Abstract
Comparative genomics of CpG dinucleotides, which are targets of DNA methyltransferases in vertebrate genomes, has been constrained by their evolutionary instability and by the effect of methylation on their mutation rates. We compared the human and chimpanzee genomes to identify DNA sequence signatures correlated with rates of mutation at CpG dinucleotides. The new signatures were used to develop robust comparative genomics of CpG dinucleotides in heterogeneous regions and to identify genomic domains that have anomalous CpG divergence rates. The data showed that there are ≈200 genomic regions where CpG distributions are far more conserved than predicted. These hyperconserved CpG domains largely coincide with domains bound by Polycomb repressive complex 2 in undifferentiated human embryonic stem cells and are almost exclusively present near genes whose products are involved in the regulation of embryonic development. Several domains were experimentally shown to be unmethylated at different developmental stages. These data indicate that particular evolutionary patterns and distinct sequence properties on scales much larger than standard transcription factor-binding sites may play an important role in Polycomb recruitment and transcriptional regulation of key developmental genes.
Keywords: comparative genomics, development, DNA methylation, epigenetics, evolution
Oxidative deamination of 5-methylcytosine (m5C) converts the base to thymine, and most m5CpG dinucleotides in vertebrate genomes are rapidly (1, 2) mutated to TpG or CpA. For example, >20% of the CpGs have diverged in the otherwise 99.2% identical human and chimp genomes. Unmethylated CpG dinucleotides are mutated at normal rates, and regions with low levels of methylation, such as CpG islands (3, 4), have consequently higher CpG contents than regions with high levels of methylation. Conservation of CpG dinucleotides may therefore be a consequence of low germ-line methylation or of purifying selection against the loss of functional CpGs. Here we show that the sequence context of a CpG can be used to accurately model its divergence rate, in accordance with recent evidence on the correlation between sequence context and methylation levels (5–7). Our model predicts that CpG divergence rates vary by a factor of 40 as a function of sequence context and show that context-aware models are essential for the analysis of CpG evolution in heterogeneous regions.
We suggest that comparative genomics offer insights into longstanding questions as to the function of DNA methylation. We demonstrate this by identifying genomic regions in which CpGs are far more conserved than expected. These hyperconserved CpG domains (HCGDs) we identify show extensive overlap with regions bound by Polycomb repressive complex 2 (PRC2) and are observed almost exclusively at genes related to the regulation of embryonic development.
Results
Sequence Context of Conserved and Diverged Primate CpG Dinucleotides.
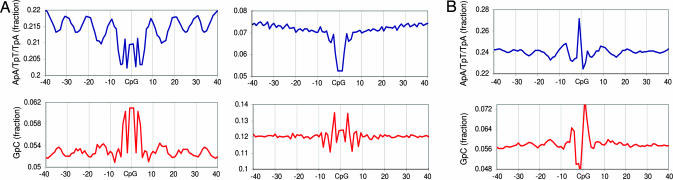
The sequence context of 8 million CpG dinucleotides that are located in nonexonic and nonrepetitive regions of the human genome was computed [supporting information (SI) Fig. 5]. The results reveal high information content in the sequences surrounding CpG dinucleotides, including a strong 10- to 10.5-bp periodicity for the densities of dinucleotides around CpGs. As shown in Fig. 1A, the ApA, TpT, and TpA dinucleotide densities are periodic and in phase with the location of the CpG (peaking at intervals of 10 bp), whereas GpC dinucleotides have a similar periodicity but a phase that peaks at 5 bp from the CpG. The 10-bp periodicities and the anticorrelation between ApA/TpT/TpA and GpC dinucleotides have been shown to correlate with nucleosome positioning in vivo (8, 9). According to the nucleosome positioning model, the ApA/TpT periodicity peaks where the DNA minor groove faces inward toward the center of curvature (10). The phasing of CpG relative to ApA and TpT suggests that CpG dinucleotides tend to be in contact with histone octamers rather than exposed on the nucleosome surface. Similar periodicities around CpG dinucleotides are observed in the mouse genome (data not shown). Because most CpG dinucleotides outside of CpG islands are methylated (11), core histones or specific histone variants are likely to be involved in the recognition of methylated CpG dinucleotides and the inhibition of transcription initiation. Alternatively, CpGs that are protected by nucleosomes may be more stable evolutionarily, possibly because they are less prone to methylation and deamination. Analysis of the sequence context around CpGs in CpG islands did not reveal strong periodicities (Fig. 1A Right). This lack of periodicity may suggest that CpG islands are not organized into nucleosome arrays or that, inside CpG islands, CpGs are protected from methylation using mechanisms other than the nucleosomes.
