Abstract
RNA interference (RNAi) has been shown to be a powerful method to study the function of genes in vivo by silencing endogenous mRNA with double-stranded (ds) RNA. Previously, we performed in vivo RNAi screening and identified 43 Drosophila genes, including 18 novel genes required for the development of the embryonic nervous system. In the present study, 22 additional genes affecting embryonic nervous system development were found. Novel RNAi-induced phenotypes affecting nervous system development were found for 16 of the 22 genes. Seven of the genes have unknown functions. Other genes found encode transcription factors, a chromatin-remodeling protein, membrane receptors, signaling molecules, and proteins involved in cell adhesion, RNA binding, and ion transport. Human orthologs were identified for proteins encoded by 16 of the genes. The total number of dsRNAs that we have tested for an RNAi-induced phenotype affecting the embryonic nervous system, including our previous study, is 7,312, which corresponds to ≈50% of the genes in the Drosophila genome.
Keywords: high-throughput screen, neural development, neural mutants
Several genetic screenings using lethal or semilethal mutations generated by P-element insertions or chemical mutagens have led to the identification of genes involved in the development of the nervous system in the Drosophila embryo. Although such strategies generally have been successful, some genes may have escaped detection because cold spots in DNA that are somewhat refractory to P-element insertion and loci that are less susceptible to ethyl methane sulfonate-induced mutations are present in the genome. In addition, it is difficult to analyze the function of a gene during early embryonic development because maternally stored transcripts can compensate for the absence of zygotic expression. Therefore, RNA interference (RNAi) can be a useful tool to overcome hurdles of conventional genetic screens.
In RNAi, a 21-nucleotide-long dsRNA molecule, small interfering RNA (siRNA), that can be delivered either directly to the cell or produced from the processing of a longer precursor by an endonuclease, Dicer, binds to a multiprotein complex (RISC). The activation of RISC leads to complementary base pairing between the antisense strand of the siRNA and the target mRNA species; this process ultimately triggers the degradation of the target mRNA molecule (1, 2). RNAi has been used successfully to screen the genomes of various species to identify genes involved in biological processes (3). Previously, we used an RNAi-based in vivo screen to identify genes required for the development of the embryonic nervous system in Drosophila (4). From a library of double-stranded (ds) RNAs corresponding to approximately one-fourth of the fruit fly genome, we identified 43 genes including 18 novel genes whose roles in embryonic nervous system development had not been described. In the present work, we report an additional 22 genes involved in embryonic nervous system development.
Results and Discussion
RNAi Screen.
Genes involved in the embryonic development of the Drosophila nervous system were identified by RNAi, which extends the results of our previous study (4). Double-stranded RNA was injected into preblastoderm embryos, and the embryos were incubated to allow development to proceed to about stage 15–16. Nervous system development was examined by staining with mAb 22C10, a monoclonal antibody that recognizes the microtubule-associated protein futsch (5). mAb 22C10 stains cell bodies and axons of all neurons in the peripheral nervous system (PNS) and a subset of neurons in the ventral nerve cord (VNC) of the central nervous system (CNS) (6). Therefore, RNAi-induced mutant phenotypes, such as disruption of the nervous system, collapse of axon tracts, fasciculation defects, or loss or gain of neurons, often can be distinguished from wild-type embryos. The staining patterns of mAb 22C10 in the VNC and the PNS of wild-type embryos are shown in Fig. 1 A and B, respectively.
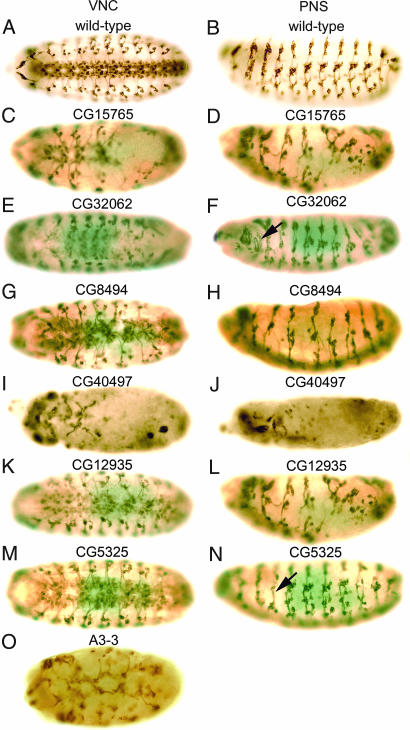
Fig. 1.
Wild-type and RNAi-induced phenotypes of novel genes with unknown functions. (A and B) Wild-type patterns of the VNC and PNS, respectively, of Drosophila embryos stained with mAb 22C10. (C–N) RNAi mutant phenotypes after injection of dsRNAs synthesized from CG15765 (C and D), CG32062 (E and F), CG8494 (G and H), CG40497 (I and J), CG12935 (K and L), CG5325 (M and N), and A3-3 (O). Ventral (A, C, E, G, I, K, M, and O) and lateral (B, D, F, H, J, L, and N) views of stage 15/16 embryos are shown. In all images, anterior is to the left, and in lateral views of embryos, dorsal is up.
