Abstract
Neuronal gene expression is tightly regulated in developing CNS. Here, we demonstrate the anti-neural function of phosphatase SCP1 (small C-terminal domain phosphatase 1) during development. We further show that the neuron-enriched microRNA miR-124 directly targets SCP1-3′ untranslated region (UTR) to suppress SCP1 expression. In developing spinal cord, expression of miR-124 and SCP1 is complementary, and miR-124 antagonism phenocopies SCP1 overexpression and vice versa. In P19 cells, miR-124 suppresses SCP1 expression and induces neurogenesis, and SCP1 counteracts this proneural activity of miR-124. Our results suggest that, during CNS development, timely down-regulation of SCP1 is critical for inducing neurogenesis, and miR-124 contributes to this process at least in part by down-regulating SCP1 expression.
Keywords: miR-124, SCP1, neural tube development, neurogenesis
In the developing CNS, dividing neural progenitors in the ventricular zone begin to express neuronal genes as they migrate laterally. Post-mitotic neurons eventually settle in the outer layer. This process of neurogenesis requires the coordinated up-regulation of neuronal genes and down-regulation of nonneuronal genes to ensure the proper timing of differentiation and number of neurons.
The transcription factor REST/NRSF suppresses neuronal genes in nonneuronal cells by binding a conserved repressor element (RE1) in neuronal gene loci and recruiting the corepressor complex containing histone deacetylases and methyl CpG-binding protein MeCP2 (Andres et al. 1999; Huang et al. 1999; Roopra et al. 2000; Ballas et al. 2001; Lunyak et al. 2002). Down-regulation of REST during transition from progenitors to post-mitotic neurons allows neuronal gene expression (Ballas et al. 2005). Small C-terminal domain phosphatase 1 (SCP1) is an anti-neural factor expressed in nonneuronal tissues, like REST, and recruited to RE1-containing neural genes by REST (Yeo et al. 2005). Phosphatase-inactive SCP1 mutant promotes neurogenesis in P19 cells by blocking the anti-neural function of REST. Thus, timely down-regulation of the anti-neural REST/SCP1 pathway is expected to be a key step in neurogenesis during CNS development. REST is subjected to both post-translational degradation and retinoic acid receptor-mediated transcriptional repression (Ballas et al. 2005). However, little is known about how SCP1 expression is regulated during development.
MicroRNAs (miRNAs), a group of noncoding small RNAs, repress gene expression by translational inhibition and destabilization of mRNAs (Bartel and Chen 2004; He and Hannon 2004). miRNA genes are transcribed as a primary miRNA transcript and processed to an ∼22-nucleotide (nt) mature miRNA. The seed region (nucleotides 2–7) of miRNAs is critical for target recognition and silencing (Doench and Sharp 2004; Lewis et al. 2005). Because miRNAs down-regulate multiple target genes simultaneously (Brennecke et al. 2005; Grun et al. 2005; Krek et al. 2005; Lewis et al. 2005), miRNAs can serve as global regulators of gene expression. Indeed, miRNAs have been implicated in many biological processes (Carrington and Ambros 2003; Ambros 2004; Alvarez-Garcia and Miska 2005).
During development, many miRNAs are expressed in neurons or specific areas of the CNS, and their roles in CNS development and direct target genes have just begun to be defined. In particular, miR-124, whose mature sequences are conserved from Caenorhabditis elegans to humans, is the most abundant miRNA in adult and embryonic CNS (Lagos-Quintana et al. 2002; Darnell et al. 2006; Deo et al. 2006; Kloosterman et al. 2006). In nonneuronal HeLa cells, misexpressed miR-124 down-regulates 174 genes expressed at lower levels in the brain (Lim et al. 2005). Although none of these genes has been validated as a direct target of miR-124, these results suggest that miR-124 may contribute to maintaining neuronal identity by suppressing nonneuronal genes in neurons.
Neurogenesis is accompanied by up-regulation of neuronal genes and down-regulation of unwanted nonneuronal genes. The opposite functions of REST/SCP1 and miR-124 in neurogenesis suggest that these two pathways are intricately orchestrated during development. Indeed, REST functions as a negative regulator of miR-124 via RE1 sites in three miR-124 genomic loci (Conaco et al. 2006). The recent computational approaches also uncovered miR-124-binding sites in the 3′ untranslated regions (UTRs) of MeCP2 and CoREST, encoding two components of the REST complex (Wu and Xie 2006). Interestingly, SCP1 was found among the 174 down-regulated genes by miR-124 in HeLa cells (Lim et al. 2005) and among up-regulated genes in miR-124-depleted cortical neurons (Conaco et al. 2006). However, it remains to be tested whether SCP1 is a direct target of miR-124.
Here, we assess the developmental functions of SCP1 and miR-124. This work demonstrates the anti-neural activity of SCP1 as well as the proneural activity of a brain-enriched miRNA miR-124 in developing CNS, and identifies SCP1 as one of miR-124’s direct targets.
