Abstract
The coactivator PGC-1α mediates key responses of skeletal muscle to motor nerve activity. We show here that neuregulin-stimulated phosphorylation of PGC-1α and GA-binding protein (GABP) allows recruitment of PGC-1α to the GABP complex and enhances transcription of a broad neuromuscular junction gene program. Since a subset of genes controlled by PGC-1α and GABP is dysregulated in Duchenne muscular dystrophy (DMD), we examined the effects of transgenic PGC-1α in muscle of mdx mice. These animals show improvement in parameters characteristic of DMD, including muscle histology, running performance, and plasma creatine kinase levels. Thus, control of PGC-1α levels in skeletal muscle could represent a novel avenue to prevent or treat DMD.
Keywords: PGC-1, neuromuscular junction, GA-binding protein, Duchenne muscular dystrophy, transcriptional regulation
Peroxisome proliferator-activated receptor γ coactivator 1α (PGC-1α) is a transcriptional coactivator of nuclear receptors and other transcription factors that regulates adaptive mitochondrial function in several other tissues including heart, brain, liver, and skeletal muscle (for review, see Handschin and Spiegelman 2006). In addition to the induction of mitochondrial biogenesis per se, PGC-1α often controls biological programs linked to the more oxidative state for a particular tissue. The PGC-1 coactivators appear to play a particularly important role in the biology of skeletal muscle. PGC-1α induces mitochondrial biogenesis when expressed in cultured muscle cells (Wu et al. 1999) and PGC-1α is expressed at highest levels in muscle beds rich in the oxidative type I and type IIa muscle fibers, such as soleus (Lin et al. 2002b). Furthermore, PGC-1α expression is induced in most muscle beds by exercise, both chronic and acute, in rodents and humans (e.g., Russell et al. 2003). PGC-1α is a key mediator of the effects of motor nerve on skeletal muscle, suggesting the possibility that it might mediate the anti-atrophic effects that nerve activity has on muscle mass. Denervation and/or disuse cause severe atrophy of skeletal muscle in both humans and rodents (Kandarian and Jackman 2006). When PGC-1α levels are maintained transgenically, denervation causes much less muscle-mass loss and maintains muscle fiber volume (Sandri et al. 2006).
The size and morphology of neuromuscular junctions (NMJ) are key regulators of many facets of skeletal muscle function. Genes encoding proteins involved in NMJ formation are thought to be controlled, at least in part, by the transcription factor GA-binding protein (GABP, alternatively called nuclear respiratory factor 2, NRF-2) (Kummer et al. 2006). Transcriptionally active GABP is a heterotetramer of the DNA-binding subunit GABPA and the transactivating subunit GABPB. PGC-1α has been shown to increase the transcript levels of GABPA and the transcriptional activity of GABP, though this original work was done in the context of mitochondrial gene expression (Mootha et al. 2004). Two lines of evidence suggested that PGC-1α might be a strong regulator of the NMJ program in a broad sense. First, PGC-1α is known to activate mitochondrial biogenesis and muscle fiber-type switching (Lin et al. 2002b), both regulated physiologically by exercise and motor-nerve activity, as is NMJ remodeling (Wilson and Deschenes 2005). Secondly, PGC-1α has been shown to increase GABP activity in transient transfections of the utrophin promoter in cultured muscle cells and in muscle in vivo (Angus et al. 2005). Whether this is through elevation of GABPA expression by PGC-1α or via physical binding and coactivation was not demonstrated. These data raise several key questions. First, is PGC-1α a dominant regulator of the broad program of NMJ formation in chronic gain and loss-of-function experiments in vivo? Second, does PGC-1α actually bind to and coactivate GABP on the NMJ genes? Finally, how can the PGC-1α/GABP complex be responsible for initiating both the mitochondrial biogenesis and NMJ gene programs in muscle fibers, when these programs are not linked in synaptic and nonsynaptic nuclei in many physiological circumstances (Ruegg 2005).
Duchenne muscular dystrophy (DMD) arises from a frameshift mutation in the X-linked gene dystrophin and leads to rapid degeneration of heart and skeletal muscle, causing disability and death by adolescence or young adulthood (Bogdanovich et al. 2004). No therapy described to date can effectively slow or halt muscle degeneration in these patients (Kapsa et al. 2003). Interestingly, several gene programs that are linked to PGC-1α expression and function are dysregulated in DMD. These include reduced capacity for mitochondrial oxidative phosphorylation (Kuznetsov et al. 1998; Timmons et al. 2005) and radical oxygen species (ROS) detoxification (Disatnik et al. 1998), abnormalities in calcium signaling (Chakkalakal et al. 2006), and irregularities in NMJ morphology in muscle damage and regeneration (Lyons and Slater 1991).
In the current study, we show that PGC-1α stimulates a broad and powerful program of NMJ-linked gene expression both in myotubes in culture and in vivo. Moreover, PGC-1α levels correlate with the number of acetylcholine receptor (AChR) clusters in myotubes and isolated single muscle fibers from transgenic animals. We show unambiguously that moderately elevated levels of PGC-1α in vivo ameliorate a murine model of Duchenne muscular dystrophy (mdx) at the biochemical, histological, and functional levels. These data support a new therapeutic approach to the muscular dystrophies through PGC-1α.
Results
PGC-1α regulates NMJ gene transcription in vitro and in vivo
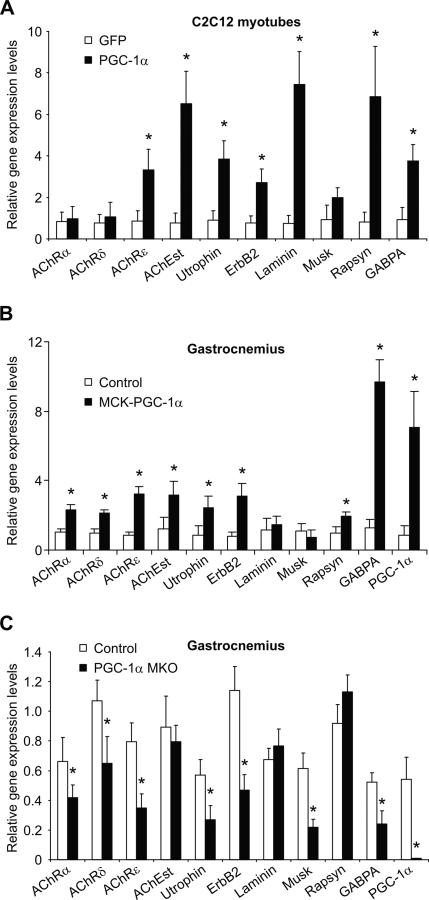
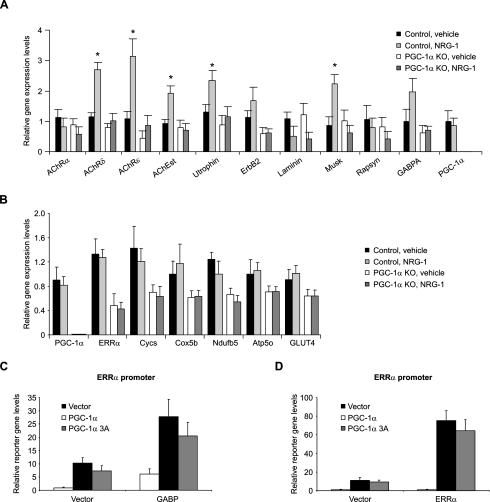
To investigate whether PGC-1α can drive expression of a broad NMJ gene program, we assessed the ability of adenoviral-expressed PGC-1α to induce expression of NMJ genes in C2C12 myotubes. After infection for 48 h with adenoviral vectors encoding PGC-1α, elevation of mRNA-encoding AChRε, AChEst, utrophin, ErbB2, laminin β2 (laminin), rapsyn, and GABPA mRNA levels were observed (Fig. 1A). The expression of these genes, all of which are related to NMJ formation and function, is increased in the range of three- to eightfold compared with control cells infected with a GFP-expressing adenovirus.