Throughout the screening procedure, we adopted highly conservative criteria of evaluation to eliminate false-positives. First, to identify candidate genes affecting nervous system development, each dsRNA was injected into at least 50–100 embryos, and only when RNAi phenotypes were found in 50% or more of the injected embryos and when results from multiple experiments by different investigators were consistent were the results considered positive. Next, to confirm the effect of RNAi on the nervous system, dsRNAs were synthesized from different regions of the candidate genes and cross-checked by injection into embryos in more than three experiments by independent investigators. From 3,998 dsRNAs, we identified 22 genes affecting the embryonic nervous system (Table 1). The effects of 16 of the genes on the embryonic nervous system have not been described previously (sections A and B in Table 1). Loss-of-function mutant phenotypes of the remaining genes were reported previously to affect the embryonic nervous system (section C in Table 1).
Table 1.
Genes identified by RNA interference
| Gene | Molecular function | Human ortholog* |
|---|---|---|
| A. With unknown functions | ||
| CG15765 | C-type lectin, sugar binding | No homolog |
| CG32062 | RNP-1 RNA-binding region | Ataxin 2-binding protein 1 |
| CG8494 | Unknown | Ubiquitin-specific protease 20 |
| CG40497 | Unknown | No homolog |
| CG12935 | Unknown | No homolog |
| CG5325 | Unknown | Pex19 |
| A3-3 | DNA binding, basic leucine zipper | FOSB |
| B. With known functions | ||
| MED25 | RNA polymerase II transcription mediator | ARC92 (MED25) |
| Tsp86D | Cell–cell adhesion, signal transduction | TSPAN33 |
| XNP | ATP-dependent helicase | ATR-X |
| pim | Sister chromatid segregation | No homolog |
| Acf1 | Chromatin assembly and remodeling | BAZ 1A |
| cact | Establishing dorsoventral polarity | NFKBIA |
| lmd | Putative zinc finger transcription factor | No homolog |
| olf186-F | Calcium release-activated calcium channel | TMEM142A |
| Imp | Regulating translation, mRNA binding | IGF2BP1 |
| C. With known nervous system development phenotypes | ||
| dmt | Nuclear localization motifs, PNS development | No homolog |
| pbl | GTP exchange factor, initiating cytokinesis | ECT2 |
| pros | Homeodomain, neuronal cell fate | PROX1 |
| crb | Adherens junctions, epithelial cell polarity | CRB1 |
| arm | β-Catenin, epithelial cell polarity | CTNB1 (catenin β1) |
| arr | Low-density lipoprotein receptor | LRP6 |
*Human orthologs shown are as reported (http://inparanoid.cgb.ki.se).
Genes with Previously Uncharacterized Nervous System Phenotypes.
Among the 16 genes with RNAi-induced nervous system phenotypes are seven genes, CG15765, CG32062, CG8494, CG40497, CG12935, CG5325, XNP, and A3-3, whose functions are uncharaterized, and nine genes, such as Mediator complex subunit 25 (MED25), Tetraspanin 86D (Tsp86D), XNP pimples (pim), ATP-dependent chromatin assembly factor large subunit (Acf1), cactus (cact), lame duck (lmd), olf186-F, and IGF-II mRNA-binding protein (Imp), whose functions have been reported; however, no roles have been reported for these genes in the development of the embryonic nervous system. RNAi-induced mutant phenotypes for these genes are shown in Figs. 1 and 2.
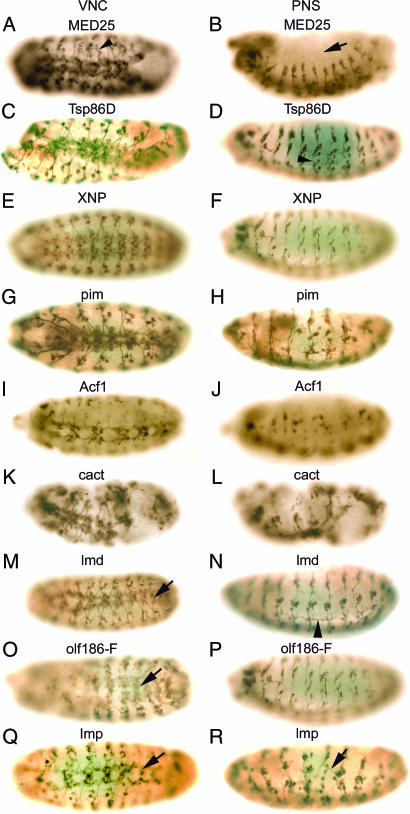
Fig. 2.
RNAi-induced phenotypes of novel genes with known functions. Embryos were injected with dsRNAs from MED25 (A and B), Tsp86D (C and D), XNP (E and F), pim (G and H), Acf1 (I and J), cact (K and L), lmd (M and N), olf186-F (O and P), and Imp (Q and R). After staining with mAb 22C10, ventral (A, C, E, G, I, K, M, O, and Q) and lateral (B, D, F, H, J, L, N, P, and R) views are shown.