Results and Discussion
Constitutive SCP1 expression attenuates neurogenesis in the developing spinal cord
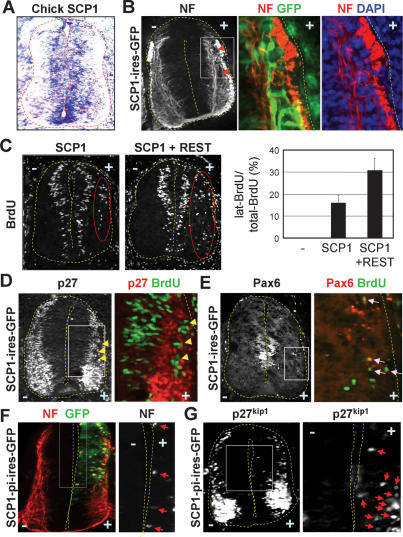
SCP1 silences neuronal genes in P19 and S2 cells (Yeo et al. 2005). To test the anti-neural function of SCP1 in the CNS, we utilized the developing chick spinal cord. Like mouse SCP1 (Yeo et al. 2005), chick SCP1 is widely expressed in multiple cell types but rapidly down-regulated in laterally located post-mitotic spinal neurons (Fig. 1A). For forced maintenance of SCP1, a vector consisting of chick β-actin promoter and the SCP1 ORF was electroporated into neuroepithelial cells along one side of the chick neural tube. Interestingly, some SCP1-misexpressing cells (i.e., GFP+ cells in Fig. 1B) settled in the lateral neuronal zone and lacked expression of neuronal markers neurofilament (NF) and TuJ (Fig. 1B; data not shown). Upon SCP1 expression, ∼16% of BrdU+-proliferating cells appeared ectopically in the lateral zone of the neural tube (Fig. 1C). These cells were not immunostained for NF (Supplementary Fig. S1A) or post-mitotic cell marker p27kip1 (Fig. 1D). SCP1 also triggered the appearance of cells still expressing progenitor markers Pax6 and Nkx6.1 in the lateral zone (Fig. 1E; Supplementary Fig. S1A). Occasionally, post-mitotic Nkx6.1+ cells were detected in the lateral zone of control side, but Nkx6.1+BrdU+ cells were seen only in the SCP1-electroporated side (Supplementary Fig. S1A, arrows). Thus, the forced maintenance of SCP1 in neuroepithelial cells interferes with the cell cycle exit of progenitors, down-regulation of progenitor genes, and the subsequent neurogenesis. This relatively subtle anti-neural effect of SCP1 is similar to that observed with REST misexpression (Paquette et al. 2000). Consistent with the cooperativeness of REST and SCP1 to silence neuronal genes in P19 cells (Yeo et al. 2005), however, coexpression of REST markedly enhanced the anti-neural phenotype of SCP1; ∼31% of BrdU+-proliferating cells were located ectopically in the lateral zone of the neural tube (Fig. 1C), which were negative for post-mitotic neuronal markers (Supplementary Fig. S1B).
Figure 1.
SCP1 as an anti-neural factor in the chick neural tube. (A) In situ hybridization on chick embryos reveals that SCP1 is widely expressed but down-regulated in the lateral neuronal zone. (B) SCP1-misexpressing cells (GFP+ cells) in the lateral zone lack the NF expression. (C) Misexpression of SCP1 leads to ectopic BrdU+-proliferating cells in the lateral zone (cells in red circle), and this is markedly enhanced by coexpression of REST. (D,E) While ectopic BrdU+ cells lack post-mitotic marker p27kip1 (arrows in D), they maintain expression of progenitor gene Pax6 (arrows in E). (F,G) Phosphatase-inactive SCP1 mutant SCP1-pi induces ectopic and precocious NF+ neuronal differentiation (arrows in F) and expression of p27kip1 (arrows in G) in the medial zone of the chick neural tube.
Expression of phosphatase-inactive SCP1 mutant SCP1-pi (Yeo et al. 2005) triggered precocious neurogenesis in the ventricular progenitor zone, as shown by more medially located NF+ and p27kip1+ post-mitotic cells (arrows in Fig. 1F,G). These data suggest that the phosphatase activity of SCP1 is required for its anti-neural activity in developing spinal cord and SCP1 antagonism in neural progenitors results in their premature neurogenesis. Thus, SCP1 is a key regulator of neurogenesis in developing CNS, and down-regulation of SCP1 is likely a prerequisite for differentiation of neural progenitors.
SCP1 is down-regulated by miR-124
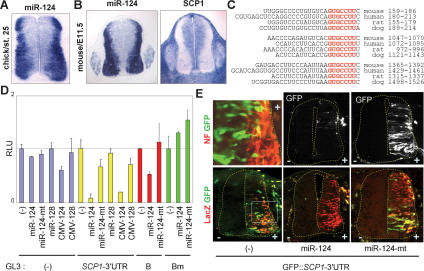
The anti-neural function of SCP1 in the developing spinal cord and its recent identification as a down-regulated transcript by miR-124 (Lim et al. 2005; Conaco et al. 2006) led us to test whether SCP1 is a direct target of miR-124. In situ hybridization with antisense miR-124 LNA oligonucleotides revealed that miR-124 expression was induced during neuronal differentiation of progenitors and maintained in a lateral neuronal domain of the developing spinal cord and dorsal root ganglion (DRG) in mouse and chick embryos but barely detected in nonneuronal cells including neural progenitors (Fig. 2A,B; Supplementary Fig. S2A,B). In contrast, SCP1 expression was largely absent in the lateral zone of the spinal cord and DRG (Fig. 2B). This complementary expression of miR-124 and SCP1 in the developing spinal cord supports the possibility that SCP1 is a target of miR-124.
Figure 2.
SCP1-3′UTR as a direct target of miR-124. (A,B) In situ detection of miR-124 in chick and mouse embryos shows that miR-124 is highly and specifically expressed in the lateral neuronal zone of the neural tube and DRG, complementary to the expression pattern of SCP1. (C) Three evolutionarily conserved miR-124 target sites are found at the 3′UTR of SCP1 mRNA. Seed match sequences are marked in red. (D) Luciferase assays with the GL3∷SCP1-3′UTR reporter in HEK293 cells. Both synthetic miR-124 RNA duplexes (miR-124) and miR-124 expression vector (CMV-124) repress luciferase expression from GL3∷SCP1-3′UTR, while the miR-124 mutant and miR-128 have no effect. GL3 vector alone is minimally affected by miRNAs. The B-segment of SCP1-3′UTR containing the second miR-124 target site is suppressed by miR-124, and mutation on the miR-124 target site in the B-segment (Bm) abolishes the repression by miR-124. (E) Electroporation of GFP∷SCP1-3′UTR directs a high level of GFP expression in the medial zone and a lower GFP expression in the NF+ lateral zone of the chick neural tube. Coelectroporation of miR-124 diminishes GFP expression from GFP∷SCP1-3′UTR, whereas miR-124-mt has no effect. CMV-LacZ was included as an electroporation indicator.