Figure 1.
PGC-1α regulates NMJ gene expression. (A) C2C12 myotubes were infected with adenoviral vectors encoding GFP and PGC-1α, respectively, on day 3 after differentiation. RNA was harvested 48 h later and relative gene expression levels were measured by real-time PCR. (*) p < 0.05 between GFP- and PGC-1α-infected cells. (B) Gastrocnemius muscle was isolated from wild-type and PGC-1α muscle-specific transgenic animals (MCK-PGC-1α). After RNA isolation, relative gene expression was analyzed by real-time PCR. (*) p < 0.05 between control and transgenic animals. (C) Control and PGC-1α muscle-specific knockout animals (PGC-1α MKO) were sacrificed, RNA was isolated from gastrocnemius, and relative gene expression levels were measured by real-time PCR. (*) p < 0.05 between control and muscle-specific knockout animals. The bars depict average gene expression normalized to 18S rRNA levels, with the error bars representing standard deviations.
The muscle-specific PGC-1α transgenic mouse represents a model of chronically elevated PGC-1α expression (Lin et al. 2002b). Importantly, this mouse model has moderate elevation of PGC-1α in most muscle beds to the level of this protein observed in muscle beds rich in oxidative fibers such as soleus. In gastrocnemius of these animals, increased transcript levels of AChRα, AChRδ, AChRε, AChEst, utrophin, ErbB2, rapsyn, and GABPA are observed compared with wild-type skeletal muscle (Fig. 1B).
PGC-1α whole-body knockout mice have a complex phenotype characterized by massive lesions in the brain and behavioral abnormalities (Lin et al. 2004; Leone et al. 2005; St-Pierre et al. 2006). In particular, these mice are hyperactive and show Huntington-like behavior. Unfortunately, many of the phenotypes resulting from ablation of PGC-1α in peripheral tissues in these mice are thus confounded by the hyperactivity. The validity of tissue-specific knockout of PGC-1α has been proven in liver-specific PGC-1α knockout mice, which have clear and expected changes in liver function (Handschin et al. 2005). Accordingly, to selectively study the post-synaptic side of NMJs in skeletal muscle and to avoid the confounding effects of genetic ablation of PGC-1α in the central nervous system, we generated a skeletal muscle-specific PGC-1α knockout animal by crossing mice with a floxed PGC-1α allele with animals that transgenically express cre recombinase under the control of the myogenin promoter and MEF2C enhancer. Similar to total knockouts, mitochondrial gene expression and exercise capacity is reduced in these mice (data not shown). Importantly, muscle-specific knockout mice have reduced expression of myosin heavy chains I and IIa and show no signs of hyperactivity, in contrast to whole body PGC-1α knockout animals (data not shown). In gastrocnemius, AChRα, AChRδ, AChRε, utrophin, ErbB2, Musk, and GABPA expression levels are reduced between 30% and 70% in the muscle-specific knockout of PGC-1α as compared with control animals (Fig. 1C). Thus, PGC-1α is required for normal expression of the NMJ gene program. Together, elevation and ablation of PGC-1α expression in skeletal muscle in vivo both have marked effects on the transcription of NMJ genes. NMJ genes are regulated slightly differently in this experimental in vitro system and in vivo. Importantly, the program of NMJ genes is consistently regulated in C2C12 cells, MCK-PGC-1α, and MKO mice (seven of 10 genes studied in C2C12, eight of 10 genes in MCK-PGC-1α, and eight of 10 genes in MKOs).
PGC-1α increases AChR clusters in myotubes
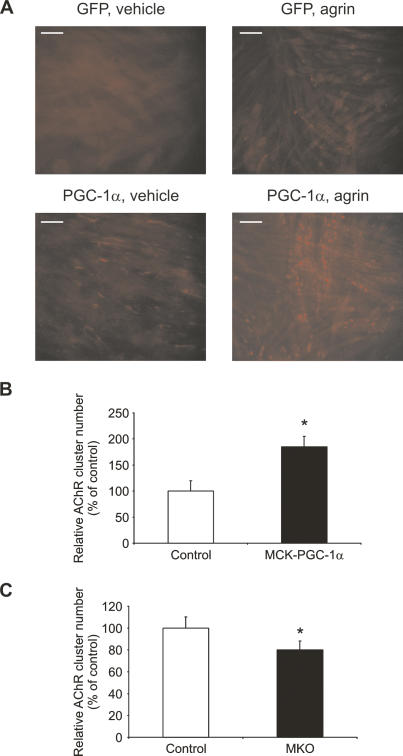
We next studied the effect of PGC-1α on AChR clustering in C2C12 myotubes induced by overnight treatment with 10 ng/mL of a recombinant, C-terminal fragment of rat agrin. C2C12 myoblasts and myotubes have very low levels of endogenous PGC-1α. Untreated, GFP-infected myotubes show only sporadic clustering of AChR on the cell membrane (Fig. 2A, top left panel). Agrin treatment of GFP-infected myotubes greatly increases the number of AChR clusters (Fig. 2A, top right panel). Surprisingly, forced expression of PGC-1α using adenoviral vectors results in a marked increase in AChR clusters, even in the absence of agrin (Fig. 2A, bottom left panel). Upon treatment with agrin, a further increase in AChR clusters was observed in PGC-1α-infected C2C12 myotubes (Fig. 2A, bottom right panel). In vehicle-treated myotubes, the clusters appear more dispersed as compared with the clusters on agrin-treated cells. This might be due to the PGC-1α-mediated increase in AChRs at the cell membrane, which are able to bind α-bungarotoxin (α-BTX). Adenoviral-encoded PGC-1α does not change endogenous agrin transcript levels in C2C12 cells (Supplementary Fig. S1). Importantly, these dispersed AChR clusters are focused by treatment with exogenous agrin. These results demonstrate that PGC-1α promotes an increase of AChR clusters on the cell membrane (see Supplementary Fig. S2 for quantification).
Figure 2.