CG15765 is a gene of unknown function encoding a protein that contains a C-type lectin domain expressed in the nervous system at late embryonic stages and is highly enriched in the adult fly brain (www.flyatlas.org). Suppression of CG15765 gene expression by RNAi resulted in a phenotype characterized by hypoplasia and disorganization of both the VNC and the PNS (Fig. 1 C and D). The level of CG15765 RNA increases markedly in 12- to 16-h-old embryos and then decreases during later embryonic development [supporting information (SI) Fig. 4A]. CG32062 encodes a protein containing a ribonucleoprotein 1 RNA recognition motif. The molecular function of this gene has not been reported; however, treatment of Drosophila cell lines with CG32062 dsRNA greatly reduced the number of cells and impaired secretion from the Golgi apparatus and Ca2+ transport (www.flyrnai.org). An enhancer trap fly line bearing an insertion within CG32062 exhibits a segmentally modulated expression pattern in the larval CNS (7). CG32062 mRNA also is expressed in the embryonic nervous system (7). Injection of CG32062 dsRNA in early embryos resulted in a decrease in neural cells in the medial region of the VNC (Fig. 1E), and some misrouted axons in the PNS (arrow in Fig. 1F). RT-PCR results show that CG32062 is zygotically expressed (SI Fig. 4B). CG8494 is a gene of unknown function encoding a protein that exhibits limited sequence homology to fat facets (faf), a Drosophila deubiquitinating enzyme that is strongly expressed in the developing CNS. Overexpression of faf in the nervous system leads to an increase in the number of synaptic boutons and synaptic branches at neuromuscular junctions (8). The human homolog of CG8494 encodes ubiquitin-specific processing protease 20 (Table 1). The neural phenotype observed after injection of CG8494 dsRNA consists of hypoplasia and disorganization of both the VNC and the PNS (Fig. 1 G and H, respectively). CG40497 encodes a protein of unknown function. Suppression of CG40497 gene expression by RNAi resulted in global hypoplasia of epidermal cells (Fig. 1 I and J). The embryos did not develop beyond stage 14, and they showed severe loss of neurons and abnormal segmental structure of the nervous system. CG12935 is a gene of unknown function. Silencing of CG12935 expression by RNAi resulted in a decrease in the number of neuromeres and severe disruption of both the VNC and the PNS (Fig. 1 K and L). RT-PCR analysis showed that CG12935 is expressed at the highest level in 4- to 8-h embryos, and expression decreases rapidly after germ band elongation (SI Fig. 4D). CG5325 encodes a protein of unknown function that is an ortholog of Caenorhabditis elegans Prx-19 and Homo sapiens Pex19, both encoding proteins essential for peroxisome biogenesis (9). Pex19 is involved in CNS development and neuron migration (10), and mutation of Pex19 results in Zellweger syndrome, which includes mental retardation (11). Down-regulation of CG5325 by injection of dsRNA into embryos resulted in disruption of the VNC (Fig. 1M), misrouting of axons (arrow in Fig. 1N), and disorganization of dorsal clusters of cells in the PNS (Fig. 1N). A3-3 encodes a protein containing a basic region-leucine zipper (bZIP) motif, similar to the fos oncogene. bZIP transcription factors bind to DNA as dimers and regulate gene expression (12). The human ortholog of A3-3 is FOSB. Suppression of A3-3 expression by RNAi resulted in severe disruption of the VNC and hypoplasia (Fig. 1O).
MED25 encodes a protein possessing RNA polymerase II transcription mediator activity. In Drosophila, 30 MED molecules have been identified by biochemical and genomic analyses (13). Although the target of MED25 has not been identified, MED25 has been shown to be required for lipopolysaccharide- or heat shock-induced gene expression (14). Treatment of Drosophila S2 cells with MED25 dsRNA resulted in a decrease in cell viability (www.flyrnai.org). The RNAi-induced MED25 phenotype includes the failure of extension of some axons from the VNC (arrowhead in Fig. 2A). All dorsal cell clusters in the PNS are missing (arrow in Fig. 2B). Tsp86D is a member of the tetraspanin superfamily, i.e., integral membrane proteins with four transmembrane domains involved in signal transduction, cell adhesion, and regulation of cellular development and proliferation (15). In Drosophila, functional redundancy or compensatory mechanisms have been observed for some of the 35 members of the tetraspanin family (16). Tsp86D is expressed in the VNC and the hypocerebral ganglion (16). Suppression of Tsp86D gene expression by RNAi resulted in abnormal formation of the VNC (Fig. 2C) and axon pathfinding defects (arrowhead in Fig. 2D) in the PNS. XNP encodes a protein containing an SNF2-related helicase domain, similar to human ATR-X, encoded by α-Thallassemia/mental retardation, X-linked ATR-X gene. Mutations of ATR-X result in mental retardation with hematologic, skeletal, and genital defects (17). Although the molecular function of XNP has not been studied, a gain-of-function screen to identify genes involved in the development of the mushroom bodies showed