Fig. 1.
Dinucleotide periodicities around CpG dinucleotides. (A) Nucleosome-like dinucleotide distributions around CpG dinucleotides. (Left) Shown are the dinucleotide densities around 8 million intergenic nonrepetitive human CpG dinucleotides that are not part of CpG islands. The data show strong periodicity of ApA/TpT/TpA dinucleotide densities peaking at 10-bp intervals from the CpG. GpCs densities are also periodic but peak at 5-bp distance from the CpG. Similar periodicities are known to be associated with sequences that are tightly bound to nucleosomes. CpG islands lack both ApA/TpT/TpA and GpC nucleosomal periodicities (Right). (B) CpGs that are diverged in chimp exhibit weaker periodicities. Shown are dinucleotide frequency data for 650,000 human CpG dinucleotides that mutated to TpG or CpA in chimp. The profiles reveal much reduced ApA/TpT/TpA and GpC periodicities and lower G + C content (see SI Fig. 7 for further analysis).
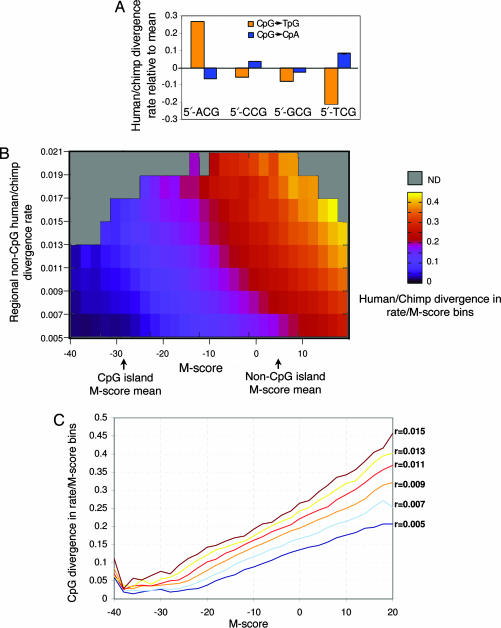
Comparison of the human and chimp genomes (12) showed that conserved and diverged CpGs are typified by distinct sequence contexts (SI Fig. 6). Analysis of the sequence contexts shows that CpG dinucleotides that have diverged between human and chimp are less likely to be embedded in nucleosomal patterns than are conserved CpGs (Fig. 1B). In general, conserved CpGs are located in contexts with higher G + C content, even when only non-CpG island loci are considered. Specific dinucleotide preferences at the positions flanking the CpG are also correlated strongly with divergence at the CpG locus. For example, CpGs located 3′ to an adenine are 60% more likely to mutate to TpG than are CpGs located 3′ to a thymine (Fig. 2A).
Fig. 2.