AChR clustering induced by PGC-1α. (A) C2C12 myotubes were infected with adenoviral GFP and PGC-1α, respectively, for 48 h. Sixteen hours before harvesting, the myotubes were treated with vehicle (PBS) or 10 ng/mL recombinant agrin. Subsequently, cells were fixed and incubated with rhodamine-labeled α-BTX, and AChR clusters were visualized by fluorescence microscopy. (B,C) The average number of NMJ counted in 20 fields per genotype was normalized to the number of nuclei in these fields, and relative acetylcholine cluster numbers were expressed as percentage of control mice. (*) p < 0.05 between muscle fibers from MCK-PGC-1α and controls as well as fibers from MKO and controls, respectively.
To investigate whether AChR cluster numbers are affected by the relative levels of PGC-1α in muscle fibers of gain- and loss-of-function animals, we isolated single muscle fibers from control, MCK-PGC-1α muscle-specific transgenic mice, and muscle-specific PGC-1α knockout animals. AChR clusters were stained with fluorescent α-BTX and their relative number normalized by the muscle fiber area as assessed by the numbers of nuclei stained with DAPI (4′,6-diamidino-2-phenylindole). Twenty different randomly chosen fields per animal were analyzed in blinded fashion and the average reported. Single muscle fibers from MCK-PGC-1α had a higher number of AChR clusters per muscle fiber area than control animals (Fig. 2B). In contrast, fewer AChR clusters were counted in PGC-1α muscle-specific knockout animals in comparison to control littermates (Fig. 2C). The increase in AChR clusters in the fibers of PGC-1α muscle-specific transgenic mice and the decrease in the fibers of muscle-specific PGC-1α knockout animals were both significant, with an increase of 85% in MCK-PGC-1α fibers and a decrease of 19% in MKO fibers (Fig. 2B,C). Representative images and additional quantification are shown in Supplementary Figure S3. Thus, PGC-1α drives the formation of AChR clusters when expressed ectopically in immortalized cell lines that are differentiated into myotubes. Moreover, the number of aneural AChR clusters in isolated single muscle fibers from MCK-PGC-1α and MKO animals correlate with the relative expression levels of PGC-1α.
PGC-1α binding to and coactivation of GABP depends on specific protein phosphorylation events
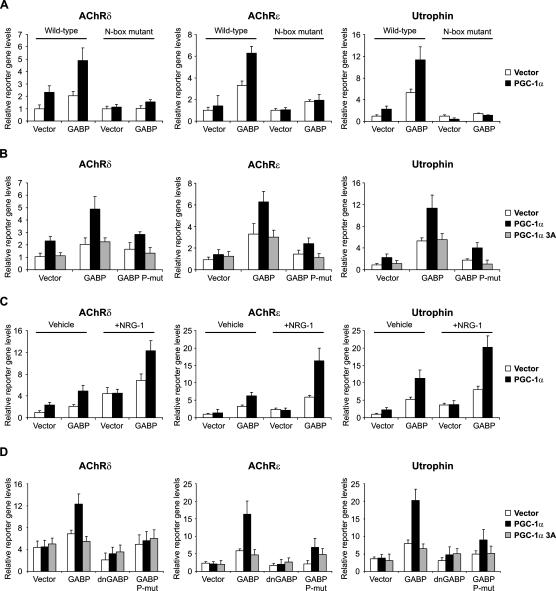
PGC-1α in skeletal muscle has been shown to induce GABPA mRNA levels and increase GABP activity in the context of transcriptional regulation of mitochondrial genes, but direct binding and coactivation was not determined (Mootha et al. 2004). Similarly, PGC-1α and GABP increase reporter gene expression driven by the utrophin promoter construct, although again, direct binding of PGC-1α to GABP could not be shown (Angus et al. 2005). GABP represents a likely candidate transcription factor recruiting PGC-1α to NMJ promoters, but whether and how these events occur is unclear. In reporter gene assays using promoter fragments from AChRδ, AChRε, and utrophin, PGC-1α stimulated GABP-mediated transcription of all three promoters (Fig. 3A). Transcriptional activity of GABP, PGC-1α, and the combination of these two proteins is completely dependent on the integrity of the N-box present in all three promoters (Fig. 3A). These results show that PGC-1α induces transcription of these NMJ genes via the N-box, the DNA-binding site for GABP. In skeletal muscle, signaling cascades initiated by trophic factors such as agrin or neuregulin-1 (NRG-1) result in phosphorylation of GABP (Bezakova and Ruegg 2003; Sunesen and Changeux 2003). Interestingly, mitogen-activated protein kinase (MAPK) has been found to phosphorylate both GABP (Flory et al. 1996) as well as PGC-1α (Puigserver et al. 2001). These phosphorylation events are critical in boosting the transcriptional activity of GABP (Sunesen et al. 2003). We therefore mutated the known p38 MAPK phosphorylation sites on GABPA (Thr 280) and GABPB1 (Ser 170 and Thr 180) (Fromm and Burden 2001) and used these phosphorylation mutants along with the PGC-1α allele with mutated MAPK phosphorylation sites (PGC-1α 3A, Thr 262, Ser 265, and Thr 298) (Puigserver et al. 2001) in reporter gene assays with the AChRδ, AChRε, and utrophin promoters. Mutation of the three phosphorylation sites on PGC-1α resulted in complete ablation of PGC-1α-mediated coactivation of GABP on all three promoters (Fig. 3B). Similarly, preventing the phosphorylation of GABPA and GABPB1 abolished transcriptional induction of the AChRδ, AChRε, and utrophin promoters by GABP and PGC-1α (Fig. 3B). Using the phosphorylation mutants for GABPA and GABPB1 individually resulted in a partially reduced transcriptional activity of the GABP/PGC-1α complex (Supplementary Fig. S4). These data underline the previously reported importance of the phosphorylation events on GABP (Sunesen et al. 2003) and furthermore show that phosphorylation of PGC-1α is equally crucial for promoting expression of these NMJ genes.
Figure 3.
PGC-1α and GABP activate NMJ gene promoters. (A) C2C12 myoblasts were transfected with wild-type and N-box mutated promoter constructs, respectively, together with expression plasmids for PGC-1α, GABPA, and GABPB1 (the combination of GABPA and GABPB1 is referred to as GABP). The cells were subsequently differentiated for 2 d and harvested, and luciferase activity was measured. (B) C2C12 myoblasts were transfected with NMJ promoter constructs together with expression plasmids for PGC-1α, GABP, PGC-1α 3A, and GABPA as well as GABPB1 alleles (collectively called GABP P-mut) with mutated phosphorylation sites. The cells were subsequently differentiated for 2 d and harvested, and luciferase activity was measured. (C) C2C12 myoblasts were transfected with NMJ promoter constructs together with expression plasmids for PGC-1α and GABP. The cells were subsequently differentiated for 2 d. Simultaneously, cells were treated with vehicle (PBS) 5 nM recombinant NRG-1. Cells were harvested and luciferase activity was measured. (D) C2C12 myoblasts were transfected with NMJ promoter constructs together with expression plasmids for PGC-1α, GABP, PGC-1α 3A, and GABPA as well as GABPB1 alleles (collectively called GABP P-mut) with mutated phosphorylation sites. The cells were subsequently differentiated for 2 d. Simultaneously, cells were treated with vehicle (PBS) 5 nM recombinant NRG-1. Cells were harvested and luciferase activity was measured. The bars depict average luciferase expression normalized to β-galactosidase activity, with the error bars representing standard deviations.
