specificity and potency of these agents (2).
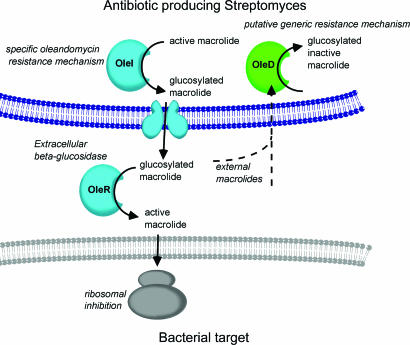
Streptomyces species that produce macrolide polyketide antibiotics also use glycosylation to inactivate these molecules and hence protect themselves from their own antimicrobial agents. In this macrolide glycosyltransferase self-resistance mechanism, intracellular glycosylation inactivates the antimicrobial agent, which is subsequently reactivated, after secretion, by an extracellular β-glycosidase (Fig. 1) (3). The glycosylation of macrolides is mediated by glycosyltransferases which transfer activated donor sugars to acceptor species (3–10). These enzymes are grouped into ≈90 sequence-based families, and to date they display only two major folds defined as GT-A and GT-B (see below; http://afmb.cnrs-mrs.fr/CAZY/). In Streptomyces antibioticus, the endogenous antibiotic oleandomycin is inactivated by two family 1 (GT-1; defined in ref. 11) glycosyltransferases, OleI and OleD, which share 45% sequence identity and catalyze glucosyl transfer with inversion of anomeric configuration from UDP-Glc to the O2 of the desosamine sugar of the antibiotic, Fig. 2 (9). OleI, which is tightly linked both genetically and biochemically to the synthesis of the macrolide antibiotic oleandomycin (7), displays a very low KM for the antibiotic, thus ensuring that high levels of the active form of the antimicrobial agent do not accumulate in the bacterium. The extracellular β-glucosidase that activates oleandomycin is encoded by oleR, which is linked to oleI and the biosynthetic operon of the antibiotic (7). Unlike OleI, which only glycosylates oleandomycin, OleD displays broad acceptor specificity and hence will inactivate a wider range of macrolide antibiotics including tylosin and erythromycin, Fig. 2 (7, 10). Such promiscuity suggests that OleD may function as a more general resistance mechanism, a view supported by the observation that oleD is not linked to the oleandomycin biosynthetic operon (12). Both OleI and OleD can use several dinucleotide sugars as the donor substrate, although, significantly, whereas OleI can mediate glycosyl transfer from both UDP-α-d-glucose and UDP-α-d-galactose, OleD can use UDP-α-d-glucose but not UDP-α-d-galactose to glycosylate macrolide antibiotics (7, 10).
Fig. 1.
The mechanism of macrolide antibiotic resistance in S. antibioticus.
Fig. 2.
The reactions catalyzed by OleI and OleD. The enzymes transfer glucose from UDP-glucose to the O2 position of the desosamine sugar appended to the macrolide backbone of macrolide antibiotics. The reaction occurs with inversion of anomeric configuration to generate the β-glucoside.
Here, we present the 3D structures of two macrolide polyketide antibiotic modifying enzymes, OleI and OleD, in complex with UDP/oleandomycin and UDP/erythromycin, respectively. The 3D structures, in conjunction with kinetic analyses of the wild-type (WT) enzymes and site-directed variants, has facilitated the dissection of the molecular mechanism by which these glycoyltransferases recognize erythromycin and oleandomycin, providing insight into how these medically significant Actinomycetes display resistance to both endogenous and exogenous antimicrobial agents. Understanding this resistance mechanism will also inform strategies for the exploitation of these enzymes for use as unique synthetic catalysts for creating modified unnatural antibiotic variants.
Results and Discussion
Structure of UDP-Glucosyltransferase OleI and OleD at 1.7-Å Resolution.
The crystal structure of OleI was solved using the single-wavelength anomalous dispersion using SeMet protein, to 1.7-Å resolution, using a P21 crystal form with two molecules of OleI in the asymmetric unit [supporting information (SI) Table 4]. The final model structure, from residues 11 to 420, has an Rcryst of 0.17 and Rfree of 0.22. OleI consists of two Rossmann-like β/α/β domains (Fig. 3) that are not tightly associated and thus displays the GT-B fold typical of several glycosyltransferase families including GT-1. In glycosyltransferase that exhibit a GT-B fold, conformational changes in the relative orientation of these domains, in addition to changes in protein loops, are associated with substrate binding (13–17). The N-terminal domain, comprising amino acids 11–235, consists of seven parallel β-strands and six associated α-helices. The C-terminal Rossmann β/α/β domain, consisting of residues Ser-243 to Leu-420, comprises six parallel β-strands that are flanked by six α-helices. The terminal helix contains a kink at position Glu-405, at the interface between the two domains, and the last 12 residues of this secondary structural element extend into the N-terminal domain. An extended surface loop, Ala-158 to Glu-180, and a very small loop consisting of Glu-302 and Val-303 are disordered. Removal of the region containing the extended loop (Δ167–185, Table 1) did not influence the catalytic activity of OleI, indicating that it does not contribute to the overall fold of the enzyme or substrate binding. One may speculate that this loop plays a role in protein–protein interaction between OleI and other partners in the oleandomycin synthetic pathway, ensuring that synthesis of the antibiotic and its cellular inactivation are tightly coupled.
Fig. 3.