Next, we used the miR-binding site search engine Target-Scan (Lewis et al. 2003), which revealed three evolutionarily conserved, potential miR-124 sites in the 3′UTR of SCP1 mRNA (Fig. 2C). To determine whether these sites are targeted by miR-124, we generated a luciferase reporter GL3∷SCP1-3′UTR, in which luciferase coding sequences are followed by an ∼1.6-kb 3′UTR of mouse SCP1 containing all three miR-124 sites. To express miR-124, we used both synthetic miR-124 RNA duplexes and the miR-124 expression vector. The latter contains an ∼320-nt miR-124-2 genomic region in the CMV vector, which includes the ∼22-nt mature miRNA sequences and ∼125 nt of genomic sequences flanking each side of the mature miR-124 sequences (Chen et al. 2004). Both miR-124 RNA duplexes and the miR-124 expression vector suppressed luciferase expression from GL3∷SCP1-3′UTR in HEK293 cells but did not affect the vector alone (Fig. 2D). Similar results were obtained with a reporter containing IQGAP1-3′UTR, a direct miR-124 target (Lim et al. 2005; data not shown). Supporting the specificity of miR-124-mediated inhibition of SCP1-3′UTR, neither the miR-128 nor miR-124 mutant (miR-124-mt), which bears two nucleotide mutations at positions 5 and 6 (Lim et al. 2005), altered luciferase expression from GL3∷SCP1-3′UTR (Fig. 2D). Segments of SCP1-3′UTR containing either the second or the third miR-124 target site also responded to miR-124, although the fold suppression by miR-124 was smaller than that for GL3-SCP1-3′UTR full-length (3′UTR segment B in Fig. 2D; data not shown). The effect of miR-124 on SCP1-3′UTR segment B required its miR-124-binding site, as mutation of this site destroyed the responsiveness to miR-124 (Bm in Fig. 2D). Thus, miR-124 specifically inhibits SCP1 expression via directly targeting the evolutionarily conserved, multiple miR-124 sites in SCP1-3′UTR.
To further test the functional interactions between miR-124 and SCP1-3′UTR in the developing spinal cord, electroporation indicator CMV-LacZ and GFP∷SCP1-3′UTR reporter, in which the CMV promoter-driven GFP gene is fused to SCP1-3′UTR, were electroporated into the chick neural tube. GFP was highly expressed in the medial zone but significantly less in the lateral neuronal domain (Fig. 2E; Supplementary Fig. S2C), in which miR-124 is highly expressed. Coelectroporation of miR-124 RNA duplexes abolished GFP expression from GFP∷SCP1-3′UTR in the neural tube, whereas miR-124-mt had little effect (Fig. 2E). Similar expression of the coelectroporated LacZ indicator in all three samples confirmed the comparable efficiency of electroporation (Fig. 2E). These results further demonstrate that SCP1-3′UTR is directly targeted by miR-124 in the developing spinal cord. These results also support that miRNAs and their target genes are preferentially expressed in neighboring tissues (Farh et al. 2005; Stark et al. 2005).
miR-124 stimulates neuronal differentiation in the developing spinal cord
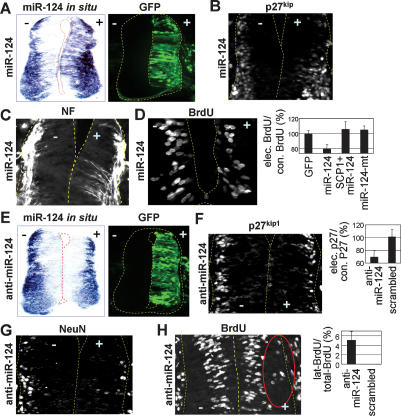
The evolutionary conservation of sequences and expression pattern for miR-124 suggests its critical role in CNS development. Our finding that miR-124 targets SCP1-3′UTR (Fig. 2) led to a proposal that miR-124 facilitates neurogenesis at least in part by blocking the anti-neural REST/SCP1 pathway. To test this idea in the developing spinal cord, we expressed miR-124 in neural progenitors of the chick neural tube. In situ hybridization confirmed overexpression of miR-124 2 d post-electroporations of miR-124 RNA duplexes (Fig. 3A). Interestingly, the p27kip1+ post-mitotic area was expanded medially (Fig. 3B). Moreover, miR-124 electroporation triggered ectopic neurogenesis in the more medial area of the neural tube, as is evident with NF staining (Fig. 3C). While the overall area occupied by BrdU+-proliferating cells did not change, the number of BrdU+ cells also decreased relative to the unelectroporated control side (Fig. 3D). In contrast, miR-124-mt RNA duplexes had no effect (Fig. 3D; data not shown). These data suggest that overexpressed miR-124 attenuates proliferation of neural progenitors and promotes their precocious neurogenesis.
Figure 3.
Proneural activity of miR-124 in the chick neural tube. (A–D) Overexpression of miR-124 in the chick spinal cord. (A) Electroporation of miR-124 RNA duplexes (with CMV-GFP as an electroporation indicator) increases miR-124 expression in the developing chick spinal cord 2 d post-electroporation as detected with in situ hybridization. (B–D) miR-124 overexpression triggers ectopic neurogenesis (p27kip1+, NF+ post-mitotic neuronal cells in B,C) in the more medial area of the neural tube, accompanied by compromised progenitor cell proliferation (D). (E–H) Inhibition of miR-124 function in the chick spinal cord. Electroporation of 2′-OMe antisense miR124 (anti-miR-124) with CMV-GFP interferes with miR-124 detection by in situ hybridization (E), decreases the expression of neuronal markers p27kip1 and NeuN in the lateral zone of the neural tube (F,G), and simultaneously triggers laterally positioned BrdU+-proliferating cells (cells in red circle in H).