NRG-1 action requires phosphorylation of PGC-1α and GABP
NRG-1 promotes phosphorylation of GABP in skeletal muscle and induces the expression of NMJ genes (Kummer et al. 2006). In reporter gene assays using the AChRδ, AChRε, and utrophin promoters, NRG-1 dramatically increases the ability of GABP and PGC-1α to promote reporter gene expression (Fig. 3C). Finally, the NRG-1-mediated boost in activation of these three promoters by PGC-1α/GABP is markedly reduced when using dominant-negative GABP. Dominant-negative GABPA lacking the C-terminal dimerization domain and dominant-negative GABPB1 lacking the C-terminal transcriptional activation signal and the homodimerization domain have been described (see Sunesen et al. 2003, and references therein). We used those in combination with the GABP and PGC-1α phosphorylation mutants to test the ability of these mutants to block NRG-1 activity on the NMJ promoters. We found that functional PGC-1α and GABP are essential for proper regulation of reporter gene transcription (Fig. 3D). Induction of endogenous NMJ genes by transfected PGC-1α in C2C12 cells is significantly blunted by addition of expression vectors for dominant-negative GABP (Supplementary Fig. S5). Together, these experiments show that PGC-1α and GABP functionally interact and drive the expression of several NMJ genes. Moreover, activation of the AChRδ, AChRε, and utrophin promoters by NRG-1 requires phosphorylation of both GABP and PGC-1α.
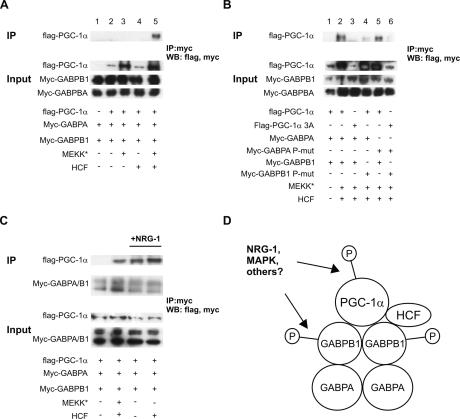
To date, no direct protein interaction between GABP and PGC-1α has been demonstrated (Scarpulla 2002; Baar 2004; Angus et al. 2005; our unpublished observations). However, our results highlighting the importance of the phosphorylation of both GABP and PGC-1α for NMJ gene expression suggested that these post-translational modifications could be important for stabilizing their physical interaction as well. Moreover, host cell factor (HCF) was found to bind to both GABPB (Vogel and Kristie 2000) and to PGC-1α (Lin et al. 2002a), and thus, could potentially serve as a linker between GABP and PGC-1α. The importance of the phosphorylation and binding of HCF in the formation of a GABP–PGC-1α complex were tested in coimmunoprecipitation assays in BOSC cells. Myc-tagged GABPA, myc-tagged GABPB1, and Flag-tagged PGC-1α were cotransfected, cell extracts were immunoprecipitated with anti-myc antibody, and subsequently blotted with anti-myc and anti-Flag antibodies. In this context, Flag-tagged PGC-1α was not associated with myc-tagged GABP (Fig. 4A, lane 2). Next, phosphorylation of PGC-1α, GABPA, and GABPB1 was induced by cotransfection of constitutively active mitogen-activated protein kinase kinase kinase (MEKK*). Transfection of MEKK* resulted in increased PGC-1α protein levels as published (Fig. 4A, lanes 3,5) (Puigserver et al. 2001). No physical interaction between PGC-1α and GABP was observed with cotransfected MEKK* (Fig. 4A, lane 3). Similarly, addition of HCF expression plasmid yielded no coimmunoprecipitation of PGC-1α with GABP (Fig. 4A, lane 4). However, cotransfection of both constitutively active MEKK* and HCF strongly promoted a physical interaction, as evidenced by detection of Flag-PGC-1α after anti-myc immunoprecipitation (Fig. 4A, lane 5).
Figure 4.
Physical interaction between PGC-1α and GABP requires phosphorylation and HCF. (A) BOSC cells were transfected with expression plasmids for Flag-tagged PGC-1α, myc-tagged GABPA and GABPB1, constitutively active MEKK (MEKK*), and HCF as indicated. Cells were harvested 48 h after transfection, 1 mg of total protein from the cell lysates was incubated with anti-myc beads, and the bound proteins as well as input (10 μg protein) were blotted with anti-Flag and anti-myc antibodies. (B) BOSC cells were transfected with expression plasmids for Flag-tagged PGC-1α, myc-tagged GABPA and GABPB1, the phosphorylation-deficient alleles Flag-PGC-1α 3A, myc-GABPA P-mut, and myc-GABPB1 P-mut, respectively, constitutively active MEKK (MEKK*), and HCF as indicated. Cells were harvested 48 h after transfection, 1 mg of total protein from the cell lysates was incubated with anti-myc beads, and the bound proteins as well as input (10 μg protein) were blotted with anti-Flag and anti-myc antibodies. (C) C2C12 myotubes were transfected with expression plasmids for Flag-tagged PGC-1α, myc-tagged GABPA and GABPB1, constitutively active MEKK (MEKK*), and HCF as indicated. The cells were differentiated for 2 d and simultaneously treated with 5 nM recombinant NRG-1. Cells were harvested, 1 mg of total protein from the cell lysates was incubated with anti-myc beads, and the bound proteins as well as input (10 μg protein) were blotted with anti-Flag and anti-myc antibodies. (D) GABPA and GABPB1 bind as heterotetramers and upon phosphorylation of GABPB1 recruit phosphorylated PGC-1α and HCF. These phosphorylation events are induced by NRG-1 signaling or constitutive MAPK activation.