The three-dimensional structures of OleI (A) and OleD (B) shown as divergent (“wall-eyed”) stereo cartoons. It can be seen that the domains of OleD lie in a more open conformation than those of OleI.
Table 1.
Kinetic parameters of WT OleI and mutants in the acceptor substrate binding site
| Enzyme | kcat, min−1 | Donor substrate*KM, μM | Acceptor substrate KM, μM† |
|---|---|---|---|
| WT | 191.4 (±15.4) | 110.7 (±8.8) | 5.9 (±1.4) |
| H25A | NA | NA | NA |
| W79A | 5.4 (±1.0) | 297.5 (32.8) | ND |
| Q83A | 164.4 (±8.0) | 268.8 (20.6) | 19.2 (±2.7) |
| M87A | 110.1 (±11.3) | 285.7 (10.9) | 33.6 (±3.0) |
| P80A | 46.7 (±5.5) | 109.1 (±11.3) | 11.3 (±1.5) |
| F90A | 1.6 (±0.1) | 38.3 (±3.4) | 34.0 (±4.2) |
| I117A | 4.1 (±0.4) | 275.5 (±35.3) | 10.1 (±1.5) |
| W120A | 10.5 (±1.9) | 472.0 (±29.4) | 21.1 (±3.2) |
| F140A | 93.8 (±7.3) | 217.4 (±15.0) | 30.0 (±2.8) |
| F146A | 28.9 (±2.3) | 360.2 (±18.9) | 54.2 (±2.2) |
| V150A | 210.8 (±9.0) | 182.6 (±15.0) | 13.3 (±2.8) |
| V153A | 177.5 (±11.3) | 160.4 (±8.2) | 6.3 (±0.7) |
| Δ167–185 | 222.1 (±21.8) | 160.2 (±7.7) | 16.0 (±2.3) |
| L207A | 46.5 (±5.6) | 340.3 (±13.8) | 12.3 (±1.6) |
| I208A | 45.5 (±4.4) | 354.5 (±24.3) | 31.5 (±4.4) |
| F266A | NA | NA | NA |
| I350A | 2.5 (±0.7) | 303.7 (±20.5) | 4.4 (±1.0) |
NA, no activity detected (<10−4 of WT); ND, KM was too high to determine.
*The donor substrate was UDP-Glc and was varied in the presence of 0.5 mM oleandomycin.
†The acceptor substrate oleandomycin was varied in the presence of 1–3 mM UDP-Glc depending on the KM of the mutant protein for the donor sugar.
OleI displays a similar fold to the other GT-1 glycosyltransferases, although sequence identity is low: DALI (18) Z scores, rms deviations, and % identity for examples of related enzymes are as follows: GtfB [Protein Data Bank (PDB) ID code liir] Z score 31.9 rms 2.8 Å (over 342 Ca atoms), 18% sequence identity; VvGT1 (PBD ID code 2clx) Z score 24.4 rms 3.4 Å (over 309 Ca atoms), 18% sequence identity; UGT71G1 (PDB ID code 2acv) Z-score 23.3, rms 3.3 Å (over 315 Cα atoms), 17% identity. The similarity between the C-terminal domains of both OleI and OleD to other GTs is higher than the N-terminal domains, which likely reflects the similarity of the nucleotide-donor substrates (which bind to the C-terminal domain), whereas the acceptor ligands, which interact primarily with the N-terminal domain, are quite different.
The interface between the two domains of the enzymes houses the active site. Electron density at the active site reveals bound UDP and oleandomycin (Fig. 4). The donor substrate is housed in a very deep pocket in the C-terminal domain (Fig. 3), whereas the acceptor, oleandomycin, is located primarily in the N-terminal domain in a wide hydrophobic pocket. In common with other GT-B glycosyltransferases, it is likely that OleI will undergo substantial conformational changes upon substrate binding, a view supported by the observation that crystals could only be produced in the presence of UDP and oleandomycin.
Fig. 4.
Observed electron density for ligands bound to OleI. A divergent stereo (divergent) view of the observed electron density (maximum-likelihood weighted 2Fobs − Fcalc contoured at 1 σ/0.5 electrons/A3) of the complex of OleI with UDP (blue electron density) and oleandomycin (red electron density) with interacting residues shown in ball-and-stick.
The crystal structure of OleD was solved, again at 1.7-Å resolution, using the N- and C-terminal domains of OleI as molecular replacement models. The two domains needed to be treated independently, reflecting not only the difference in domain orientation between OleI and OleD but also the difference between the two OleD molecules in the asymmetric unit. The structure, determined in the presence of UDP and the acceptor erythromycin, has been traced from residues 7 to 400. Again, the structure forms a classic GT-B fold of two “Rossmann-like” β/α/β domains, which comprise residues 7–213 (plus C-terminal α helix 384–399) and 218–384, respectively (Fig. 3). The overall folds of OleI and OleD are extremely similar, with DALI comparisons reporting a Z score of 44.9, an rms of 3.8 Å (over 375 Cα atoms), and 47% sequence identity. The structure of OleD exemplifies the flexibility in domain orientation of GT-B fold enzymes. Despite being crystallized as the equivalent dead-end complex of UDP and acceptor, OleD crystallizes in a much more “open” form, in which the N- and C-terminal domains are 10–15° more widely separated (Fig. 3). Consequently, UDP in OleD is 6–7 Å more distant from the acceptor, when the equivalent acceptors of OleD and OleI are overlapped. Given that the domain orientation of OleI and the comparative positions of nucleotide and acceptor are essentially as observed for the Michaelis complex of the grape enzyme VvGT1, the domains of OleD must undergo further conformational changes for catalysis to occur (17). Despite this, identification of the location of the substrates and reaction products in both OleD and OleI allows the description of the enzyme–ligand interactions, described below.