To test whether SCP1 is a key target for miR-124 to trigger neurogenesis, we coelectroporated the miR-124 and SCP1 expression vector lacking SCP1-3′UTR. This prevented miR-124 from reducing progenitor proliferation, resulting in laterally positioned BrdU+ cells, like SCP1 alone (Fig. 3D; Supplementary Fig. S3). Next, we generated the miR-124-sensitive SCP1 expression vector by including the SCP1-3′UTR in the downstream region of the SCP1 coding sequences. This SCP1∷SCP1-3′UTR construct, without or with miR-124, failed to induce ectopic BrdU+ cells in the lateral spinal cord (Supplementary Fig. S3). These results suggest that SCP1 is a key target of miR-124 during developmental conversion of progenitors to neurons.
To further define the miR-124 function in neurogenesis, we inhibited miR-124 function with 2′-O-methyl (2′-OMe) antisense miR-124 oligoribonucleotides, which specifically inactivate their target miRNA through irreversible binding (Meister et al. 2004). Electroporations of 2′-OMe antisense miR-124 to the chick neural tube noticeably decreased miR-124 detection in the lateral zone (Fig. 3E). Interestingly, this resulted in reduced neuronal differentiation, as monitored by immunostaining with the post-mitotic marker p27kip1 (Fig. 3F) and neuronal marker NeuN (Fig. 3G). Moreover, inhibition of the miR-124 function triggered laterally positioned BrdU+-proliferating cells (Fig. 3H). In contrast, 2′-OMe-scrambled miR-124 had no effect (Fig. 3F,H). These results of miR-124 inhibition are similar to those of forced SCP1 expression in the neural tube (Fig. 1B–E), supporting the antagonistic regulations between SCP1 and miR-124 in the developing neural tube. Interestingly, these phenotypes were augmented by cotransfection of GFP∷SCP1-3′UTR, although GFP∷SCP1-3′UTR alone had no detectable effect (data not shown), suggesting that overexpression of SCP1-3′UTR may help sequester miR-124, as it contains miR-124 sites.
Our results in the chick neural tube show that miR-124 facilitates neurogenesis in the developing spinal cord, which is likely required for neural progenitors to acquire robust neuronal phenotypes in a timely manner. However, the overall strength of the proneural activity of overexpressed miR-124 is relatively subtle. This could be due to an already sufficient level of endogenous miR-124. Alternatively, miR-124 may require additional factors/signals to trigger robust neurogenesis, because it is unlikely to be a single determinant for neurogenesis in the developing CNS.
SCP1 antagonizes miR-124-mediated neurogenesis of P19 cells
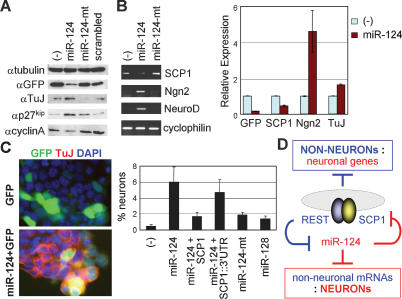
To further assess the neurogenic function of miR-124 in a more homogenous cellular system, we utilized P19 cells, which lack endogenous miR-124 but undergo neuronal differentiation upon either aggregation plus retinoic acid treatment or expression of proneural basic helix–loop–helix (bHLH) factors (Jones-Villeneuve et al. 1982; Farah et al. 2000). During neurogenesis of P19 cells, REST and SCP1 are down-regulated and miR-124 is induced (Yeo et al. 2005; Conaco et al. 2006). When examined 3 d post-transfection, miR-124 expression appeared to promote cell cycle exit of P19 cells, as shown by down-regulated proliferation marker cyclinA and simultaneously induced post-mitotic marker p27kip1 (Fig. 4A). This reduced the number of transfected cells, as evident from a lower expression of the transfection indicator GFP in P19 cells transfected with miR-124 than those with miR-124-mt or scrambled RNA duplexes (Fig. 4A,B). Intriguingly, despite the decreased number of transfected cells, miR-124 markedly induced expression of two proneural bHLH factors Ngn2 and NeuroD and neuronal marker TuJ, while reducing expression of SCP1 (Fig. 4A,B). To examine P19 cell differentiation at a cellular level, we performed immunostaining. Expression of miR-124, but not miR-124-mt or miR-128, induced expression of TuJ and triggered neurite outgrowth (Fig. 4C; Supplementary Fig. S4). In contrast, miR-124 had no effect on cell death in P19 cells (data not shown). We noted that neurogenesis triggered by miR-124 expression is relatively modest in comparison to expression of proneural bHLH factors in P19 cells (Yeo et al. 2005), similar to the effect of miR-124 in the developing spinal cord (Fig. 3). Nonetheless, these findings clearly establish a neurogenic potential of miR-124 in P19 cells.
Figure 4.
miR-124-induced neurogenesis is antagonized by SCP1 in P19 mouse embryonic cells. (A) Western analyses show that, in P19 cells, miR-124 expression induces post-mitotic cell marker p27kip1 and neuronal marker TuJ, while suppressing proliferation marker cyclinA. Notably, transfection indicator GFP expression is lower in miR-124-transfected cells. (B) RT–PCR assays reveal that expression of miR-124, but not miR-124-mt, reduces SCP1 and induces proneural bHLH factors Ngn2 and NeuroD and neuronal marker TuJ. Quantitative RT–PCR results are as shown. (C) Neuronal differentiation analyses in transfected P19 cells. miR-124 expression triggers TuJ+ neuronal differentiation, which is compromised by cotransfection of the miR-124-resistant SCP1 vector but not by the miR-124-sensitive SCP1 vector containing SCP1-3′UTR (SCP1∷ 3′UTR). Percentage of neuronal differentiation is measured by percentage of TuJ+ neurons among GFP+-transfected cells. (D) Negative feedback loop between the anti-neural REST/SCP1 and proneural miR-124 pathways contributes to a rapid and efficient transition of cellular phenotypes between neural progenitors and post-mitotic neurons (see text). Blue and red denote the regulatory pathways in nonneurons and neurons, respectively.