Having established the importance of phosphorylation and presence of HCF for physical interaction between PGC-1α and GABP, we further tested whether phosphorylation of GABPA, GABPB1, or PGC-1α is important for the interaction. Substitution of wild-type PGC-1α with the PGC-1α 3A allele completely abolished the interaction between PGC-1α and GABP (Fig. 4B, lane 2 vs. lane 3). Similarly, mutation of the phosphorylation sites of GABPB1 resulted in greatly diminished coimmunoprecipitation of PGC-1α and GABP (Fig. 4B, lane 4). In contrast, phosphorylation of GABPA is not important for this interaction (Fig. 4B, lane 5). As predicted, cotransfection of phosphorylation mutants for GABPA, GABPB1, and PGC-1α did not result in any interaction (Fig. 4B, lane 6). In all of these experiments, we used constitutively active MEKK* to induce phosphorylation of GABP and PGC-1α. To see whether the interaction between PGC-1α and GABP can be recapitulated in a more physiological context, coimmunoprecipitation experiments were performed in C2C12 myotubes treated with NRG-1. Interestingly, NRG-1 promoted coimmunoprecipitation of PGC-1α with GABP even in the absence of exogenous HCF (Fig. 4C, lane 3 with NRG-1 vs. lane 1 without NRG-1 treatment). However, addition of HCF further stimulated the interaction (Fig. 4C, lane 4). Our data demonstrate that PGC-1α and GABP are found in a complex following phosphorylation of both PGC-1α and GABPB1, and that HCF is an important part of the protein complex (Fig. 4D). Phosphorylation of the GABPA subunit has been reported to be absolutely indispensable for transcriptional activity and changes in intranuclear mobility of the GABP complex (Sunesen et al. 2003). In contrast, the function of the GABPB phosphorylation remained unclear (Sunesen et al. 2003). Our findings suggest that GABPB1 phosphorylation is needed for the recruitment of PGC-1α, perhaps via the interaction with HCF that exclusively binds to GABPB and not GABPA (Vogel and Kristie 2000). The phosphorylation of PGC-1α and GABP could furthermore control the specificity of the coactivation and direct the protein complex to NMJ gene promoters. Thus, in primary myotubes derived from wild-type animals, NRG-1 significantly induced expression of AChRδ, AChRε, AChEst, utrophin, and Musk (Fig. 5A). In marked contrast, this induction was completely lost in PGC-1α knockout myotubes. In contrast to NMJ genes, none of the PGC-1α target genes involved in metabolic pathways is regulated by NRG-1 in wild-type and PGC-1α knockout primary myotubes, respectively (Fig. 5B). These genes have previously been shown to be under transcriptional control of GABP and ERRα coactivated by PGC-1α (Mootha et al. 2004). Finally, the phosphorylation-deficient PGC-1α 3A protein has no transcriptional activity on NMJ promoters, and this protein is unable to interact with GABP in this context (Fig. 5C). Surprisingly, PGC-1α 3A is as efficient in increasing GABP activity on the ERRα promoter as wild-type PGC-1α (Fig. 5C). Wild-type PGC-1α and PGC-1α 3A both coactivate ERRα on the ERRα promoter to the same extent (Fig. 5D). In addition, wild-type PGC-1α and the PGC-1α 3A protein were also equally potent in coactivating the nuclear respiratory factor 1 in reporter gene assays (Supplementary Fig. S6). These results clearly show that increase of mitochondrial genes by coactivation of GABP by PGC-1α is different from the induction of NMJ genes, in that it requires phosphorylation of PGC-1α and GABP. Moreover, phosphorylation stimulated by NRG-1 only elevates NMJ genes, but not mitochondrial or other PGC-1α target genes, in skeletal muscle cells. Thus, NRG-1 directs modifications of GABP and PGC-1α and induces a very specific subset of GABP/PGC-1α target genes in skeletal muscle fibers.
Figure 5.
Specificity of NRG-1-induced NMJ gene expression via PGC-1α. (A,B) Primary muscle cells were isolated from control and PGC-1α knockout animals. After differentiation into myotubes, cells were treated with vehicle (PBS) or 5 nM recombinant NRG-1 for 48 h. RNA was isolated from harvested cells and relative gene expression levels of NMJ genes (A) and other PGC-1α target genes (B) were determined by real-time PCR. (*) p < 0.05 between vehicle and NRG-1 treated cells. The bars depict average gene expression normalized to 18S rRNA levels, with the error bars representing standard deviations. (C,D) C2C12 myoblasts were transfected with a reporter plasmid containing the ERRα promoter and expression plasmids for PGC-1α, GABPA and GABPB1 (called GABP), ERRα, and the phosphorylation-deficient PGC-1α 3A. The cells were subsequently differentiated for 2 d and harvested, and luciferase activity controlled by PGC-1α and GABP (C) and PGC-1α and ERRα (D) was measured. The bars depict average luciferase expression normalized to β-galactosidase activity, with the error bars representing standard deviations.
PGC-1α ameliorates muscular dystrophy in sedentary DMD (mdx) mice
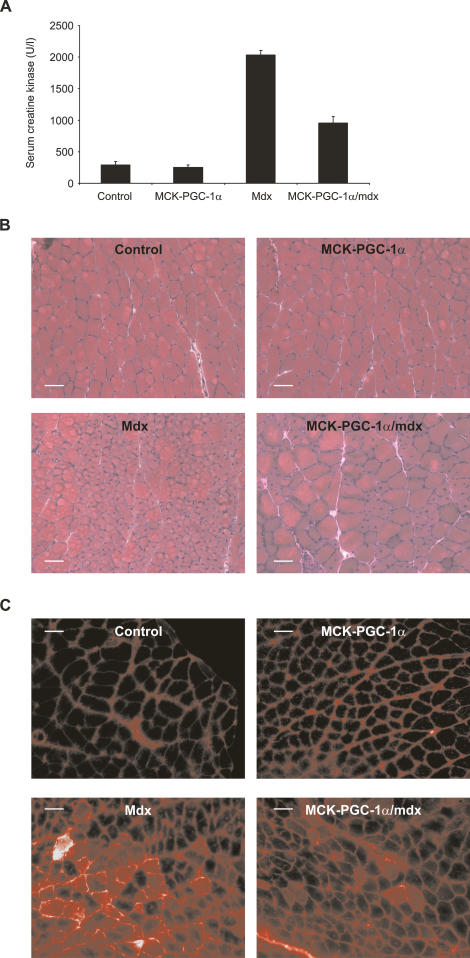
Many genes controlled by PGC-1α are dysregulated in DMD (see Discussion section for an overview). We therefore examined the effect of transgenic expression of PGC-1α on the disease phenotype of mdx mice by interbreeding with MCK-PGC-1α muscle-specific transgenic animals. The main pathology in mdx mice is detected at the age of 4–6 wk. We thus analyzed our animals at 5 wk of age. Introducing the PGC-1α transgene into the mdx background reduced serum creatine kinase levels by ∼55% (Fig. 6A). As compared with control and MCK-PGC-1α transgenic mice, haematoxylin and eosin (H&E) staining of cross-sections of mdx gastrocnemius muscles show a large proportion of muscle fibers with centrally located nuclei (Fig. 6B). Muscle fibers with centrally located nuclei are a hallmark of regenerating muscle and their relative number is proportional to muscle damage in mdx mice (e.g., for review, see Briguet et al. 2004). In MCK-PGC-1α/mdx mice, the relative numbers of muscle fibers with centrally located nuclei are reduced by 44% as compared with mdx animals (Fig. 6B; Supplementary Fig. S7). Intraperitoneal injection of Evans blue is commonly used to assess skeletal muscle fiber lesions, since this dye will only penetrate damaged fibers. As expected, no damaged fibers were found in gastrocnemius of control or MCK-PGC-1α transgenic mice (Fig. 6C). A substantial proportion of muscle fibers in mdx mice is damaged, as indicated by penetration of Evans blue (Fig. 6C). In contrast, expression of the PGC-1α transgene in skeletal muscle of mdx mice reduces the fraction of damaged muscles by ∼55% (Fig. 6C; Supplementary Fig. S6). These results clearly demonstrate that elevation of PGC-1α levels in skeletal muscle has a beneficial effect on the muscular dystrophy in mdx mice through a reduction in muscle fiber damage. It is conceivable that this improvement is at least in part mediated by increased utrophin levels in MCK-PGC-1α/mdx mice compared with mdx animals (Supplementary Fig. S8).