M-scores, regional mutation rates, and their effect on CpG divergence rates. (A) Flanking nucleotides predict CpG mutation rate. Shown are the fractions of human intergenic nonrepetitive CpGs that are aligned with a chimp TpG (blue) or CpA (orange), grouped according to the 5′ nucleotide. Ninety-five percent confidence intervals are shown. CpGs with a 5′ adenine are mutated 60% more rapidly than are CpGs with 3′ thymine. (B) M-scores. The M-score probabilistically summarizes the sequence context of each CpG dinucleotide to allow prediction of the CpG divergence probability. The model is constructed by comparative analysis of the sequences around conserved and diverged CpGs; it implicitly and systematically takes into account the G + C content, predicted nucleosome affinity as shown in Fig. 1, and the identity of the nucleotides immediately flanking the CpG. CpGs were binned according to their M-scores (x axis) and the background mutation rate of non-CpGs in the 20-kb window around them (regional rate, y axis; see Methods). The CpG divergence probability (fraction of CpGs in either human or chimp that were diverged between the species) is color-coded for each bin. CpG divergence is shown to increase independently with either the regional mutation rate or the M-score. The average M-scores for CpGs in CpG islands and outside of islands are marked for reference. (C) Dynamic range of M-score predictions. Shown are cross-sections of the 2D B image, depicting the increase in CpG divergence probability as a function of the M-score for fixed regional mutation rate levels (r). The M-score is shown to be linearly correlated with the divergence probability within almost 2 orders of magnitude.
Predicting CpG Divergence from the Sequence.
We used comparisons of sequence context of conserved and diverged CpGs to identify sequence correlates of CpG divergence rates and to determine whether the slow divergence rate of CpGs in CpG islands represents a binary effect that partitions CpGs into two evolutionary regimes (fast or slow evolving), or whether the sequence contexts surrounding CpGs determine a continuum of divergence levels. To distinguish between these two possibilities, we developed a probabilistic model that computes a mutability score (M-score) for each CpG dinucleotide by summarizing information from the flanking dinucleotides in the −200 to +200 range (SI Fig. 7 and Methods). Low M-scores (low rates of divergence) are associated with high G + C content and/or high predicted nucleosome affinities; high M-scores are associated with high A + T content and/or low nucleosome affinity. We predict the divergence probability of a CpG by empirically estimating the fraction of diverged CpGs from a large set of CpGs with similar M-scores in genomic regions that exhibit similar non-CpG divergence rates (Fig. 2 A and Methods). The M-score provides a continuum of predicted divergence probabilities and is generalizing the current partition of genomic sequences into CpG islands and nonisland regions (SI Fig. 8), eliminating the need of arbitrary thresholds of G + C content, CpG density, and length (3). Indeed, divergence is shown to quantitatively correlate with both the M-score and regional mutation rate (Fig. 2 B and C). The increase in divergence is gradual and linear, from low probabilities (≈0.01) at M-score values that are deep in the CpG island range (−40) to higher probabilities in the intermediate island/nonisland range (−20 to −10) to very high probabilities (0.4) for CpGs with M-scores >10. We discuss some possible evolutionary and functional implications of the M-score in SI Note 1 and SI Figs. 9 and 10 and show that M-score values strongly correlate with experimentally determined methylation levels. The M-score normalizes background probabilities of CpG divergence based on sequence context to allow identification of genomic regions that are evolving at anomalous rates. Normalization against sequence context is crucial because, as shown above, CpG divergence probabilities can span almost 2 orders of magnitude even within a small genomic region (i.e., the introns and promoter region of a single gene).
Identification of HCGDs.
A new statistical score, termed context-based CpG analysis of divergence (COCAD), compares the actual rate of human–chimp CpG divergence to the rate predicted by M-scores and regional non-CpG divergence rates (Fig. 3A). The null hypothesis used by COCAD assumes that CpGs are evolving independently from each other with rates that are determined by their sequence contexts. Negating this null hypothesis for a genomic interval is an indication for some regional modulation of CpG divergence rates that is not predicted by our current model. For example, a large CpG island would achieve a significant COCAD score only if the CpG dinucleotides in the island are evolving more slowly than CpG dinucleotides in other CpG islands of equivalent length and dinucleotide composition. The global COCAD score distribution (SI Fig. 11) reveals that the model predicts CpG divergences within statistical reason (−3 <Z score <3) for 95% of the genome, suggesting that for most of the genome, sequence context and regional divergence rates predict CpG divergence accurately. Divergence in the rest of the genome is hypothesized to be affected by additional factors, including (but not restricted to) changes in methylation level and selection.
Fig. 3.