that overexpression of XNP resulted in fewer neurons in the mushroom bodies (18). Embryos injected with XNP dsRNA exhibited disruption of the VNC (Fig. 2E) and reduction of the intersegmental and segmental nerves (Fig. 2F). pim is a protein involved in sister chromatid segregation during mitosis (19, 20). A high level of maternal pim transcript is present in the early embryo with mitotically synchronized cell division before cellularization; after that, pim transcripts remain distributed throughout the embryo and become restricted to the developing nervous system where cells proliferate in late embryos (20), consistent with the RT-PCR results showing that pim is highly expressed during early stages of development and decreases rapidly at later stages (SI Fig. 4H). Injection of pim dsRNA in early embryos resulted in a loss of neurons in the VNC and a loss of neurons and severe disruption of the PNS (Fig. 2 G and H). We observed a similar RNAi-induced mutant phenotype by injection of dsRNA corresponding to three rows (4), which also is required for sister chromatid separation (21). pim protein associates in vivo with three rows forming a complex (21). The Acf1 gene encodes a large subunit of the ATP-using chromatin assembly and remodeling factor (ACF) required for the assembly of histone–DNA complexes into periodic nucleosome arrays (22). Acf1 contains two PHD fingers and a bromodomain, and it associates with Imitation of Switch (ISWI), a homolog of yeast SWI2/SNF2 and a small subunit of ACF (22, 23). The expression of Acf1 was observed in a subset of cells in the midline primordium region of the CNS (24). Embryos injected with Acf1 dsRNA exhibited missing segments (Fig. 2 I and J), loss of neurons, and disruption of the architecture of the VNC (Fig. 2I) and severe hypoplasia and disorganization of the PNS (Fig. 2J). Although Acf1 genetic mutants develop normally during embryogenesis (22), the severe RNAi phenotype we observed can be explained by the depletion of maternally stored Acf1 mRNA by RNAi. cactus, a homolog of vertebrate NFkBIA, contains seven ankyrin repeats and possesses transcription regulator activity (25). cactus is involved in the innate immune response through the Toll signaling pathway (26) and also is required for the establishment of dorsal-ventral polarity by direct interaction with Dorsal, a homolog of vertebrate NFκB (27, 28). A loss-of-function mutation of cactus leads to ventralized embryos lacking dorsal cuticle and embryonic lethality (27). cactus is necessary for the normal function of the larval neuromuscular system, and loss-of-function cactus mutants have altered bouton numbers and neurotransmitter release at neuromuscular junctions (29). Reduction of cactus gene expression in embryos by RNAi resulted in severe hypoplasia and overall disruption of both the VNC and the PNS (Fig. 2 K and L). lmd encodes a putative zinc finger transcription factor related to the vertebrate GLI superfamily (30). lmd is expressed exclusively in mesodermal cells in a Wingless- and Notch-dependent manner and acts as a key regulator of myogenesis (30). The earliest expression of lmd is observed in the primordium of the caudal visceral mesoderm at stage 10, and strong expression is predominantly in fusion-competent myoblasts at stage 12 (30), consistent with our RT-PCR result showing that the highest expression of lmd is in 4- to 12-h-old embryos (SI Fig. 4I). A loss-of-function mutation of lmd leads to a complete lack of myoblast fusion and to embryonic lethality (30, 31). We observed that many embryos injected with lmd dsRNA exhibited relatively normal development of the CNS; however, some embryos had abnormal VNC morphology (arrow in Fig. 2M). Injected embryos exhibited a more severe altered phenotype in the PNS, such as disorganization and misrouting with axons crossing segmental boundaries in the ventrolateral region (arrowhead in Fig. 2N). olf186-F is a protein encoding Ca2+ release-activated Ca2+ channel modulator-1 (CRACM1) identified by a genome-wide RNAi-based screen in Drosophila cells for genes affecting calcium influx across the plasma membrane (32). RNAi phenotypes of Drosophila embryos injected with olf186-F dsRNA exhibit defects in the organization of the VNC (arrow in Fig. 2O) and severe disruption and hypoplasia of the PNS (Fig. 2P). Imp is an IGF-II mRNA-binding protein similar to zipcode-binding protein (ZBP-1). A gain-of-function screen showed that Imp is involved in axon pathfinding and synapse formation in the neuromuscular system of Drosophila larva (33). Norga et al. (34) also reported that a P-element insertion in Imp results in an increase in the number of adult sensory bristles. Maternally derived Imp mRNA is distributed evenly in the whole embryo at early stages, and in later stages, Imp is expressed exclusively in the developing CNS (35). We confirmed the maternal expression of Imp by RT-PCR (SI Fig. 4K). Injection of Imp dsRNA in early embryos resulted in loss of some segments, loss of neurons, and axon pathfinding defects in both the VNC and the PNS (arrows in Fig. 2 Q and R).
Genes with Known Nervous System Phenotypes.