Donor Sugar Binding Site of OleI and OleD.
Uracil recognition.
The uridine moiety is buried in a deep pocket in both OleI and OleD. The uracil base stacks against the aromatic side chain of Trp-312 in OleI and Trp-290 in OleD, Fig. 4 and SI Fig. 5, explaining why hydrophobic residues are conserved at this position in GT-1 enzymes (17, 19). In OleI, the O2 of uridine makes a hydrogen bond with the side chain of Gln-245 and O4 interacts with both the hydroxyl of Ser-290 and the backbone N of Val-313, whereas N3 of the uracil ring makes a hydrogen bond with the carbonyl of Val-313. The nucleotide base makes similar hydrogen bonds with OleD. The interaction between uracil and the OleI residues Ser-290 and Gln-245, however, have little influence on catalytic activity, as S290A and Q245A (Table 2) display similar activity to WT OleI. Similar interactions are observed in other GT-1 enzymes that are not highly related to OleI and OleD. Thus, in the Medicago terpenoid glycosyltransferase, UGT71G1, the equivalent residues to OleI Trp-312 (Trp-339) and Val-313 (Ala-340) interact with the nucleotide base (19), whereas in VvGT1, the corresponding amino acids are Trp-332 and Ala-333 (17). The minor influence on activity displayed by the Q245A mutation is consistent with the observation that O2 of uracil is not generally exploited as a specificity determinant (reviewed in ref. 20) by other GT-B fold glycosyltransferases; although, in MurG, Arg-164 might make a productive interaction with this carbonyl group (16).
Table 2.
Kinetic parameters of WT OleI and mutants in the donor–substrate binding site
| Enzyme | kcat, min−1 | Donor substrate KM, μM | Acceptor substrate KM, μM† |
|---|---|---|---|
| WT | 191.4 (±15.4) | 110.7 (±8.8) | 5.9 (±1.4) |
| WT* | 24.0 (±4.1) | 500.3 (±88.6) | ND |
| N27A | 25.8 (±1.9) | 450.1 (±24.2) | 10.2 (±1.5) |
| R242A | 118.8 (±1.0) | 183.2 (±24.0) | 8.8 (±0.9) |
| Q245A | 150.0 (±26.5) | 639.1 (±101) | 4.9 (±1.2) |
| S264A | 0.8 (±0.1) | 59.5 (±2.9) | 2.1 (±0.6) |
| S290A | 152.2 (±24.0) | 451.1 (±44.7) | 5.5 (±1.0) |
| H328A | 14.0 (±1.5) | 277.7 (±25.9) | 11.2 (±2.5) |
| S333A | 67.4 (±5.6) | 3,760.2 (±496.3) | 44.6 (±4.3) |
| E336A | NA | NA | NA |
| Q349A | 185.3 (±1.4) | 143.1 (±15.8) | 7.0 (±1.1) |
| E352A | 1.9 (±0.2) | 449.6 (±50.7) | 4.0 (±0.8) |
| E352D | 0.75 (±0.2) | 400.8 (55.5) | ND |
| E352D* | NA | NA | NA |
| Q353A | 9.9 (±1.6) | 632.2 (±52.7) | 8.8 (±1.2) |
| N356A | 47.6 (±5.1) | 90.9 (±6.6) | 10.0 (±1.6) |
ND, not determined; NA, no activity detected (<10−4 of WT). The donor substrate was UDP-Glc unless asterisked, in which case UDP-Gal was used.
†The acceptor substrate oleandomycin was varied in the presence of 1–3 mM UDP-Glc depending on the KM of the mutant protein for the donor sugar.
Ribose recognition.
In OleI O2 and O3 of the nucleotide sugar, ribose, hydrogen bond with the carboxylate of Glu-336 (Fig. 4), whereas O2 may also interact with the Nε2 of Gln-315. Similar interactions are observed between the ribose component of the donor substrate and OleD. The identity of these residues, and their interactions with the donor substrate, are highly conserved in more distantly related GT-1 enzymes (17, 19), suggesting that these amino acids create a near-perfect “ribose pocket.” Furthermore, the lack of activity displayed by E336A (Table 2) confirms the importance of the interaction between the ribose sugar and the carboxylate side chain of Glu-336 in donor substrate binding. In OleI, but not in OleD or the other known GT-1 structures, the O3 of the ribose also makes a hydrogen bond with the Nδ1 of Asn-27 from the acceptor domain, and the importance of this interaction is reflected by the low catalytic activity displayed by the N27A mutant (Table 2). Intriguingly, in the structurally unrelated GT-A enzyme mannosylglycerate synthase (21), the interaction between O2 and O3 of the dinucleotide ribose and Glu-11 (equivalent to OleI Glu-336) does not play a key role in substrate binding. Futhermore, in GtfD and GtfA, GT-1 enzymes that use nucleoside thymidine in the donor substrate, Glu-336 is replaced by aliphatic residues and the deoxyribose of TDP does not make direct hydrogen bonds with these glycosyltransferases (4, 6). To compensate for the relatively weak interaction with the nucleotide sugar these enzymes may bind more tightly to the thymine base, which is sandwiched between a hydrophobic residue and a buried ion pair (6).