Figure 6.
PGC-1α ameliorates muscle dystrophy in sedentary mdx mice. (A) Five-week-old control, PGC-1α muscle-specific transgenic (MCK-PGC-1α), mdx, and PGC-1α muscle-specific transgenic animals in the mdx background (MCK-PGC-1α/mdx) were sacrificed and serum creatine kinase activity was determined. The bars depict the average serum creatine kinase levels, and error bars represent standard deviations. (B) H&E-stained histology of gastrocnemius from control, PGC-1α muscle-specific transgenic (MCK-PGC-1α), mdx, and PGC-1α muscle-specific transgenic animals in the mdx background (MCK-PGC-1α/mdx). (C) Five-week-old control, PGC-1α muscle-specific transgenic (MCK-PGC-1α), mdx, and PGC-1α muscle-specific transgenic animals in the mdx background (MCK-PGC-1α/mdx) were injected i.p. with 1% v/w Evans blue solution and sacrificed 16 h later. Dye incorporation into muscle fibers was visualized by fluorescence microscopy.
PGC-1α improves muscular dystrophy in physically challenged mdx mice
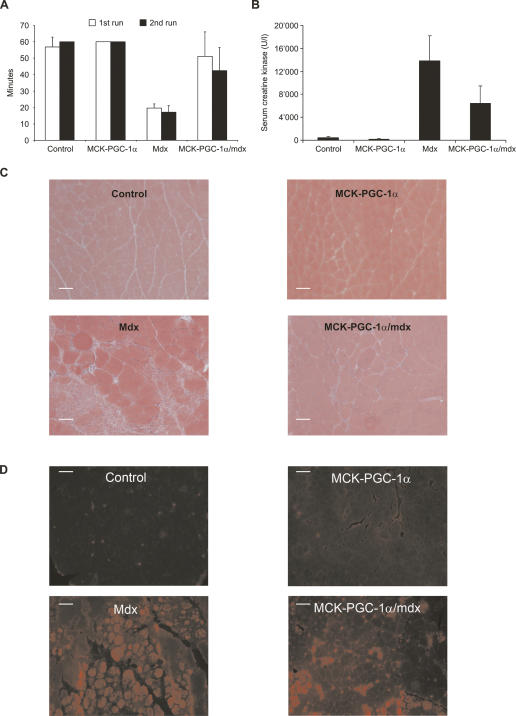
In contrast to human DMD patients, mdx mice have a relatively mild muscular dystrophy that is most noticeable at the age of 4–6 wk and is later offset by compensatory up-regulation of utrophin and massive muscle regeneration (Bulfield et al. 1984). Dystrophin provides mechanical reinforcement to the sarcolemma, and its absence increases the susceptibility of muscle fibers to contractile stress. However, mdx mice challenged with eccentric physical exercise constitute a more reliable model of the human disease than sedentary mdx animals (Brussee et al. 1997). Thus, wild-type, MCK-PGC-1α, mdx, and MCK-PGC-1α/mdx were run on a treadmill for 1 h at 15° downhill. Subsequently, this exercise protocol was repeated 24 h after the first run. Importantly, this exercise protocol is not exhaustive for wild-type animals. Accordingly, wild-type and MCK-PGC-1α animals had no problem running 1 h on two consecutive days (Fig. 7A). In contrast, mdx mice barely make it through 50% of each run (Fig. 7A). MCK-PGC-1α/mdx animals perform almost as well as control mice in the first run; they showed a decreased performance in the second run, though their performance in the second run is still significantly better than that of mdx mice in the first or the second run (Fig. 7A).
Figure 7.
PGC-1α improves muscle function in exercised mdx mice. (A) Twelve-week-old control, PGC-1α muscle-specific transgenic (MCK-PGC-1α), mdx, and PGC-1α muscle-specific transgenic animals in the mdx background (MCK-PGC-1α/mdx) were run for 1 h on a treadmill at a 15° downhill angle (first run). This exercise protocol was repeated after a resting period of 24 h (second run). The bars depict average running times, with the error bars representing standard deviations. (B) Two hours after the first run, blood was drawn from the mice and serum creatine kinase activity was determined. The bars depict average serum creatine kinase levels, with the error bars representing standard deviations. (C) Mice were sacked after the second run and gastrocnemius sections were stained with H&E. (D) Before sacking, Evans blue was injected i.p. and histological sections from diaphragms were analyzed by fluorescence microscopy.
Two hours after the first run, serum creatine kinase activity was determined, which is a reflection of the muscle damage induced by the eccentric exercise. Serum creatine kinase levels of mdx animals were about sevenfold higher that those in sedentary mdx mice (Fig. 7B vs. Fig. 6B). The MCK-PGC-1α/mdx mice had a 50% reduction in creatine kinase levels after exercise, similar to the reduction observed in sedentary animals (Fig. 7B). Histological analysis of quadriceps muscles from mice sacrificed after the second run revealed a lower number of necrotic lesions in the MCK-PGC-1α/mdx mice as compared with mdx animals (Fig. 7C). Moreover, a greater number of large and noncentrally nucleated fibers are found in MCK-PGC-1α/mdx than mdx mice (Fig. 7C; Supplementary Fig. S7). Similarly, fiber damage as determined by Evans blue uptake is lower in MCK-PGC-1α/mdx diaphragms versus mdx diaphragms (Fig. 7D; Supplementary Fig. S7). Thus, the moderately elevated PGC-1α in skeletal muscle results in an improvement in fiber damage, fiber necrosis, and a decrease in serum creatine kinase levels in a mouse model of DMD to an extent that results in vastly improved exercise tolerance. Together, these effects of ectopic expression of PGC-1α show a clear and substantial improvement in the structure and function of skeletal muscle in this disease model.
Discussion
The size and morphology of NMJs evolves in development and changes according to the relative degree of physical activity and the fiber-type distribution (Wilson and Deschenes 2005; Hughes et al. 2006). NMJ formation and plasticity are controlled by the trophic factors agrin and NRG-1 released from the motor neuron (Kummer et al. 2006). Agrin stimulates clustering of AChRs at the site of the NMJ and promotes transcription of NMJ-specific genes. NRG-1 is sufficient to induce transcript levels of many NMJ genes, although recent reports showed that NRG-1 signaling is not required for NMJ formation in vivo (Escher et al. 2005; Jaworski and Burden 2006). Importantly, agrin and NRG-1 signaling both converge on the transcription factor GABP (Briguet and Ruegg 2000). The two GABP subunits, GABPA and GABPB, are subsequently phosphorylated and bind to N-box elements in the promoters of NMJ genes. We now show that PGC-1α is a powerful transcriptional regulator of the NMJ gene program, at least in part by a bona fide coactivation of GABP. Moreover, we demonstrate that the relative levels of PGC-1α are important for the formation of AChR clusters in isolated muscle fibers. The changes in NMJ gene expression and in relative AChR cluster numbers induced by PGC-1α in the basal (nonexercised) state are moderate. Moreover, NMJ formation per se can clearly happen in vivo in the absence of PGC-1α. As observed for PGC-1α in skeletal muscle in general, this coactivator does not regulate basal functions as much as adaptive changes in response to exercise and other physiological alterations. The rapid changes in PGC-1α levels upon physical exercise and inactivity (e.g., Pilegaard et al. 2003; Russell et al. 2003) imply that PGC-1α is probably very important for adaptive changes in the NMJ gene program in certain contexts. In addition, as shown here, the changes induced by PGC-1α are striking in mice with an mdx background.