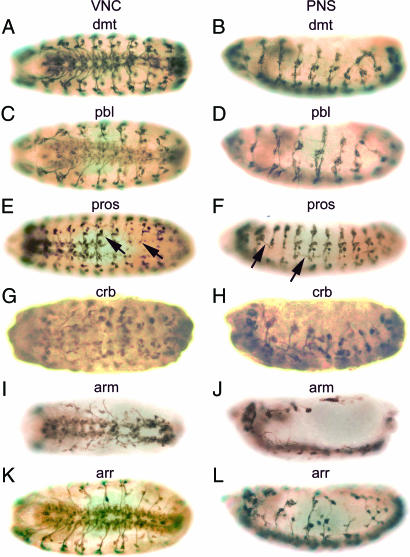
RNAi-induced phenotypes for six genes, dalmatian (dmt), pebble (pbl), prospero (pros), crumbs (crb), armadillo (arm), and arrow (arr), whose roles in the development of the embryonic nervous system have been described previously, are shown in Fig. 3. dmt encodes a putative nuclear protein that is not related to any known protein. Expression of dmt increases during germ band extension and becomes more restricted to the PNS, VNC, and embryonic brain during stage 12 (36). Embryos carrying hypomorphic mutant alleles of dmt display a mild phenotype, such as infrequent loss of neurons and occasional pathfinding defects in the PNS (37); whereas more severe alleles exhibit a general disorganization of the PNS (34). RNAi-induced phenotypes exhibited disruption of the VNC (Fig. 3A) and severe hypoplasia and misrouting of neurons in the PNS (Fig. 3B), consistent with genetic mutant phenotypes. pbl encodes a GTP-exchange factor for Rho 1 GTPase that is an essential regulator of cytokinesis (38). High levels of maternally provided pbl transcripts are present in preblastoderm embryos, and the highest protein levels are in nuclei of dividing cells, such as embryonic neuroblasts (39). These results are consistent with our RT-PCR analysis showing the highest level of expression of pbl during very early embryogenesis (SI Fig. 4M). Embryos homozygous for pbl alleles display gross defects in axon trajectories, loss of neurons, and fasciculation defects in the PNS, leading to embryonic lethality (40). A gain-of-function screen also revealed that pbl is required for controlling motor axon guidance and synaptogenesis (33). Injection of pbl dsRNA in embryos resulted in an abnormal VNC (Fig. 3C), loss of neurons, misrouting of axons, and fasciculation defects in the PNS (Fig. 3D), in agreement with genetic mutant phenotypes. pros is a transcription factor containing a homeodomain and is involved in neuronal fate determination and the formation of axons (41). During neuroblast (NB) division in early embryogenesis, pros is localized basally in NBs and is segregated asymmetrically to the smaller daughter cell [ganglion mother cell (GMC)] (42, 43). pros is expressed in a subset of NBs and GMCs but not in VNC neurons (41, 44). Homozygous mutant alleles display defects in axon pathfinding in both the CNS and PNS, leading to aberrant neuronal connections (41, 45). We observed a similar RNAi-induced phenotype after injection of pros dsRNA into embryos. Intersegmental and segmental nerves often are missing or are not properly extended in the VNC (arrows in Fig. 3E), and sensory axons exhibit aberrant connections or pathfinding defects in the PNS (arrows in Fig. 3F). crb is an integral membrane protein containing EGF-like repeats and is localized exclusively in the apical membranes of epithelial cells during cellularization and also is expressed in many tissues, including NBs and the embryonic PNS (46). crb is involved in the biogenesis of adherens junctions composed of cadherin–catenin complexes (47, 48) and is required for the establishment of epithelial cell polarity (49). The RNAi-induced mutant phenotype of crb exhibits global hypoplasia of the VNC and severe disruption of the PNS including defects in neuromere organization and axon genesis (Fig. 3 G and H). arm is a homolog of vertebrate catenin β-1 and is involved in many biological processes during embryogenesis, such as cell–cell adhesion, establishment/maintenance of cell polarity, and NB fate determination depending on the Wg/Wnt signaling pathway (for a review, see ref. 50). arm binds to α-catenin, E-cadherin, and shotgun (51) at adherens junctions in the apical membrane of the epidermis, and it regulates cell polarity. arm is expressed in many tissues, including the embryonic CNS and larval brain where arm is enriched in fiber tracks of axons (52). Homozygous arm mutant alleles exhibit strong segment polarity defects and the absence of embryonic cuticle, whereas hypomorphic mutant alleles display defects in NB fate determination and axon development (53). Embryos injected with arm dsRNA exhibit severe RNAi-induced phenotypes, such as hypoplasia and overall disruption both of the CNS and the PNS (Fig. 3 I and J). Epidermal cells are not formed (Fig. 3I). We also observed decreases in the size of embryos resulting from segment polarity defects (data not shown). This phenotype resembles the shotgun RNAi-dependent phenotype shown in our previous study (4). arr is an essential factor in the Wg/Wnt signaling pathway functioning in embryonic segmentation, limb development, and CNS organization (54). Loss of arr expression disrupts midgut morphogenesis and the nervous system (55). The mutant embryos exhibit disorganized neuropile, gaps in the longitudinal nerves, tangled and misguided commissural nerves in the CNS, and reduced and mispositioned sensory neurons in the PNS (55). RNAi-dependent suppression of arr gene expression resulted in disorganization in the VNC (Fig. 3K) and severe hypoplasia and pathfinding defects in the PNS (Fig. 3L), which are similar to the genetic mutant phenotypes.
Fig. 3.