Phosphate recognition.
In OleI the α-phosphate O1 of the donor substrate interacts with both the backbone N and the hydroxyl side chain of Ser-333 (Fig. 4) and the main chain amide of Gly-332. The Nε2 of His-328 makes polar contacts with the O3 and O2 of the α and β phosphate, respectively, whereas the amide and OH of Ser-264 hydrogen bonds to the O2 of the β phosphate. Although Ser-333, Ser-264, and His-328 are highly conserved in GT-1 glycosyltransferases, indicating that these amino acids play a key role in nucleotide binding, in OleD, Ser-333 is replaced with Gly. In OleD the Nε2 of His-306 and the backbone N of Gly-311 and Ser-241 make identical interactions with the nucleotide phosphates to the equivalent residues in OleI (His-328, Ser-333, and Ser-264). To probe the role of O phosphate coordination in catalysis, the activity of the OleI mutants S264A, H328A, and S333A was assessed. The observation that the mutations all cause a substantial reduction in catalytic efficiency when the donor substrate is varied (Table 2) supports the view that the side chains of Ser-264, His-328, and Ser-333 make productive interactions with the diphosphate of UDP-Glc. The S333A mutation results in a substantial increase (34-fold) in the KM for UDP-Glc, and although the H328A mutants also displays a modest increase in this kinetic parameter, S264A has a KM for the donor substrate that is 2-fold lower than WT OleI.
Glucose recognition.
The high structural similarity in the glucose donor site of GT-1 enzymes allows placement of the Glc moiety from the UDP-2FGlc/VvGT1 complex (17) into the OleI and OleD active sites. The modeled glucose in the donor substrate is predicted to make several critical interactions with the proteins. Nε2 of Gln-353 OleI and Gln-331 of OleD should form hydrogen bonds with both the O2 and O3 of glucose, whereas O3 and O4 of the hexose is predicted to interact with the carboxylate oxygens of OleI Glu-352 (Fig. 4). In OleD, however, the corresponding residue Asp-330 (SI Fig. 5) is disordered, and thus its interaction with the donor sugar is difficult to assess. Whereas OleI Gln-353 is invariant in GT-1 enzymes, Glu-352 is often replaced with aspartate in other family 1 members (4–6, 17, 19), or Gln in the closely related MurG enzyme from family GT-28 (16, 22). Indeed, it has been suggested (16) that Glu/Asp-Gln (E/D-Q) is a critically conserved motif at positions equivalent to OleI 352–353. A picture emerges, therefore, that perhaps residues equivalent to 352–353 in OleI contribute strongly to donor specificity. In the plant GT-1, UGT71G1, and the Escherichia coli GT-28, MurG, the equivalent residues to Glu-352 and Gln-353 (Glu-381/Gln-382 in UGT71G1 and Gln-288/Gln-289 in MurG, respectively) make similar interactions with the donor sugar (although MurG uses UDP-GlcNAc as the donor substrate rather than UDP-Glc), reinforcing the important role these two amino acids play in substrate recognition. Support for this view is provided by the catalytic activities displayed by OleI E352A and Q353A, which are 425- and 106-fold lower, respectively, than the WT enzyme (Table 2) and the observation that the E381A (UGT71G1) and D330A (OleD) mutations completely inactivate these enzymes (19) (Table 3), whereas the Q289A amino acid substitution in MurG causes a 275-fold reduction in catalytic efficiency (16). O6 of the sugar is not used as a crucial specificity determinant as UDP-Xyl, which lacks C6–O6, was shown to function as a donor substrate (10).
Table 3.
Kinetic parameters of OleD
| Enzyme | Substrate | kcat, min−1 | Donor substrate KM, μM* | kcat/KM, μM−1 min−1 | Acceptor substrate KM, μM† | kcat/KM, μM−1 min−1 |
|---|---|---|---|---|---|---|
| Wild type | Oleandomycin and UDP-Glc | 339.3 (±28.6) | 121.7 (±11.5) | 2.8 | 3.2 (±0.6) | 107.2 |
| Wild type | Erythromycin and UDP-Glc | 13.8 (±1.8) | 673.0 (±77.0) | 0.02 | 3.5 (±0.3) | 4.0 |
| Wild type | Tylosin and UDP-Glc | 212.9 (±20.9) | 603.1 (±46.0) | 0.35 | 8.2 (±1.6) | 23.7 |
| D330E | Oleandomycin and UDP-Glc | 90.8 (±5.0) | 472.8 (±50.7) | 0.19 | ND | ND |
| D330E | Oleandomycin and UDP-Gal | NA | NA | NA | NA | NA |
| D330A | Olendomycin and UDP-Glc | NA | NA | NA | NA | NA |
ND, not determined; NA, no activity detected (<10−4 of WT).
*Acceptor substrate fixed at 0.5 mM for erythromycin and tylosin; 0.05 mM for oleandomycin. Note that oleandomycin causes substrate inhibition >0.05 mM with a Ki of 177.3 μM.
†UDP-Glc fixed at 1 mM for oleandomycin and 3 mM for tylosin and erythromycin.