Induction of GABPA transcript levels and increase of GABP activity by PGC-1α in the context of mitochondrial genes has previously been shown (Mootha et al. 2004). However, we here show for the first time that PGC-1α physically interacts with and coactivates GABP. Importantly, our results give an indication as to how PGC-1α and GABP specifically regulate NMJ gene expression in synaptic nuclei. PGC-1α is required for the effects of NRG-1 that promotes phosphorylation of GABP to increase transcription of NMJ genes. Furthermore, we found that the interaction between PGC-1α and GABP is highly dependent on the phosphorylation state of PGC-1α and GABPB1, the transactivating subunit of GABP. Phosphorylation of GABPA has previously been reported to change the mobility of the GABP complex in the nucleus (Sunesen et al. 2003). The significance of the GABPB1 phosphorylations has not been clear to date (Sunesen et al. 2003). Here, we propose that these post-translational modifications on GABPB1 control the recruitment of PGC-1α to GABP. In addition, PGC-1α requires the presence of HCF in the PGC-1α–GABP protein complex. Additional members of the transcriptional PGC-1α/HCF/GABP complex and the exact domains mediating these interactions will be identified in future studies. In all of the interactions studied to date, PGC-1α directly binds to the transcription factor it coactivates. Why coactivation of GABP by PGC-1α requires these additional steps is not clear. However, it is plausible that the requirement for phosphorylations and the presence of HCF may control the interaction between PGC-1α and GABP specifically in synaptic nuclei where NMJ genes are expressed. Unfortunately, due to the lack of suitable tools, we were not able to study the significance of the phosphorylation of endogenous PGC-1α and GABP. Obviously, characterizing the post-translational modifications of the endogenous proteins in skeletal muscle in vivo will contribute to our understanding of the signaling cascades involved in NMJ remodeling in the future.
PGC-1α muscle-specific transgenic mice in an mdx background have lower creatine kinase levels, reduced necrotic lesions, decreased fiber damage, and increased muscle function. NRG-1 treatment, which improves the function of mdx mice (Krag et al. 2004), has been proposed to work through up-regulation of utrophin, since NRG-1 has no beneficial effect in dystrophin/utrophin double-knockout animals (Krag et al. 2004) and transgenic expression of utrophin ameliorates muscle damage in mdx animals (Tinsley et al. 1998). One obvious link between the PGC-1α-mediated changes in NMJ gene expression and amelioration of muscle dystrophy in mdx mice is the induction of utrophin mRNA and protein (Fig. 1; Supplementary Fig. S9). Elevation of utrophin has been proposed to constitute a therapeutic approach to DMD (Miura and Jasmin 2006). However, although the outcome of crossing muscle-specific PGC-1α transgenic animals into a dystrophin/utrophin double-knockout background is unknown, there is strong evidence suggesting that PGC-1α has beneficial effects on DMD that go well beyond simple increase of utrophin expression. First, PGC-1α has an anti-atrophic effect on muscle fibers in sciatic nerve denervation (Sandri et al. 2006). The exact mechanism of this effect is unknown, but it is clear that PGC-1α inhibits the function of the proatrophic transcription factor FoxO3 and blunts induction of the atrogenes atrogin-1, MuRF-1, and cathepsin-L (Sandri et al. 2006). Recent data have shown that dystrophin disruption as observed in DMD results in down-regulation of Akt signaling, subsequent activation of FoxO3, and elevation of the atrogenes (Acharyya et al. 2005). Second, PGC-1α is a potent regulator of mitochondrial biogenesis and oxidative metabolism in skeletal muscle (Wu et al. 1999; Lin et al. 2002b). The skeletal muscles of mdx mice have impaired oxidative phosphorylation and mitochondrial function (Kuznetsov et al. 1998; Timmons et al. 2005). Moreover, in skeletal muscle of DMD patients, a general metabolic crisis is precipitated by a large-scale down-regulation of nuclear-encoded mitochondrial genes (Chen et al. 2000). Third, PGC-1α is a direct target of calcium/calmodulin signaling in skeletal muscle (Wu et al. 2002; Handschin et al. 2003). Disruption of calcium/calmodulin signaling in mdx mice exacerbates the dystrophic phenotype (Chakkalakal et al. 2006). Fourth, mdx show abnormalities in NMJ morphology in fiber damage and regeneration (Lyons and Slater 1991) that might be prevented by the PGC-1α-mediated induction of NMJ genes. Finally, reactive oxygen species (ROS) may be causally linked to the muscle damage in DMD (Disatnik et al. 1998). The normal cellular response to ROS challenge relies on PGC-1α, which is a potent driver of the gene program for ROS defense (St-Pierre et al. 2006).
In summary, PGC-1α most likely improves muscle dystrophy by multiple molecular pathways. Thus, an increase in PGC-1α levels or PGC-1α activity and recruitment to specific transcription factors such as GABP represent a promising and novel strategy for the treatment of DMD, the related Becker muscular dystrophy, and other neuromuscular diseases.
Materials and methods
Plasmids and reagents
Promoter constructs for AChRδ (770 base pair [bp]), AChRε (210 bp), and utrophin (1.2 kb) were amplified by PCR and subcloned into the pGL3 basic luciferase reporter gene plasmid (Promega), similar to published constructs (Koike et al. 1995; Duclert et al. 1996; Gramolini et al. 1998). Mutations of the N-box DNA elements in these promoters were obtained by PCR-mediated site-directed mutagenesis. Phosphorylation mutants of GABPA (Thr 280) and GABPB1 (Ser 170 and Thr 180) were obtained by site-directed mutagenesis replacing the threonines and serines with alanines. Myc tags were C-terminally attached to GABPA and N-terminally attached to GABPB1 by PCR. Dominant-negative GABPA and GABPB1 alleles lacking amino acids 400–454 and 330–383, respectively, were obtained by PCR. Constitutively active MEKK3 was used to induce phosphorylation of PGC-1α and GABP. All constructs were verified by sequencing. Recombinant rat C-terminal agrin and recombinant human NRG-1 extracellular domain were purchased from R&D Systems. Tetramethylrhodamine-labeled α-BTX was obtained from Invitrogen.