RNAi-induced phenotypes of known genes whose functions have been described in the development of the embryonic nervous system. Embryos were injected with dsRNAs from dmt (A and B), pbl (C and D), pros (E and F), crb (G and H), arm (I and J), and arr (K and L) and were stained with mAb 22C10. The photographs were taken from ventral (A, C, E, G, I, and K) and lateral (B, D, F, H, J, and L) views.
In summary, genes found in this work encode transcription factors, a chromatin remodeling factor, membrane receptors, signaling molecules, and other proteins involved in various biological processes. We observed similar RNAi-dependent phenotypes in genes involved in the same pathway, such as arr, a receptor of Wnt/Wg, and arm, a key regulator of the Wnt/Wg signaling pathway. Because of RNAi, both phenotypes display severe hypoplasia and axon pathfinding defects during the development of the embryonic nervous system. In addition, down-regulation by RNAi of CG8494, which encodes a putative deubiquitinating protease, and pbl, encoding a substrate for ubiquitin E3 ligase (56), resulted in disruption of the nervous system, emphasizing the importance of ubiquitin-dependent protein degradation in the development of the nervous system. Comparison of the phenotypes identified from our RNAi screen with the corresponding mutant phenotypes obtained in genetic screens showed that RNAi-induced mutant phenotypes resemble genetic mutant phenotypes, indicating that RNAi can be used efficiently to identify genes that are involved in the development of the embryonic nervous system of Drosophila.
Materials and Methods
dsRNA Synthesis.
For the primary RNAi screen to select candidate genes required for embryonic nervous system development, we synthesized dsRNAs from the Unigene 1.0 cDNA library as described previously (4). After selection of candidate genes, dsRNA again was synthesized but from a different region of the gene, to eliminate false-positives and to determine whether RNAi-induced mutant phenotypes were reproducible. For this second dsRNA synthesis, two rounds of PCR amplifications were performed. The primers were designed by Primer3 Input (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi). The specificity of the amplified fragments was confirmed by NCBI Blast Searches (www.ncbi.nlm.nih.gov/BLAST), and the primer sequences, size of dsRNAs, and the cDNA or genomic regions used for template are shown in SI Table 2. For the first PCR amplification, either genomic DNA or cDNA synthesized from total RNA from wild-type adult flies was used. For the second PCR step, primers were used containing 18 bp of nested inner sequences of the first amplified fragment and a T7 RNA polymerase promoter sequence at the 5′ end. After PCR, in vitro transcription was catalyzed by T7 RNA polymerase (Takara Bio, Otsu, Shiga, Japan). The RNA products were treated with DNase I (Takara Bio) and then purified with mini-Quick Spin RNA columns (Roche Applied Science, Indianapolis, IN). After annealing sense and antisense RNA, the concentration of dsRNA was measured and adjusted to 1 μg/μl in 5 mM KCl/0.1 mM NaH2PO4, pH 6.8, containing 5% food color (green; McCormick, Baltimore, MD).
Embryo Injection and Immunohistochemistry.
Drosophila melanogaster, Oregon R strain (wild type) was used for dsRNA injection. Embryo collection, injection, and immunostaining were performed as described previously (4).
Supplementary Material
Acknowledgments
We thank Sachiko Higashi, Kazumi Furuhara, Kimie Iwasaki, and Yoshie Ichikawa for technical assistance.
Abbreviations
- NB
neuroblast
- PNS
peripheral nervous system
- VNC
ventral nerve cord.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0611687104/DC1.
References
- 1.Hannon GJ. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- 2.Sharp PA. Genes Dev. 2001;15:485–490. doi: 10.1101/gad.880001. [DOI] [PubMed] [Google Scholar]
- 3.Friedman A, Perrimon N. Curr Opin Genet Develop. 2004;14:470–476. doi: 10.1016/j.gde.2004.07.010. [DOI] [PubMed] [Google Scholar]