A difference in donor substrate specificity between OleI and OleD is that only OleI can use UDP-Gal as a donor sugar albeit with an efficiency ≈30 times less than with UDP-Glc (Table 2). This finding is consistent with the almost identical environment for the donor sugar when compared with, for example, the glycosyltransferase VvGT1, which also uses UDP-Gal (≈10% of the rate of UDP-Glc; ref. 17). This variation in substrate recognition does not simply reflect the nature of the carboxylic amino acid at the position equivalent to OleI 352 (Glu in OleI and Asp in OleD, SI Fig. 5) as the residue in VvGT1 is an Asp and the OleD mutant D330E is unable to utilize UDP-Gal, although it retains 7% of its activity against UDP-Glc (Table 3). Indeed, the OleI mutant E352D displays no activity for either UDP-Glc or UDP-Gal (Table 2). The flexible conformation adopted by Asp-330 in the absence of donor sugar is consistent with the capacity of OleD to utilise either acid amino acid in donor substrate recognition (although a carboxylate at position 330 is important as the D330A mutant is inactive, Table 3), whereas the steric constraints imposed on the equivalent OleI residue, Glu-352, appear to be more significant. Similar unsuccessful, attempts to modulate the Glc vs. Gal specificity have been reported elsewhere on other GT-1 enzymes (17, 23).
The Acceptor Binding Site of OleI and OleD.
OleI has been shown to use only oleandomycin as the acceptor substrate, whereas OleD was able to use a range of macrolide antibiotics as acceptor substrates (7, 10). Kinetic analysis of OleD (Table 3) revealed that oleandomycin causes substrate inhibition at concentrations >0.05 mM (Ki 177.3 μM for oleandomycin inhibition; Table 3), and the enzyme displays ≈4- and ≈25-fold lower catalytic efficiency for tylosin and erythromycin, respectively, compared with oleandomycin. The crystal structures of OleI and OleD in complex with oleandomycin (OleI) and erythromycin (OleD), respectively, enabled the structural basis for the different acceptor specificities displayed by these enzymes to be explored.
Oleandomycin makes numerous hydrophobic, but only limited polar interactions with OleI (Fig. 4). The epoxide ring sits in a hydrophobic cradle formed by Phe-90 and Trp-120. C1, C13, C31, C33, and C35 of the macrocyclic ring (substrate nomenclature is in SI Fig. 6) make van der Waals contacts with Leu-207, Ala-86, and Phe-146, whereas C29 interacts with both Phe-140 and Thr-139. The dimethyl group of the sugar β-d-desosamine (in a classical 4C1 chair conformation) makes several hydrophobic contacts with Trp-79. Other residues that are likely to make van der Waals contacts with the antibiotic are Pro-80, Met-87, Ile-117, Trp-120, Val-150, Val-153, Ile-208, Phe-266, and Ile-350. The importance of these residues in oleandomycin recognition was explored using a mutagenesis strategy. The data (Table 1) show that mutation of Trp-79, Phe-90, Ile-117, Trp-120, Phe-146, Phe-266, or Ile-350 results in a substantial reduction in catalytic efficiency, demonstrating that these residues play a significant role in the formation of a productive enzyme complex with the acceptor substrate. The limited polar interactions between oleandomycin and OleI include hydrogen bonds between the backbone amides of Ala-351 and Glu-352 and O6 (O4 of the sugar component l-oleandrose), and the Nε2 of Gln-83 and O11 and O12 of the macrocyclic ring, whereas O2 of the sugar β-d-desosamine (the site of glycosylation; O22 in the labeling of oleandomycin) is within hydrogen bonding distance (2.7 Å) of His-25. Surprisingly, mutation of Gln-83 results in only a modest reduction (4-fold) in catalytic efficiency indicating that the formation of productive complexes with the antibiotic is dominated by hydrophobic interactions. The H25A mutant displays no catalytic activity (Table 1), consistent with its proposed role as the catalytic base that activates the acceptor substrate by abstracting a proton from the O2 of β-d-desosamine (SI Fig. 7 and SI Text). The catalytic function of His-25 is consistent with the observation that this residue is invariant in the 50 glycosyltransferases that display closest sequence identity to OleI, and the equivalent histidine in the GT-1 enzyme UGT71G1 (His-22) and the GT-28 biocatalyst MurG (His-18) also comprise the respective catalytic base (6, 16, 19). By contrast, in the GT-1 glycosyltransferase, GtfB, the putative base D13A mutant retained ≈10% activity, leading the authors to suggest that an alternative catalytic base (Asp-332) is located in the C-, rather than the N-terminal domain (5). Although this may be possible, it could simply reflect the lesser requirement for activation of the aromatic hydroxyl of the GtfB acceptor, which would have a lower pKa and hence be a better nucleophile than a sugar hydroxyl.