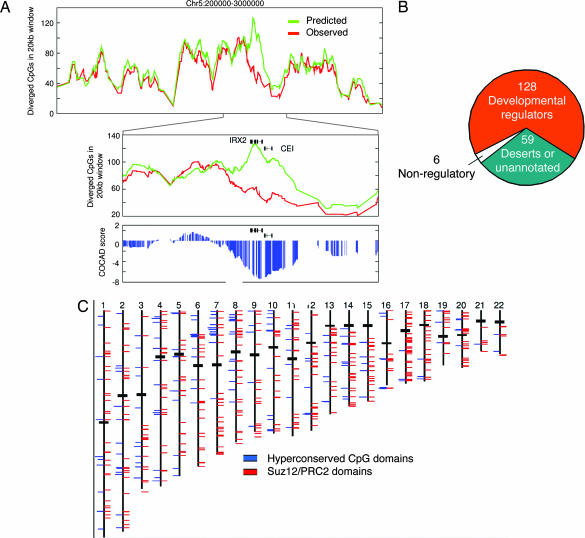
HCGDs. (A) The COCAD assay. Shown are the observed (red) and predicted (green) number of human or chimp CpGs that are not conserved in a 20-kb sliding window at part of chromosome 5p. The prediction is based on the M-score of each CpG and can vary significantly depending on the sequence context of the CpG. An anomalously low CpG divergence at the Iroquois homeobox protein 2 (IRX2) and CEI genes is shown (Center), and the difference between observed and predicted divergence is transformed to a statistical score (blue, Lower). Genomic loci with COCAD scores lower than −5 are defined as HCGDs. (B) HCGDs are located near genes for key developmental regulators. Of the 134 HCGDs that are near characterized genes, 128 are near genes related to regulation of developmental processes. (C) Chromosomal distribution of HCGDs (blue) and PRC2 domains (red).
As shown in Fig. 3B, ≈200 nonoverlapping genomic regions had COCAD scores below −5 (corrected P < 10−6; see SI Table 1 for an annotated list). We termed these regions HCGDs. Of the 200 HCGDs, 59 are located within sparsely annotated genomic regions, many of which harbor conserved regions and uncharacterized CpG islands. Of the remaining 134 HCGDs, 128 (95%) overlap genes that encode known or putative developmental regulators, nearly all of which are known transcription factors. The list of HCGDs includes almost 100 key developmental loci (HOX clusters, FOX genes, TBX genes, and more), as well as developmental signaling genes and genes implicated in cancer and differentiation of immune cells. As shown in Fig. 4, HCGDs frequently contain multiple regions annotated as CpG islands, many of which are not associated with the 5′ exon of a gene. The screen for HCGDs is not biased toward large CpG islands (which may also be overrepresented near developmental regulators), because the observed divergence at HCGDs is much lower than predicted by the M-score, which corrects for the slow divergence at CpG islands. HCGDs do not correspond to regions of general hyperconservation (13), because the COCAD assay controls for regional mutation rates and the regions immediately flanking HCGDs were found to have background conservation values similar to those inside the domain (SI Fig. 12), whereas conservation of CpG distributions was observed only inside the domains.
Fig. 4.