- 4.Ivanov AI, Rovescalli AC, Pozzi P, Yoo S, Mozer B, Li HP, Yu SH, Higashida H, Guo V, Spencer M, Nirenberg M. Proc Natl Acad Sci USA. 2004;101:16216–16221. doi: 10.1073/pnas.0407188101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hummel T, Krukkert K, Roos J, Davis G, Klämbt C. Neuron. 2000;26:357–370. doi: 10.1016/s0896-6273(00)81169-1. [DOI] [PubMed] [Google Scholar]
- 6.Fujita SC, Zipursky SL, Benzer S, Ferrus A, Shotwell SL. Proc Natl Acad Sci USA. 1982;79:7929–7933. doi: 10.1073/pnas.79.24.7929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bajpai R, Sambrani N, Stadelmayer B, Shashidhara LS. Gene Expr Patterns. 2004;5:113–121. doi: 10.1016/j.modgep.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 8.DiAntonio A, Haghighi AP, Portman SL, Lee JD, Amaranto AM, Goodman CS. Nature. 2001;412:449–452. doi: 10.1038/35086595. [DOI] [PubMed] [Google Scholar]
- 9.Heiland I, Erdmann R. FEBS J. 2005;272:2362–2372. doi: 10.1111/j.1742-4658.2005.04690.x. [DOI] [PubMed] [Google Scholar]
- 10.Suzuki Y, Shimozawa N, Imamura A, Fukuda S, Zhang Z, Orii T, Kondo N. J Inherit Metab Dis. 2001;24:151–165. doi: 10.1023/a:1010310816743. [DOI] [PubMed] [Google Scholar]
- 11.Matsuzono Y, Kinoshita N, Tamura S, Shimozawa N, Hamasaki M, Ghaedi K, Wanders RJ, Suzuki Y, Kondo N, Fujiki Y. Proc Natl Acad Sci USA. 1999;96:2116–2121. doi: 10.1073/pnas.96.5.2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baranger AM. Curr Opin Chem Biol. 1998;2:18–23. doi: 10.1016/s1367-5931(98)80031-8. [DOI] [PubMed] [Google Scholar]
- 13.Kim YJ, Lis JT. Trends Biochem Sci. 2005;30:245–249. doi: 10.1016/j.tibs.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 14.Kim TW, Kwon YJ, Kim JM, Song YH, Kim SN, Kim YJ. Proc Natl Acad Sci USA. 2004;101:12153–12158. doi: 10.1073/pnas.0401985101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stipp CS, Kolesnikova TV, Hemler ME. Trends Biochem Sci. 2003;28:106–112. doi: 10.1016/S0968-0004(02)00014-2. [DOI] [PubMed] [Google Scholar]
- 16.Fradkin LG, Kamphorst JT, DiAntonio A, Goodman CS, Noordermeer JN. Proc Natl Acad Sci USA. 2002;99:13663–13668. doi: 10.1073/pnas.212511099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gibbons RJ, Bachoo S, Picketts DJ, Aftimos S, Asenbauer B, Bergoffen J, Berry SA, Dahl N, Fryer A, Keppler K, et al. Nat Genet. 1997;17:146–148. doi: 10.1038/ng1097-146. [DOI] [PubMed] [Google Scholar]
- 18.Nicolai M, Lasbleiz C, Dura JM. J Neurobiol. 2003;57:291–302. doi: 10.1002/neu.10277. [DOI] [PubMed] [Google Scholar]
- 19.Leismann O, Herzig A, Heidmann S, Lehner CF. Genes Dev. 2000;14:2192–2205. doi: 10.1101/gad.176700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stratmann R, Lehner CF. Cell. 1996;84:25–35. doi: 10.1016/s0092-8674(00)80990-3. [DOI] [PubMed] [Google Scholar]
- 21.Lee LA, Orr-Weaver TL. Annu Rev Genet. 2003;37:545–578. doi: 10.1146/annurev.genet.37.110801.143149. [DOI] [PubMed] [Google Scholar]
- 22.Fyodorov DV, Blower MD, Karpen GH, Kadonaga JT. Genes Dev. 2004;18:170–183. doi: 10.1101/gad.1139604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ito T, Levenstein ME, Fyodorov DV, Kutach AK, Kobayashi R, Kadonaga JT. Genes Dev. 1999;13:1529–1539. doi: 10.1101/gad.13.12.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kearney JB, Wheeler SR, Estes P, Parente B, Crews ST. Dev Biol. 2004;275:473–492. doi: 10.1016/j.ydbio.2004.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Geisler R, Bergmann A, Hiromi Y, Nusslein-Volhard C. Cell. 1992;71:613–621. doi: 10.1016/0092-8674(92)90595-4. [DOI] [PubMed] [Google Scholar]
- 26.Lemaitre B, Nicolas E, Michaut L, Reichhart JM, Hoffmann JA. Cell. 1996;86:973–983. doi: 10.1016/s0092-8674(00)80172-5. [DOI] [PubMed] [Google Scholar]
- 27.Schupbach T, Wieschaus E. Genetics. 1989;121:101–117. doi: 10.1093/genetics/121.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Whalen AM, Steward R. J Cell Biol. 1993;123:523–534. doi: 10.1083/jcb.123.3.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beramendi A, Peron S, Megighian A, Reggiani C, Cantera R. Neuroscience. 2005;134:397–406. doi: 10.1016/j.neuroscience.2005.04.046. [DOI] [PubMed] [Google Scholar]