OleD binds to erythromycin in a similar hydrophobic-dominated pocket (SI Fig. 8) although the enzyme displays a noncatalytically competent open conformation, which may reflect the relatively poor activity of the glycosyltransferase when using this macrolide as the acceptor substrate (Table 3). Interestingly, the conformation adopted by erythromycin bound to OleD or bacterial ribosomes is essentially identical (24) (SI Fig. 9 and SI Text), suggesting that the proteins recognise a minimum energy conformation of the macrolide antibiotic. Despite their similarity, the “on-enzyme” conformations of oleandomycin (on OleI) and erythromycin (on OleD) are significantly different (SI Fig. 8), which reflects the different chemistry of the two compounds (Fig. 2 and SI Fig. 6) The major differences between the macrolide antibiotics are as follows: considering the macrolide skeleton only, erythromycin contains a hydroxyl attached to C12 and C6, an additional methyl group appended to C36, whereas the epoxide group of oleandomycin is replaced with a methyl moiety. Both compounds display the same desosamine sugar, but in erythromycin the second sugar is not l-oleandrose but l-cladinose, which contains an additional methyl and inverted configuration at C16 (equating to C3 of the sugar itself). It is not surprising, therefore, that the hydrophobic residues that comprise the acceptor binding site are similar but not completely conserved between OleI and OleD. Although the OleI residues Trp-79, Phe-90, Ile-117, Val-150, and Phe-66 are invariant in the two enzymes, Trp-120 and Phe-146 are replaced with tyrosine in OleD, and Met-87, Ile-120, Leu-207, and Ile-350 are substituted with other aliphatic residues, whereas the amino acids at positions equivalent to the OleI residues 80, 86, 139, 153, 208, and 355 are quite different in these GT-1 glycosyltransferases (SI Fig. 5). The only obvious structural changes between OleI and OleD, which affects hydrogen bonding to the macrolide backbone are Ile-208 in OleI (which is Ser-184 in OleD) that hydrogen bonds to the additional OH at C12 in erythromycin, and Leu-68 in OleD, which is equivalent to Gln-83 in OleI (the interactions made by Gln-83 are discussed above). The introduction of the I208S mutation into OleI, however, did not influence catalytic activity or the enzyme's tight specificity for oleandomycin (data not shown). The primary feature of the two antibiotics that contribute to their different conformations, and most likely to the difference in specificity of the two enzymes, is the position of the l-cladinose (erythromycin) and l-oleandrose (oleandomycin) sugars, which are markedly different between these compounds (SI Fig. 8).
Two main features of the macrolide antibiotics contribute to the different positioning of the l-cladinose/l-oleandose sugar unit, which both display a 1C4 conformation, typical of l-sugars. The first is the sugar itself, in which l-cladinose not only contains an additional methyl group but is also opposite in configuration at this center. However, it is most likely not the sugar itself that leads to its different conformation but the additional hydroxyl group at C6 of the macrolide backbone. This hydroxyl would make a steric clash with the cladinose if it was located in the same position as oleandose in oleandomycin (Fig. 5). It would seem that the subtle changes of the macrolide backbone give rise to a significant change in the position of the “second” sugar moiety. Furthermore, it is the position of this sugar that likely governs the markedly different specificities of OleI and OleD. Within this context, it is interesting that clarithromycin is one of the few macrolides not to be glycosylated by OleD (8), despite containing the same sugar moieties as erythromycin. The hydroxyl at C6 of clarithromycin is methylated, and because substitution at C6 influences the position of cladinose (see above), it is likely that the methyl group forces this sugar to adopt a conformation that cannot be accommodated by OleD.
With the exception of the Ile–Ser change discussed above, the overlap of the acceptor species and their environments in OleI and OleD shows that the major structural differences in the acceptor sites of the catalytic centres lie in two β-α units and their connecting loop [corresponding to 65–84(OleD)/70–89(OleI) and 326–340(OleD)/346–362(OleI)]. Although an acceptor-based overlap must be viewed with some caution, given that major conformational changes are possible, even necessary in the case of OleD, one can say that these two structural elements differ in their sequence (SI Fig. 5), in their interaction not only with each other but also with other regions of their respective scaffolds and are in markedly different positions in the two structures (Fig. 5). Of note, the loop from 350 to 352 in OleI makes considerable steric clashes with the l-cladinose moiety of erythromycin, when it is overlapped with oleandomycin. The subtle but complex structural basis for the differences in antibiotic recognition precludes the facile use of protein engineering to alter acceptor substrate specificity in OleI and OleD. Indeed, exchanging the OleI specificity loop Lys-71 to Leu-89 with the corresponding OleD sequence (Gly-66 to Pro-84) identified above reduces activity but does not alter specificity, whereas the introduction of the mutations Q83L, I350A, I208S, A142G, M355G, T354F, and M355G/T354F into OleI did not confer measurable erythromycin glycosylation activity (data not shown).
Discussion
Resistance to macrolide antibiotics is often achieved, in the endogenous host, by glycosylation, which prevents binding to their cellular target, the ribosome. By resolving the crystal structure of the glycosyltransferases that catalyze macrolide glycosylation, in tandem with a rational design strategy to explore the functional significance of active site residues, this report brings insights into the structural and mechanistic basis for this important biological process.
In OleI and OleD, oleandomycin and erythromycin, respectively, are located in a hydrophobic dish within the active site. Site-direced mutagenesis demonstrates that aromatic residues play a pivotal role in macrolide recognition and thus the two enzymes interact with these substrates primarily through hydrophobic interactions and van der Waals forces. Thus, despite containing 13 oxygens and 1 nitrogen, OleD does not appear to use these polar atoms as important specificity determinants. Intriguingly, the major interactions between erythromycin and the 23S RNA component of the ribosome are also dominated by hydrophobic interactions; the apolar face of the lactone ring contains three methyl groups that insert into a hydrophobic pocket formed by A2100, A2099, and G2646, whereas the polar face of the macrolide ring is solvent exposed (24).