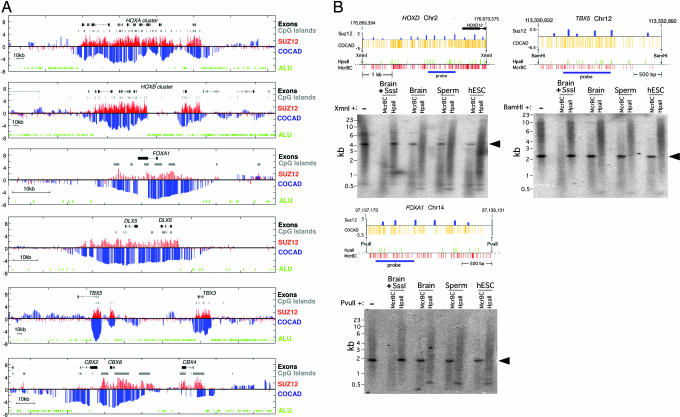
Hyperconserved CpG domains correspond to unmethylated PRC2-binding domains. (A) Genomic regions around several key developmental genes (drawn to scale; exons are marked as thick lines). For each genomic region, we plot the Suz12 raw ChIP-binding ratio (red) alongside the COCAD CpG conservation score (blue). Also plotted are Alu elements (green) and CpG islands (gray). The COCAD and Suz12 profiles, although obtained by unrelated methods, show strong similarity. The observed correlation is particularly significant because for the large majority of the genome, both the COCAD scores and Suz12 ChIP intensities are not significantly different from 0 (see http://www.pnas.org/cgi/content/full/0609746104/DC1SI Text). (B) HCGDs are unmethylated in human ES cells, sperm, and brain. Resistance to McrBC (a methylation-dependent endonuclease complex) and sensitivity to HpaII (a methylation-inhibited restriction endonuclease) show that regions with low COCAD scores and high PRC2 association are unmethylated at all stages tested. Distributions of McrBC and HpaII recognition sequences are shown. Prior methylation of DNA at all CpG dinucleotides with SssI DNA methyltransferase renders DNA resistant to HpaII and sensitive to McrBC, whereas DNA from human ES cells, sperm, and brain is sensitive to HpaII and resistant to McrBC and therefore unmethylated at all or nearly all CpG dinucleotides at the regions indicated. The hybridization background is an artifact of the very high G + C contents of the probe and target sequences. Arrowheads indicate unmethylated domains in DNA from brain, sperm, and human ES cells.
CpG Conservation Predicts PRC2-Binding Sites.
The distribution of HCGDs was compared with genome-wide binding profiles of the Suz12 component of PRC2 in human ES cells (14). Fig. 4A shows that PRC2-binding profiles strongly resemble the profiles of HCGDs, even though COCAD scores are based only on evolutionary dynamics and Suz12-binding profiles were determined experimentally. Overall, 69% of the HCGDs are within 10 kb of a high-significance PRC2-binding locus, and HCGDs near genes that encode developmental regulators overlap a PRC2 domain at 85% of the loci (and for 95% of the genes, several HCGDs cover clusters of related genes). This is a much higher fraction than the overall enrichment of PRC2 sites near developmental regulators (14–17). The strong association between PRC2 binding and domains of hyperconserved CpGs is further demonstrated by the presence of PRC2 domains near 63% of the 59 HCGDs that are not adjacent to well characterized genes; the expected value is <1% (P < 10−100). Furthermore, PRC2 domains associated with HCGDs are much larger than average PRC2 domains [6,073 vs. 1,988 bp; P < 10−22 (Kolmogorov-Smirnov test)]. Although most HCGDs and PRC2-binding domains contain CpG islands, and many contain multiple islands, their overlap cannot be explained by a common but uncorrelated bias toward CpG islands, because the majority of CpG islands lack both CpG conservation and PRC2 binding domains (SI Fig. 13). Furthermore, it was formerly noted that HOX clusters have much lower densities of Alu transposons than expected (18). As shown in Fig. 4A, this is also true of other genomic regions associated with PRC2-binding and HCGDs even when only single genes are involved.
To gain preliminary insights into the epigenetic status of CpGs at HGCDs, we measured methylation levels in three conserved regions near the HOXD, TBX5, and FOXA1 loci. We performed methylation analysis by the method of Rollins et al. (11) on DNA from human embryonic stem cells (hESC), brain, and sperm. Regions with COCAD scores less than −5 were found to be unmethylated. This, as well as indirect computational evidence (SI Note 2 and Fig. 14), suggests that a large factor in the slow rate of CpG divergence in HGCDs is the lack of methylation in the germ line rather than selection against mutated CpG dinucleotides.