To test whether SCP1 suppression is critical for the proneural function of miR-124 in P19 cells, we monitored miR-124-mediated neurogenesis following cotransfection of the expression vector for SCP1-ORF or miR-124-sensitive SCP1∷SCP1-3′UTR. While constitutive expression of SCP1-ORF significantly compromised TuJ expression, inclusion of SCP1-3′UTR blocked SCP1 from suppressing the ability of miR-124 to induce TuJ expression in P19 cells (Fig. 4C). Thus, miR-124 likely induces neurogenesis in P19 cells at least in part through directly down-regulating the anti-neural factor SCP1. However, SCP1 expression did not completely abolish the neurogenic potential of miR-124, suggesting that SCP1 is not the sole target of miR124, and complete interference of the miR-124 function involves additional miR-124 targets.
Neurogenesis in developing CNS is regulated by complex molecular events. Here, we demonstrate the anti-neural function of SCP1 in the developing CNS and the importance of its timely down-regulation for proper CNS development. In particular, SCP1 antagonism in neural progenitors with the phosphatase-inactive SCP1 mutant promotes precocious neurogenesis. Although SCP1 is known to dephosphorylate Ser 5 of the C-terminal domain of RNA polymerase II (Yeo et al. 2003; Zhang et al. 2006), the identity of phosphatase substrate(s) mediating the anti-neural function of SCP1 is unclear.
Our findings indicate that miR-124 targets the anti-neural factor SCP1 during neurogenesis. Accordingly, miR-124 shows a modest but specific ability to induce neuronal gene expression in both P19 cells and the developing spinal cord. Moreover, miR-124 antagonism in the neural tube results in proliferating cells in the lateral post-mitotic zone accompanied by reduction of neuronal markers, supporting the idea that miR-124 participates actively in neurogenesis. The proneural function of miR-124 could also involve up-regulation of proneural bHLH factor Ngn2, likely through suppression of the Ngn2 repressor (Fig. 4B). As miR-124 expression is maintained throughout adulthood, miR-124 may also play additional roles in mature neurons, which could not be addressed in our study due to the nature of the transient expression of miR-124 or 2′-OMe antisense miR-124 oligonucleotides.
SCP1 is one of the key direct targets of miR-124, as the miR-124-resistant SCP1 expression construct lacking SCP1-3′UTR attenuates the neurogenic potential of miR-124 in P19 cells and the chick neural tube. These results, combined with the recent report that REST transcriptionally inhibits miR-124 genes (Conaco et al. 2006), suggest a negative feedback loop between REST/SCP1 and miR-124 for a rapid transition between neural progenitors and post-mitotic neurons (Fig. 4D). In nonneuronal cells including neural progenitors, the REST/SCP1 complex transcriptionally represses expression of miR-124 and other neuronal genes. As the REST level decreases during neurogenesis, miR-124 expression is derepressed, and subsequently, miR-124 post-transcriptionally suppresses multiple anti-neural factors including SCP1, resulting in further inhibition of the anti-neural pathway by REST/SCP1. This regulatory loop may represent a key mechanism to sense the intricate balance between proneural and anti-neural cues during development, to coordinate robust neuronal gene expression, and to confer neuronal identity in a timely manner. In Drosophila, miR-124 is also expressed exclusively in neuronal cells as they begin to differentiate (Stark et al. 2005). A single Drosophila dSCP gene is involved in neuronal gene silencing as knockdown of dSCP leads to up-regulation of neural genes (Yeo et al. 2005). Thus, the negative feedback loop between anti-neural SCP and proneural miR-124 pathways may represent an evolutionarily conserved developmental strategy. Interestingly, Drosophila has a functional ortholog of REST, Tramtrack 88 (ttk88) (Dallman et al. 2004), and Drosophila miR-124 promoter contains a consensus ttk88 response element (Conaco et al. 2006). However, analyses of the current databases revealed that 3′UTR of dSCP1 does not appear to have miR-124-binding sites (data not shown).
Overall, our studies suggest that the brain-enriched miRNAs can play key roles in coordinating vertebrate CNS development, and establish a novel evolutionarily conserved strategy to keep the balance between miRNAs and their transcriptional regulatory programs.
Materials and methods
In ovo electroporations, BrdU incorporation assays, immunostaining, and in situ hybridization
These assays were done as described (Lee and Pfaff 2003). Forty micromolar miR-124 or miR-124-mt RNA duplex, 2′-OMe antisense or 2′-OMe-scrambled miR-124 oligoribonucleotides (IDT) were injected into a Hamburger and Hamilton (HH) stage 13 chick neural tube. Locked nucleic acid (LNA)-modified miR124 oligonucleotide probe (Exiqon) was labeled with digoxigenin according to the supplier’s protocol (Roche) and used for in situ hybridization as described (Kloosterman et al. 2006). At least five embryos and 50 sections were analyzed for individual immunostaining and in situ hybridization results.
Luciferase assays
HEK293 cells, grown in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS), were transfected using SuperFect (Qiagen), harvested 3 d post-transfection, and assayed for luciferase activity with normalization for β-galactosidase activity. All transfections were independently repeated at least three times. The final 100 nM RNA duplexes were used for transfection.