OleI and OleD present unusual differences in donor and acceptor substrate specificity. OleI is very specific for the macrolide oleandomycin, yet will tolerate UDP-Gal in addition to the favored UDP-Glc donor (Table 2). In contrast, OleD can utilize UDP-Glc but not UDP-Gal as the donor substrate, although it is able to glucosylate other macrolide antibiotics, notably tylosin and erythromycin, in addition to oleandomycin (Table 3). The differences in the plasticity of donor and acceptor specificity of the highly related glycosyltransferases OleI and OleD indicate that hybrids of these enzymes could be used to synthesize novel bioactive molecules. For example, a recent study has shown that the galactosylation of macrolide antibiotics can increase the targeting of these antimicrobial agents to Gram-negative pathogens (10), and thus increasing acceptor specificity of OleI may increase the range of macrolides that can be decorated with galactosyl residues. Even in the light of 3D structure, however, attempts to increase acceptor and donor substrate plasticity in OleI and OleD has not been possible through simple loop swaps and single amino acid changes, indicating that engineering strategies will need to be more radical encompassing further rounds of rational design in concert with forced protein evolution. Indeed, successful examples of engineering the specificity of GT-B fold enzymes are very limited (see ref. 25 for review), which may reflect conformational changes in these enzymes during catalysis that have not been revealed by the static crystal structures of these glycosyltransferases in complex with their substrates in the ground state. Nevertheless, the crystal structures of OleI and OleD in complex with oleandomycin and erythromycin, reported here, will inform and direct these future rational design and forced protein evolution programmes that are aimed at engineering acceptor and donor substrate specificity in these key synthetic enzymes to generate novel bioactive molecules.
Materials and Methods
Plasmids, Bacterial Strains, and Growth Media.
The E. coli strains used, culture conditions used to express OleD and OleI, and the construction of pMP1 and pMP2 have been described (10). The plasmid pDB1 was constructed by amplifying the oleD gene from pMP2 by PCR using primers that contain 5′ NcoI and XhoI sites and cloned into NcoI/XhoI digested pET32b (Novagen, Nottingham, U.K.). The plasmid encodes a thioredoxin fusion partner followed by an internal His6 tag and an enterokinase cleavage site. The culture conditions used by Flint et al. (21) were used to produce selenomethionine containing OleI.
Site-Directed Mutagenesis.
QuikChange mutagenesis (Stratagene, La Jolla, CA) was used to introduce small amino acid substitutions into OleI and OleD using pMP1 and pMP2 as template DNA, respectively.
Purification of OleI and OleD.
For enzyme assays, OleI and OleD were purified by immobilized metal ion affinity chromatography (IMAC) (10). For crystallization the two enzymes were further purified as described in SI Text.
Enzyme Assay.
The activity of OleI and OleD were determined by using a linked enzyme activity assay described by Gosselin et al. (26). The 500-μl reaction (carried out at 27°C) consisted of 20 mM Tris·HCl buffer, pH 8.0, containing 13 mM MnCl2, 1 mg/ml BSA, 0.7 mM potassium phosphoenolpyruvate, 0.15 mM NADH, 1.8 units of pyruvate kinase, and 3.6 units of lactate dehydrogenase. The amount of enzyme varied between 10 nM and 5 μM. To determine the kinetic parameters for the donor sugar with OleI, the concentration of oleandomycin was 0.5 mM, and UDP-glucose was varied from 0.2 to 5–10 times the KM. Similar amounts were used for OleD with erythromycin and tylosin, although with oleandomycin, the concentration was reduced 50 μM to prevent inhibition. To determine the KM and kcat for the acceptor substrate, the concentration of the donor sugar was fixed at 1–3 mM, depending on the KM for the enzyme variant, whereas the concentration of the antibiotic was varied from 0.5 to 10 times the KM value. To ensure that release of UDP was by glycosyl transfer and not hydrolysis of UDP-glucose, the release of free glucose was determined using the glucose detection kit from Megazyme. Note that these assay conditions are different than those used by Yang et al. (10), which are suboptimal for both glycosyltransferases.
Crystallization and Data Collection and Structure Solution.
OleI and OleD were crystallized as described in SI Text. The structure of OleI was determined by using the single-wavelength anomalous dispersion method at a wavelength optimized for the ƒ″ signal of the selenium. Ten Se sites were found by using SHELXD (27). Heavy-atom phasing was performed with MLPHARE with noncrystallographic symmetry averaging using DM (both CCP4 suite). Automated model building and refinement using REFMAC (28) and ARP/wARP (29) resulted in a model with a crystallographic R of 0.25 (Rfree, 0.20), which was completed with manual correction using COOT (30) and refinement with REFMAC. OleD crystallized in a P21 crystal form, distinct from the above, but again with two molecules in the asymmetric unit. The N- and C-terminal domains of OleI model were used as the molecular replacement models using PHASER (31). The structure was rebuilt with COOT and refined using REFMAC. Structural figures were drawn with MOLSCRIPT (32) and BOBSCRIPT (33).
Supplementary Material
Acknowledgments
This work was funded by the Biotechnology and Biological Sciences Research Council (BBSRC). G.J.D. is the holder of a Royal Society–Wolfson Merir Award.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. J.C. is a guest editor invited by the Editorial Board.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 2IYA and 2IYF).
This article contains supporting information online at www.pnas.org/cgi/content/full/0607897104/DC1.