Discussion
In this work, we establish basic molecular-level understanding of the ways by which patterns of CpG dinucleotides evolve in mammalian genomes. According to the results, the evolution of CpG distributions is driven by a complex combination of a context-dependent mutational process, variation in germ-line methylation levels, and selection against loss of functional CpGs. The context-dependent mutational process renders CpGs in mutation-favoring sequence contexts up to 40 times more mutable than CpGs in mutation-resistant contexts. Changes in germ-line methylation levels may be responsible for much of this variability by increasing the rate of 5-methylcytosine deamination in highly methylated regions. Selection on functional CpGs also plays a major role and is likely to constrain a substantial subset of the genomic CpGs (SI Fig. 10). All three factors are together shaping a challenging evolutionary landscape, contributing to the still-elusive functional role of DNA methylation in central processes of vertebrate biology.
The pressures that shape CpG distributions are not readily identified by standard comparative genomics methods. We therefore developed methods to model sequence contexts and to formulate their affect on CpG evolution, deriving a detailed model that predicts correctly the evolution of CpGs in >95% of the nonrepetitive genome. Using this model, we identified a set of HCGDs and studied their properties in detail. We envision a new generation of evolutionary models that could capture both the neutral and functional consequence of higher levels of genomic organization. The link we report between CpG dinucleotides and the nucleosome positioning pattern (Fig. 1) underlines the complex and interdependent nature of the epigenetic effects on the genome and leads toward highly integrated evolutionary models geared toward expressing this complexity.
The sources of sequence specificity for the assembly of Polycomb repressive complexes and the interactions between them and other factors remain largely unknown. The anomalous conservation of CpG distributions at PRC2-binding domains may have several implications. Polycomb complexes may be shielding regulatory domains near key regulators against aberrant DNA methylation. Alternatively (or in addition), CpG conservation at HGCDs may suggest that the recognition of long regions rich in unmethylated CpG dinucleotides is involved in the recruitment of PRCs. In any case, the depletion of Alu elements from HCGDs/PRC2 domains, together with recent experimental evidence (19), suggests that these loci may be under selection to maintain their higher level organization, indicating that sequence properties on scales much larger than standard transcription factor-binding sites may play an important role in gene silencing. Such highly organized regions are rare, except in the vicinity of important regulators of embryonic patterning that are complexed with PRC2 in undifferentiated human ES cells. As more experimental and evolutionary data become available, the mechanisms by which Polycomb repressive complexes are directed to specific loci will be further revealed.
Methods
Data Collection.
Genomic sequence, features, and alignments were downloaded from the University of California, Santa Cruz, genome browser site (20) and from refs. 14 and 21. For additional information see SI Text.
Probabilistic Modeling of CpG Divergence and the M-Score Model.
All nonrepetitive intergenic CpGs in the human genome were partitioned into four groups: chimp-conserved, chimp-plus-deaminated (CpG→TG), chimp-minus-deaminated (Cp→CpA), and other. For each group, the dinucleotide counts at each position relative to the CpG (−200 to +200) were collected. Denote the densities of a dinucleotide d at relative position i by:
p(d,i) for conserved CpGs
p+(d,i) for plus strand deaminated CpGs
p−(d,i) for minus strand deaminated CpGs.
Basically, we assume that the sequence context of conserved CpGs is characterized by the dinucleotide distribution p(d, i), and that the sequence context of plus (minus) strand deaminated CpGs is characterized by the dinucleotide distribution p+(d, i) (p-(d, i)). The M-score for a CpG at position i inside sequence context s is defined by summing up log odds:
Plus strand deamination:
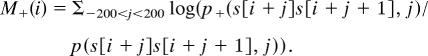 |
Minus strand deamination:
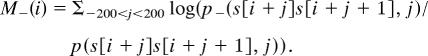 |
Summing up >200 values provides similar results to those reported here. In principle it is possible to transform the M-score log odds directly into posterior deamination probabilities. Alternatively, as done here, one can use the M-score, together with additional factors (here the regional mutation rate) to construct an empirical background hypothesis for the rate of evolution of CpG distribution (see below).
Computing Regional Mutation Rates.
Regional mutation rates were computed by counting human–chimp conserved and diverged nonrepetitive, non-CpG nucleotides in windows of 20 kb. Low-quality alignments (divergence >10%) were excluded from the analysis. Rates were computed separately for intronic and intergenic regions. Windows with <500 intergenic (intronic) nonrepetitive and aligned nucleotides were excluded from the analysis.