RT–PCR, Western blotting, and cell differentiation assays
P19 cells, grown in αMEM supplemented with 10% FBS, were transfected with plasmids and/or RNA duplexes using Lipofectamine 2000 (Invitrogen) and analyzed for gene expression and cell differentiation 3 d post-transfection. Total RNA was extracted with Trizol (Invitrogen) and RT was performed using SuperScript III (Invitrogen). The levels of mRNA were determined using quantitative RT–PCR (Mx3000P; Stratagene).
Acknowledgments
We thank Sam Pfaff, David Moore, and Huda Zoghbi for discussions, and Dustin Baldridge for constructing miR-expression vectors. This work was supported by grants from NIH (R01 NS054941), PEW, March of Dimes Foundations, and MRDDRC P30 HD24064.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1519107
References
- Alvarez-Garcia I., Miska E.A., Miska E.A. MicroRNA functions in animal development and human disease. Development. 2005;132:4653–4662. doi: 10.1242/dev.02073. [DOI] [PubMed] [Google Scholar]
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Andres M.E., Burger C., Peral-Rubio M.J., Battaglioli E., Anderson M.E., Grimes J., Dallman J., Ballas N., Mandel G., Burger C., Peral-Rubio M.J., Battaglioli E., Anderson M.E., Grimes J., Dallman J., Ballas N., Mandel G., Peral-Rubio M.J., Battaglioli E., Anderson M.E., Grimes J., Dallman J., Ballas N., Mandel G., Battaglioli E., Anderson M.E., Grimes J., Dallman J., Ballas N., Mandel G., Anderson M.E., Grimes J., Dallman J., Ballas N., Mandel G., Grimes J., Dallman J., Ballas N., Mandel G., Dallman J., Ballas N., Mandel G., Ballas N., Mandel G., Mandel G. CoREST: A functional corepressor required for regulation of neural-specific gene expression. Proc. Natl. Acad. Sci. 1999;96:9873–9878. doi: 10.1073/pnas.96.17.9873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballas N., Battaglioli E., Atouf F., Andres M.E., Chenoweth J., Anderson M.E., Burger C., Moniwa M., Davie J.R., Bowers W.J., Battaglioli E., Atouf F., Andres M.E., Chenoweth J., Anderson M.E., Burger C., Moniwa M., Davie J.R., Bowers W.J., Atouf F., Andres M.E., Chenoweth J., Anderson M.E., Burger C., Moniwa M., Davie J.R., Bowers W.J., Andres M.E., Chenoweth J., Anderson M.E., Burger C., Moniwa M., Davie J.R., Bowers W.J., Chenoweth J., Anderson M.E., Burger C., Moniwa M., Davie J.R., Bowers W.J., Anderson M.E., Burger C., Moniwa M., Davie J.R., Bowers W.J., Burger C., Moniwa M., Davie J.R., Bowers W.J., Moniwa M., Davie J.R., Bowers W.J., Davie J.R., Bowers W.J., Bowers W.J., et al. Regulation of neuronal traits by a novel transcriptional complex. Neuron. 2001;31:353–365. doi: 10.1016/s0896-6273(01)00371-3. [DOI] [PubMed] [Google Scholar]
- Ballas N., Grunseich C., Lu D.D., Speh J.C., Mandel G., Grunseich C., Lu D.D., Speh J.C., Mandel G., Lu D.D., Speh J.C., Mandel G., Speh J.C., Mandel G., Mandel G. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell. 2005;121:645–657. doi: 10.1016/j.cell.2005.03.013. [DOI] [PubMed] [Google Scholar]
- Bartel D.P., Chen C.Z., Chen C.Z. Micromanagers of gene expression: The potentially widespread influence of metazoan microRNAs. Nat. Rev. Genet. 2004;5:396–400. doi: 10.1038/nrg1328. [DOI] [PubMed] [Google Scholar]
- Brennecke J., Stark A., Russell R.B., Cohen S.M., Stark A., Russell R.B., Cohen S.M., Russell R.B., Cohen S.M., Cohen S.M. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington J.C., Ambros V., Ambros V. Role of microRNAs in plant and animal development. Science. 2003;301:336–338. doi: 10.1126/science.1085242. [DOI] [PubMed] [Google Scholar]
- Chen C.Z., Li L., Lodish H.F., Bartel D.P., Li L., Lodish H.F., Bartel D.P., Lodish H.F., Bartel D.P., Bartel D.P. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- Conaco C., Otto S., Han J.J., Mandel G., Otto S., Han J.J., Mandel G., Han J.J., Mandel G., Mandel G. Reciprocal actions of REST and a microRNA promote neuronal identity. Proc. Natl. Acad. Sci. 2006;103:2422–2427. doi: 10.1073/pnas.0511041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallman J.E., Allopenna J., Bassett A., Travers A., Mandel G., Allopenna J., Bassett A., Travers A., Mandel G., Bassett A., Travers A., Mandel G., Travers A., Mandel G., Mandel G. A conserved role but different partners for the transcriptional corepressor CoREST in fly and mammalian nervous system formation. J. Neurosci. 2004;24:7186–7193. doi: 10.1523/JNEUROSCI.0238-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell D.K., Kaur S., Stanislaw S., Konieczka J.K., Yatskievych T.A., Antin P.B., Kaur S., Stanislaw S., Konieczka J.K., Yatskievych T.A., Antin P.B., Stanislaw S., Konieczka J.K., Yatskievych T.A., Antin P.B., Konieczka J.K., Yatskievych T.A., Antin P.B., Yatskievych T.A., Antin P.B., Antin P.B. MicroRNA expression during chick embryo development. Dev. Dyn. 2006;235:3156–3165. doi: 10.1002/dvdy.20956. [DOI] [PubMed] [Google Scholar]