References
- 1.Mendez C, Salas JA. Trends Biotechnol. 2001;19:449–456. doi: 10.1016/s0167-7799(01)01765-6. [DOI] [PubMed] [Google Scholar]
- 2.Douthwaite S. Clin Microbiol Infect. 2001;7:11–17. [PubMed] [Google Scholar]
- 3.Vilches C, Hernandez C, Mendez C, Salas JA. J Bacteriol. 1992;174:161–165. doi: 10.1128/jb.174.1.161-165.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mulichak AM, Losey HC, Lu W, Wawrzak Z, Walsh CT, Garavito RM. Proc Natl Acad Sci USA. 2003;100:9238–9243. doi: 10.1073/pnas.1233577100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mulichak AM, Losey HC, Walsh CT, Garavito RM. Structure (London) 2001;9:547–557. doi: 10.1016/s0969-2126(01)00616-5. [DOI] [PubMed] [Google Scholar]
- 6.Mulichak AM, Lu W, Losey HC, Walsh CT, Garavito RM. Biochemistry. 2004;43:5170–5180. doi: 10.1021/bi036130c. [DOI] [PubMed] [Google Scholar]
- 7.Quiros LM, Aguirrezabalaga I, Olano C, Mendez C, Salas JA. Mol Microbiol. 1998;28:1177–1185. doi: 10.1046/j.1365-2958.1998.00880.x. [DOI] [PubMed] [Google Scholar]
- 8.Quiros LM, Carbajo RJ, Brana AF, Salas JA. J Biol Chem. 2000;275:11713–11720. doi: 10.1074/jbc.275.16.11713. [DOI] [PubMed] [Google Scholar]
- 9.Quiros LM, Salas JA. J Biol Chem. 1995;270:18234–18239. doi: 10.1074/jbc.270.31.18234. [DOI] [PubMed] [Google Scholar]
- 10.Yang M, Proctor MR, Bolam DN, Errey JC, Field RA, Gilbert HJ, Davis BG. J Am Chem Soc. 2005;127:9336–9337. doi: 10.1021/ja051482n. [DOI] [PubMed] [Google Scholar]
- 11.Coutinho PM, Deleury E, Davies GJ, Henrissat B. J Mol Biol. 2003;328:307–317. doi: 10.1016/s0022-2836(03)00307-3. [DOI] [PubMed] [Google Scholar]
- 12.Hernandez C, Olano C, Mendez C, Salas JA. Gene. 1993;134:139–140. doi: 10.1016/0378-1119(93)90189-a. [DOI] [PubMed] [Google Scholar]
- 13.Buschiazzo A, Ugalde JE, Guerin ME, Shepard W, Ugalde RA, Alzari PM. EMBO J. 2004;23:3196–3205. doi: 10.1038/sj.emboj.7600324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Campbell RE, Mosimann SC, Tanner ME, Strynadka NC. Biochemistry. 2000;39:14993–5001. doi: 10.1021/bi001627x. [DOI] [PubMed] [Google Scholar]
- 15.Gibson RP, Tarling CA, Roberts S, Withers SG, Davies GJ. J Biol Chem. 2004;279:1950–1955. doi: 10.1074/jbc.M307643200. [DOI] [PubMed] [Google Scholar]
- 16.Hu Y, Chen L, Ha S, Gross B, Falcone B, Walker D, Mokhtarzadeh M, Walker S. Proc Natl Acad Sci USA. 2003;100:845–849. doi: 10.1073/pnas.0235749100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Offen W, Martinez-Fleites C, Yang M, Kiat-Lim E, Davis BG, Tarling CA, Ford CM, Bowles DJ, Davies GJ. EMBO J. 2006;25:1396–1405. doi: 10.1038/sj.emboj.7600970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holm L, Sander C. J Mol Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 19.Shao H, He X, Achnine L, Blount JW, Dixon RA, Wang X. Plant Cell. 2005;17:3141–3154. doi: 10.1105/tpc.105.035055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu Y, Walker S. Chem Biol. 2002;9:1287–1296. doi: 10.1016/s1074-5521(02)00295-8. [DOI] [PubMed] [Google Scholar]
- 21.Flint J, Taylor E, Yang M, Bolam DN, Tailford LE, Martinez-Fleites C, Dodson EJ, Davis BG, Gilbert HJ, Davies GJ. Nat Struct Mol Biol. 2005;12:608–614. doi: 10.1038/nsmb950. [DOI] [PubMed] [Google Scholar]
- 22.Ha S, Walker D, Shi Y, Walker S. Protein Sci. 2000;9:1045–1052. doi: 10.1110/ps.9.6.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kubo A, Arai Y, Nagashima S, Yoshikawa T. Arch Biochem Biophys. 2004;429:198–203. doi: 10.1016/j.abb.2004.06.021. [DOI] [PubMed] [Google Scholar]
- 24.Tu D, Blaha G, Moore PB, Steitz TA. Cell. 2005;121:257–270. doi: 10.1016/j.cell.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 25.Hancock SM, Vaughan MD, Withers SG. Curr Opin Chem Biol. 2006;10:509–519. doi: 10.1016/j.cbpa.2006.07.015. [DOI] [PubMed] [Google Scholar]
- 26.Gosselin S, Alhussaini M, Streiff MB, Takabayashi K, Palcic MM. Anal Biochem. 1994;220:92–97. doi: 10.1006/abio.1994.1303. [DOI] [PubMed] [Google Scholar]
- 27.Schneider TR, Sheldrick GM. Acta Crystallogr D. 2002;58:1772–1779. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- 28.Murshudov GN, Vagin AA, Dodson EJ. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 29.Perrakis A, Morris R, Lamzin VS. Nat Struct Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 30.Emsley P, Cowtan K. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 31.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Acta Crystallogr D. 2005;61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 32.Kraulis PJ. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 33.Esnouf RM. J Mol Graphics Model. 1997;15:132–134. doi: 10.1016/S1093-3263(97)00021-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.