Computing Empirical CpG Divergence Rates in Bins of Regional Rate and M-Score (Fig. 2B).
CpGs dinucleotide in the aligned human and chimp genome were grouped into 2D bins according to their regional mutation rate and M-score, using mutation rate bins of size 0.002 and M-score bins of size 2. M-scores were computed from the human sequence (computing M-scores from the chimp sequence provide very similar results). For each bin, the joint distribution of human and chimp dinucleotides was assessed and denoted by:
 |
The joint distribution was reconstructed separately for intergenic and intronic sequences, with very similar results, intron being slightly more conserved. To gain accuracy, the two distributions (intergenic and intronic) were used separately (see below). Fig. 2B represents the CpG divergence rate in intergenic bins by plotting:
The COCAD Assay.
The COCAD assay is a simple heuristic application of the M-score model and the Qb empirical distributions. After extensive experimentation with principled maximum-likelihood-based models (which will be described elsewhere), the empirical approach was preferred as being conservative and robust. The empirical approach does not attempt to reconstruct the ancestral sequence or to model the irreversible deamination process explicitly. Instead, the COCAD background hypothesis assumes that CpGs are evolving independently once their M-score and regional divergence rates are given. The divergence probabilities are computed by using the Qb distributions, and the assay is analyzing only genomic positions with a CpG in either the human or chimp genome. It is thus ignoring positions that possibly lose a CpG in both lineages and rely on the relative proximity of the chimp and human genome to increase the probability that the vast majority of CpGs in the human–chimp ancestral genome were conserved in at least one of the species. The COCAD assay tests the neutral hypothesis in a sliding window (here of size 20 kb). In a given window, all loci bearing a CpG in either the human or chimp genomes are being considered. For each such CpG, the observed divergence equals 1 if the CpG was not conserved between human and chimp and zero otherwise. The divergence probability for that CpG is computed by looking up the joint distribution of the bins defined by the locus's regional mutation rate and plus- and minus-strand M-scores. Note that we are heuristically averaging the estimates from the two-stranded M-scores, and that these are typically very similar. Denote the appropriate bins as pb for the plus-strand M-score and mb for the minus-strand M-score. The divergence probability is defined as:
where ppCG = (ΣdQpb(CG,d) + ΣdQpb(d,CG) − Qpb(CG,CG)) (fraction of positions with at least one CpGs in the positive mscore bin) and pmCG = (ΣdQmb(CG,d) + ΣdQmb(d,CG) − Qmb(CG,CG)) (fraction of positions with at least one CpGs in the negative mscore bin).
The Q distributions are intergenic or intronic according to the genomic context, and the summation is done over all d dinucleotides. The COCAD score equals the Z score of the sum of observed divergences for all CpGs in the window, given the total expected divergence and assuming the variance to be the sum of individual CpG variances [p(1 − p), where p is the CpG divergence probability]. To use the Z score for normal estimation of P values, one has to consider windows with sufficiently high expected divergence (e.g., more than six), which is almost always the case for 20-kb windows and the divergence rates typical to the human–chimp lineage.
Methylation Profiling.
Regions within the hyperconserved domains were selected for Southern blot analysis based on high COCAD score, high Suz12 binding, and substantial numbers of McrBC and HpaII sites. High-molecular-weight DNA was subjected to two rounds of digestion with McrBC or HpaII, followed by digestion with a methylation-insensitive enzyme so that the region could be visualized as a discrete band (BamHI for TBX5, PvuII for FOXA1, and XmnI for HOXD). The samples were also digested with an additional methylation-insensitive enzyme that did not cut within the region but reduced the background on the blots (SphI for Tbx5 and XbaI for FoxA1 and HoxD). Control samples were prepared by methylating the DNA at all CpG dinucleotides with M.SssI before digestion. Details of the methods can be found in Rollins et al. (11).