- Deo M., Yu J.Y., Chung K.H., Tippens M., Turner D.L., Yu J.Y., Chung K.H., Tippens M., Turner D.L., Chung K.H., Tippens M., Turner D.L., Tippens M., Turner D.L., Turner D.L. Detection of mammalian microRNA expression by in situ hybridization with RNA oligonucleotides. Dev. Dyn. 2006;235:2538–2548. doi: 10.1002/dvdy.20847. [DOI] [PubMed] [Google Scholar]
- Doench J.G., Sharp P.A., Sharp P.A. Specificity of microRNA target selection in translational repression. Genes & Dev. 2004;18:504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farah M.H., Olson J.M., Sucic H.B., Hume R.I., Tapscott S.J., Turner D.L., Olson J.M., Sucic H.B., Hume R.I., Tapscott S.J., Turner D.L., Sucic H.B., Hume R.I., Tapscott S.J., Turner D.L., Hume R.I., Tapscott S.J., Turner D.L., Tapscott S.J., Turner D.L., Turner D.L. Generation of neurons by transient expression of neural bHLH proteins in mammalian cells. Development. 2000;127:693–702. doi: 10.1242/dev.127.4.693. [DOI] [PubMed] [Google Scholar]
- Farh K.K., Grimson A., Jan C., Lewis B.P., Johnston W.K., Lim L.P., Burge C.B., Bartel D.P., Grimson A., Jan C., Lewis B.P., Johnston W.K., Lim L.P., Burge C.B., Bartel D.P., Jan C., Lewis B.P., Johnston W.K., Lim L.P., Burge C.B., Bartel D.P., Lewis B.P., Johnston W.K., Lim L.P., Burge C.B., Bartel D.P., Johnston W.K., Lim L.P., Burge C.B., Bartel D.P., Lim L.P., Burge C.B., Bartel D.P., Burge C.B., Bartel D.P., Bartel D.P. The widespread impact of mammalian microRNAs on mRNA repression and evolution. Science. 2005;310:1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- Grun D., Wang Y.L., Langenberger D., Gunsalus K.C., Rajewsky N., Wang Y.L., Langenberger D., Gunsalus K.C., Rajewsky N., Langenberger D., Gunsalus K.C., Rajewsky N., Gunsalus K.C., Rajewsky N., Rajewsky N. microRNA target predictions across seven Drosophilaspecies and comparison to mammalian targets. PLoS Comput. Biol. 2005;1:e13. doi: 10.1371/journal.pcbi.0010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L., Hannon G.J., Hannon G.J. MicroRNAs: Small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- Huang Y., Myers S.J., Dingledine R., Myers S.J., Dingledine R., Dingledine R. Transcriptional repression by REST: Recruitment of Sin3A and histone deacetylase to neuronal genes. Nat. Neurosci. 1999;2:867–872. doi: 10.1038/13165. [DOI] [PubMed] [Google Scholar]
- Jones-Villeneuve E.M., McBurney M.W., Rogers K.A., Kalnins V.I., McBurney M.W., Rogers K.A., Kalnins V.I., Rogers K.A., Kalnins V.I., Kalnins V.I. Retinoic acid induces embryonal carcinoma cells to differentiate into neurons and glial cells. J. Cell Biol. 1982;94:253–262. doi: 10.1083/jcb.94.2.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloosterman W.P., Wienholds E., de Bruijn E., Kauppinen S., Plasterk R.H., Wienholds E., de Bruijn E., Kauppinen S., Plasterk R.H., de Bruijn E., Kauppinen S., Plasterk R.H., Kauppinen S., Plasterk R.H., Plasterk R.H. In situ detection of miRNAs in animal embryos using LNA-modified oligonucleotide probes. Nat. Methods. 2006;3:27–29. doi: 10.1038/nmeth843. [DOI] [PubMed] [Google Scholar]
- Krek A., Grun D., Poy M.N., Wolf R., Rosenberg L., Epstein E.J., MacMenamin P., da Piedade I., Gunsalus K.C., Stoffel M., Grun D., Poy M.N., Wolf R., Rosenberg L., Epstein E.J., MacMenamin P., da Piedade I., Gunsalus K.C., Stoffel M., Poy M.N., Wolf R., Rosenberg L., Epstein E.J., MacMenamin P., da Piedade I., Gunsalus K.C., Stoffel M., Wolf R., Rosenberg L., Epstein E.J., MacMenamin P., da Piedade I., Gunsalus K.C., Stoffel M., Rosenberg L., Epstein E.J., MacMenamin P., da Piedade I., Gunsalus K.C., Stoffel M., Epstein E.J., MacMenamin P., da Piedade I., Gunsalus K.C., Stoffel M., MacMenamin P., da Piedade I., Gunsalus K.C., Stoffel M., da Piedade I., Gunsalus K.C., Stoffel M., Gunsalus K.C., Stoffel M., Stoffel M., et al. Combinatorial microRNA target predictions. Nat. Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M., Rauhut R., Yalcin A., Meyer J., Lendeckel W., Tuschl T., Rauhut R., Yalcin A., Meyer J., Lendeckel W., Tuschl T., Yalcin A., Meyer J., Lendeckel W., Tuschl T., Meyer J., Lendeckel W., Tuschl T., Lendeckel W., Tuschl T., Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- Lee S.K., Pfaff S.L., Pfaff S.L. Synchronization of neurogenesis and motor neuron specification by direct coupling of bHLH and homeodomain transcription factors. Neuron. 2003;38:731–745. doi: 10.1016/s0896-6273(03)00296-4. [DOI] [PubMed] [Google Scholar]