- 30.Duan H, Skeath JB, Nguyen HT. Development (Cambridge, UK) 2001;128:4489–4500. doi: 10.1242/dev.128.22.4489. [DOI] [PubMed] [Google Scholar]
- 31.Ruiz-Gomez M, Coutts N, Suster ML, Landgraf M, Bate M. Development (Cambridge, UK) 2002;129:133–141. doi: 10.1242/dev.129.1.133. [DOI] [PubMed] [Google Scholar]
- 32.Zhang SL, Yeromin AV, Zhang XH, Yu Y, Safrina O, Penna A, Roos J, Stauderman KA, Cahalan MD. Proc Natl Acad Sci USA. 2006;103:9357–9362. doi: 10.1073/pnas.0603161103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kraut R, Menon K, Zinn K. Curr Biol. 2001;11:417–430. doi: 10.1016/s0960-9822(01)00124-5. [DOI] [PubMed] [Google Scholar]
- 34.Norga KK, Gurganus MC, Dilda CL, Yamamoto A, Lyman RF, Patel PH, Rubin GM, Hoskins RA, Mackay TF, Bellen HJ. Curr Biol. 2003;13:1388–1396. doi: 10.1016/s0960-9822(03)00546-3. [DOI] [PubMed] [Google Scholar]
- 35.Nielsen J, Cilius Nielsen F, Kragh Jakobsen R, Christiansen J. Mech Dev. 2000;96:129–132. doi: 10.1016/s0925-4773(00)00383-x. [DOI] [PubMed] [Google Scholar]
- 36.Prokopenko SN, He Y, Lu Y, Bellen HJ. Genetics. 2000;156:1691–1715. doi: 10.1093/genetics/156.4.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Salzberg A, Prokopenko SN, He Y, Tsai P, Pal M, Maroy P, Glover DM, Deak P, Bellen HJ. Genetics. 1997;147:1723–1741. doi: 10.1093/genetics/147.4.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prokopenko SN, Brumby A, O'Keefe L, Prior L, He Y, Saint R, Bellen HJ. Genes Dev. 1999;13:2301–2314. doi: 10.1101/gad.13.17.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prokopenko SN, Saint R, Bellen HJ. Mech Dev. 2000;90:269–273. doi: 10.1016/s0925-4773(99)00233-6. [DOI] [PubMed] [Google Scholar]
- 40.Abdelilah-Seyfried S, Chan YM, Zeng C, Justice NJ, Younger-Shepherd S, Sharp LE, Barbel S, Meadows SA, Jan LY, Jan YN. Genetics. 2000;155:733–752. doi: 10.1093/genetics/155.2.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Doe CQ, Chu-LaGraff Q, Wright DM, Scott MP. Cell. 1991;65:451–464. doi: 10.1016/0092-8674(91)90463-9. [DOI] [PubMed] [Google Scholar]
- 42.Hirata J, Nakagoshi H, Nabeshima Y, Matsuzaki F. Nature. 1995;377:627–630. doi: 10.1038/377627a0. [DOI] [PubMed] [Google Scholar]
- 43.Kaltschmidt JA, Davidson CM, Brown NH, Brand AH. Nat Cell Biol. 2000;2:7–12. doi: 10.1038/71323. [DOI] [PubMed] [Google Scholar]
- 44.Matsuzaki F, Koizumi K, Hama C, Yoshioka T, Nabeshima Y. Biochem Biophys Res Commun. 1992;182:1326–1332. doi: 10.1016/0006-291x(92)91878-t. [DOI] [PubMed] [Google Scholar]
- 45.Gao FB, Brenman JE, Jan LY, Jan YN. Genes Dev. 1999;13:2549–2561. doi: 10.1101/gad.13.19.2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tepass U, Theres C, Knust E. Cell. 1990;61:787–799. doi: 10.1016/0092-8674(90)90189-l. [DOI] [PubMed] [Google Scholar]
- 47.Grawe F, Wodarz A, Lee B, Knust E, Skaer H. Development (Cambridge, UK) 1996;122:951–959. doi: 10.1242/dev.122.3.951. [DOI] [PubMed] [Google Scholar]
- 48.Tepass U. Dev Biol. 1996;177:217–225. doi: 10.1006/dbio.1996.0157. [DOI] [PubMed] [Google Scholar]
- 49.Lu B, Roegiers F, Jan LY, Jan YN. Nature. 2001;409:522–525. doi: 10.1038/35054077. [DOI] [PubMed] [Google Scholar]
- 50.Miller JR, Moon RT. Genes Dev. 1996;10:2527–2539. doi: 10.1101/gad.10.20.2527. [DOI] [PubMed] [Google Scholar]
- 51.Pai LM, Kirkpatrick C, Blanton J, Oda H, Takeichi M, Peifer M. J Biol Chem. 1996;271:32411–32420. doi: 10.1074/jbc.271.50.32411. [DOI] [PubMed] [Google Scholar]
- 52.Patel NH, Schafer B, Goodman CS, Holmgren R. Genes Dev. 1989;3:890–904. doi: 10.1101/gad.3.6.890. [DOI] [PubMed] [Google Scholar]
- 53.Loureiro J, Peifer M. Curr Biol. 1998;8:622–632. doi: 10.1016/s0960-9822(98)70249-0. [DOI] [PubMed] [Google Scholar]
- 54.Wehrli M, Dougan ST, Caldwell K, O'Keefe L, Schwartz S, Vaizel-Ohayon D, Schejter E, Tomlinson A, DiNardo S. Nature. 2000;407:527–530. doi: 10.1038/35035110. [DOI] [PubMed] [Google Scholar]
- 55.Li K, Kaufman TC. Cell. 1996;85:585–596. doi: 10.1016/s0092-8674(00)81258-1. [DOI] [PubMed] [Google Scholar]
- 56.Reiter LT, Seagroves TN, Bowers M, Bier E. Hum Mol Genet. 2006;15:2825–2835. doi: 10.1093/hmg/ddl225. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.