Supplementary Material
Acknowledgments
We thank A. Abeliovich (Columbia University) for his generous gift of DNA from human ES cells and J. Widom, E. Nili-Gal Yam, R Sorek, I. Gat Viks, D. Pe'er, J. Edwards, K. Anderson, E. Bernstein, E. Segal, E. Siggia, and M. Krieger for discussions and/or critical reading of the manuscript. A.T. was supported by the Rothschild foundation, and T.H.B. was supported by grants from the National Institutes of Health.
Abbreviations
- PRC2
Polycomb repressive 2 complex
- M-score
mutability score
- HCGD
hyperconserved CpG domain
- COCAD
context-based CpG analysis of divergence.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0609746104/DC1.
References
- 1.Siepel A, Haussler D. Mol Biol Evol. 2004;21:468–488. doi: 10.1093/molbev/msh039. [DOI] [PubMed] [Google Scholar]
- 2.Arndt PF, Hwa T. Bioinformatics. 2005;21:2322–2328. doi: 10.1093/bioinformatics/bti376. [DOI] [PubMed] [Google Scholar]
- 3.Takai D, Jones PA. Proc Natl Acad Sci USA. 2002;99:3740–3745. doi: 10.1073/pnas.052410099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saxonov S, Berg P, Brutlag DL. Proc Natl Acad Sci USA. 2006;103:1412–1417. doi: 10.1073/pnas.0510310103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bock C, Paulsen M, Tierling S, Mikeska T, Lengauer T, Walter J. PLoS Genet. 2006;2:e26. doi: 10.1371/journal.pgen.0020026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Das R, Dimitrova N, Xuan Z, Rollins RA, Haghighi F, Edwards JR, Ju J, Bestor TH, Zhang MQ. Proc Natl Acad Sci USA. 2006;103:10713–10716. doi: 10.1073/pnas.0602949103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Handa V, Jeltsch A. J Mol Biol. 2005;348:1103–1112. doi: 10.1016/j.jmb.2005.02.044. [DOI] [PubMed] [Google Scholar]
- 8.Satchwell SC, Drew HR, Travers AA. J Mol Biol. 1986;191:659–675. doi: 10.1016/0022-2836(86)90452-3. [DOI] [PubMed] [Google Scholar]
- 9.Segal E, Fondufe-Mittendorf Y, Chen L, Thastrom A, Field Y, Moore IK, Wang JP, Widom J. Nature. 2006;442:772–778. doi: 10.1038/nature04979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Widom J. Q Rev Biophys. 2001;34:269–324. doi: 10.1017/s0033583501003699. [DOI] [PubMed] [Google Scholar]
- 11.Rollins RA, Haghighi F, Edwards JR, Das R, Zhang MQ, Ju J, Bestor TH. Genome Res. 2006;16:157–163. doi: 10.1101/gr.4362006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.The Chimpanzee Sequencing Analysis Consortium. Nature. 2005;437:69–87. [Google Scholar]
- 13.Bejerano G, Pheasant M, Makunin I, Stephen S, Kent WJ, Mattick JS, Haussler D. Science. 2004;304:1321–1325. doi: 10.1126/science.1098119. [DOI] [PubMed] [Google Scholar]
- 14.Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K. Genes Dev. 2006;20:1123–1136. doi: 10.1101/gad.381706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 17.Squazzo SL, O'Geen H, Komashko VM, Krig SR, Jin VX, Jang SW, Margueron R, Reinberg D, Green R, Farnham PJ. Genome Res. 2006;16:890–900. doi: 10.1101/gr.5306606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M., FitzHugh W, et al. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 19.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 20.Hinrichs AS, Karolchik D, Baertsch R, Barber GP, Bejerano G, Clawson H, Diekhans M, Furey TS, Harte RA, Hsu F, et al. Nucleic Acids Res. 2006;34:D590–D598. doi: 10.1093/nar/gkj144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eckhardt F, Lewin J, Cortese R, Rakyan VK, Attwood J, Burger M, Burton J, Cox TV, Davies R, Down TA, et al. Nat Genet. 2006;38:1378–1385. doi: 10.1038/ng1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.