- Lewis B.P., Shih I.H., Jones-Rhoades M.W., Bartel D.P., Burge C.B., Shih I.H., Jones-Rhoades M.W., Bartel D.P., Burge C.B., Jones-Rhoades M.W., Bartel D.P., Burge C.B., Bartel D.P., Burge C.B., Burge C.B. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- Lewis B.P., Burge C.B., Bartel D.P., Burge C.B., Bartel D.P., Bartel D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Lim L.P., Lau N.C., Garrett-Engele P., Grimson A., Schelter J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M., Lau N.C., Garrett-Engele P., Grimson A., Schelter J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M., Garrett-Engele P., Grimson A., Schelter J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M., Grimson A., Schelter J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M., Schelter J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M., Bartel D.P., Linsley P.S., Johnson J.M., Linsley P.S., Johnson J.M., Johnson J.M. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Lunyak V.V., Burgess R., Prefontaine G.G., Nelson C., Sze S.H., Chenoweth J., Schwartz P., Pevzner P.A., Glass C., Mandel G., Burgess R., Prefontaine G.G., Nelson C., Sze S.H., Chenoweth J., Schwartz P., Pevzner P.A., Glass C., Mandel G., Prefontaine G.G., Nelson C., Sze S.H., Chenoweth J., Schwartz P., Pevzner P.A., Glass C., Mandel G., Nelson C., Sze S.H., Chenoweth J., Schwartz P., Pevzner P.A., Glass C., Mandel G., Sze S.H., Chenoweth J., Schwartz P., Pevzner P.A., Glass C., Mandel G., Chenoweth J., Schwartz P., Pevzner P.A., Glass C., Mandel G., Schwartz P., Pevzner P.A., Glass C., Mandel G., Pevzner P.A., Glass C., Mandel G., Glass C., Mandel G., Mandel G., et al. Corepressor-dependent silencing of chromosomal regions encoding neuronal genes. Science. 2002;298:1747–1752. doi: 10.1126/science.1076469. [DOI] [PubMed] [Google Scholar]
- Meister G., Landthaler M., Dorsett Y., Tuschl T., Landthaler M., Dorsett Y., Tuschl T., Dorsett Y., Tuschl T., Tuschl T. Sequence-specific inhibition of microRNA- and siRNA-induced RNA silencing. RNA. 2004;10:544–550. doi: 10.1261/rna.5235104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquette A.J., Perez S.E., Anderson D.J., Perez S.E., Anderson D.J., Anderson D.J. Constitutive expression of the neuron-restrictive silencer factor (NRSF)/REST in differentiating neurons disrupts neuronal gene expression and causes axon pathfinding errors in vivo. Proc. Natl. Acad. Sci. 2000;97:12318–12323. doi: 10.1073/pnas.97.22.12318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roopra A., Sharling L., Wood I.C., Briggs T., Bachfischer U., Paquette A.J., Buckley N.J., Sharling L., Wood I.C., Briggs T., Bachfischer U., Paquette A.J., Buckley N.J., Wood I.C., Briggs T., Bachfischer U., Paquette A.J., Buckley N.J., Briggs T., Bachfischer U., Paquette A.J., Buckley N.J., Bachfischer U., Paquette A.J., Buckley N.J., Paquette A.J., Buckley N.J., Buckley N.J. Transcriptional repression by neuron-restrictive silencer factor is mediated via the Sin3–histone deacetylase complex. Mol. Cell. Biol. 2000;20:2147–2157. doi: 10.1128/mcb.20.6.2147-2157.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark A., Brennecke J., Bushati N., Russell R.B., Cohen S.M., Brennecke J., Bushati N., Russell R.B., Cohen S.M., Bushati N., Russell R.B., Cohen S.M., Russell R.B., Cohen S.M., Cohen S.M. Animal MicroRNAs confer robustness to gene expression and have a significant impact on 3′UTR evolution. Cell. 2005;123:1133–1146. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- Wu J., Xie X., Xie X. Comparative sequence analysis reveals an intricate network among REST, CREB and miRNA in mediating neuronal gene expression. Genome Biol. 2006;7:R85. doi: 10.1186/gb-2006-7-9-r85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo M., Lin P.S., Dahmus M.E., Gill G.N., Lin P.S., Dahmus M.E., Gill G.N., Dahmus M.E., Gill G.N., Gill G.N. A novel RNA polymerase II C-terminal domain phosphatase that preferentially dephosphorylates serine 5. J. Biol. Chem. 2003;278:26078–26085. doi: 10.1074/jbc.M301791200. [DOI] [PubMed] [Google Scholar]
- Yeo M., Lee S.K., Lee B., Ruiz E.C., Pfaff S.L., Gill G.N., Lee S.K., Lee B., Ruiz E.C., Pfaff S.L., Gill G.N., Lee B., Ruiz E.C., Pfaff S.L., Gill G.N., Ruiz E.C., Pfaff S.L., Gill G.N., Pfaff S.L., Gill G.N., Gill G.N. Small CTD phosphatases function in silencing neuronal gene expression. Science. 2005;307:596–600. doi: 10.1126/science.1100801. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Kim Y., Genoud N., Gao J., Kelly J.W., Pfaff S.L., Gill G.N., Dixon J.E., Noel J.P., Kim Y., Genoud N., Gao J., Kelly J.W., Pfaff S.L., Gill G.N., Dixon J.E., Noel J.P., Genoud N., Gao J., Kelly J.W., Pfaff S.L., Gill G.N., Dixon J.E., Noel J.P., Gao J., Kelly J.W., Pfaff S.L., Gill G.N., Dixon J.E., Noel J.P., Kelly J.W., Pfaff S.L., Gill G.N., Dixon J.E., Noel J.P., Pfaff S.L., Gill G.N., Dixon J.E., Noel J.P., Gill G.N., Dixon J.E., Noel J.P., Dixon J.E., Noel J.P., Noel J.P. Determinants for dephosphorylation of the RNA polymerase II C-terminal domain by Scp1. Mol. Cell. 2006;24:759–770. doi: 10.1016/j.molcel.2006.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]