Mdx animals
C57BL/10ScSn-Dmdmdx/J mice (mdx mice) (Bulfield et al. 1984) were purchased from Jackson Laboratories. These mice were crossed with the previously described muscle-specific PGC-1α transgenic animals (Lin et al. 2002b). To control variations of different genetic backgrounds (MCK-PGC-1α: C57Bl/6J; mdx: C57Bl/10ScSnJ), only littermates from the breeding were used in sufficient number per experimental group. Sedentary mice were analyzed at the age of 5 wk, eight animals per group. For the exercise experiment, mice at the age of 12 wk were used, six animals per group.
PGC-1α muscle-specific knockout animals
The generation of mice with a floxed PGC-1α allele has been described (Lin et al. 2004). To obtain muscle-specific knockout animals, mice with a loxed PGC-1α allele were crossed with animals transgenically expressing cre recombinase under the control of the myogenin promoter and MEF2C enhancer (a generous gift from Rhonda Bassel-Duby and Eric N. Olson, University of Texas Southwestern Medical Center, Dallas, TX; Li et al. 2005). However, male mice with the floxed PGC-1α allele are infertile because of the presence of the neomycin cassette. Thus, muscle-specific knockout animals could only be obtained in mice with one floxed and one knockout allele of PGC-1α (lox/−, myogenin-cre). As control mice, we used a mixed population of different mice heterozygous for PGC-1α (lox/−; +/−; +/−, myogenin-cre). A detailed description of the muscle-specific knockout animals will be published elsewhere. Muscle-specific knockout animals and MCK-PGC-1α animals were sacrificed at the age of 6 wk to analyze gene expression; six animals per genotype were used.
Cell culture
C2C12 cells were cultured in DMEM (Invitrogen) with 10% FBS (GIBCO) and differentiated in DMEM with 2% horse serum (GIBCO) for 3 d. Differentiated myotubes were infected with adenoviral vectors and cells were harvested after 48 h. Primary muscle cells were isolated from whole hind-leg muscles by enzymatic digestion with collagenase/dispase (Roche) and serial plating on collagen-coated culture dishes (BD Biosciences). These cells were cultured in Ham’s F-10 medium (Invitrogen) supplemented with 20% FBS (Hyclone) and 2.5 ng/mL basic fibroblast growth factor (Invitrogen). Differentiation was induced by addition of DMEM with 5% horse serum.
Single muscle fiber culture and NMJ quantification
Single muscle fibers were prepared as described (Rosenblatt et al. 1995). Briefly, extensor digitorum longus (EDL) muscle was removed from 6-wk-old animals by cutting the tendons. The muscle was placed in 0.2% collagenase solution and incubated for 60 min at 35°C. Single muscle fibers were liberated from the EDL by shearing using heat-polished Pasteur pipettes in DMEM supplemented with 10% horse serum (GIBCO). The fibers were washed in PBS and fixed in 1.5% paraformaldehyde for 10 min. Subsequently, the fibers were stained with α-BTX in PBS for 30 min, washed twice in PBS with 20 mM glycine, and transferred to a glass slide. The fibers were embedded in SlowFade Gold anti-fade reagent with DAPI (Invitrogen) and analyzed by fluorescent microscopy. At a 60× magnification, 20 fields containing muscle fibers were randomly chosen per animal, and the number of NMJs as well as the number of nuclei were counted. For the histogram depicting the relative number of AChR clusters per muscle fiber, 50 muscle fibers per genotype were analyzed and AChR were counted.
Transient transfections
C2C12 myoblasts were transfected with Lipofectamine 2000 at 80% confluency and subsequently differentiated for 2 d. In some experiments, cells were treated for 2 d with vehicle or 5 nM NRG-1. Then, cells were harvested, and luciferase activity was measured and normalized to β-galactosidase levels.
Coimmunoprecipitation
BOSC cells were transfected by calcium phosphate precipitation (Promega). Two days after transfection, cells were harvested, proteins were isolated, and 1 mg of protein extract was used for immunoprecipitation with anti-myc beads (Santa Cruz Biotechnology). Proteins were subsequently eluted from the beads and analyzed by Western blotting using anti-Flag (Sigma) and anti-myc (Santa Cruz Biotechnology) antibodies along with 10 μg of input. C2C12 cells were transfected by calcium phosphate precipitation at 80% confluency, differentiated for 2 d, and simultaneously treated with vehicle or 5 nM NRG-1. Then, cells were harvested and processed as described above.
AChR clustering in vitro
C2C12 cells were differentiated for 3 d and infected with adenoviral vectors encoding GFP and PGC-1α, respectively. After 1.5 d, cells were treated with vehicle or 10 ng/mL agrin for 16 h. Cells were fixed in 3% formaldehyde, incubated with α-BTX to stain NMJs, and analyzed by fluorescence microscopy.
Histological analysis, Evans blue injection
Tissues were dissected, embedded in OCT compound (VWR), dehydrated, embedded in paraffin, and sectioned. The sections were subsequently stained with H&E. Evans blue dye (1% solution) was i.p.-injected at a concentration of 1% volume per gram of body weight. After 16 h, mice were sacrificed and skeletal-muscle frozen histological sections were prepared. Evans blue incorporation was analyzed by fluorescence microscopy. Evans blue-penetrated muscle fibers and fibers with centrally located nuclei were counted in 20 randomly chosen sections in blinded fashion and were normalized by the total number of muscle fibers per field.
RNA isolation and real-time PCR analysis
Total RNA from cells or tissues was prepared using the Trizol reagent (GIBCO). Before reverse transcription, 1 μg of total RNA was treated with DNase I (Roche) and then treated with MMLV reverse transcriptase (Invitrogen). This cDNA was subsequently analyzed by semiquantitative real-time PCR using the ΔΔCt method on an Applied Biosystems Real-Time PCR System 7300. Relative gene expression levels were normalized to 18S rRNA expression.
Serum creatine kinase assay
Mouse blood was collected and serum-isolated using heparin-coated collection tubes (BD Biosciences). Serum creatine kinase activity was then determined with the Creatine Kinase-SL Assay Kit according to the manufacturer’s protocol (Diagnostic Chemicals Limited).
Treadmill exercise
Animals ran on a treadmill tilted 15° downhill until exhaustion or for maximal 60 min. The running protocol consisted of 5 min at 10 m/min, 5 min at 15 m/min, 5 min at 20 m/min, and 45 min at 25 m/min. The run was repeated 24 h after the first run.
Western blotting
Protein extract from gastrocnemius (100 μg) was run on a 6% polyacrylamide gel, transferred to PVDF membrane, and blotted for utrophin using an anti-utrophin antibody (Santa Cruz Biotecnology).
Acknowledgments
We thank Pere Puigserver and Peter Sicinski for helpful comments on the manuscript. C.H. is supported by a Scientist Career Development Grant of the Muscular Dystrophy Association, the Swiss National Science Foundation Professorship PP00A-110746, and the University Research Priority Program “Integrative Human Physiology” of the University of Zurich. This work is supported by NIH grants DK54477 and DK61562 to B.M.S.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1525107
References
- Acharyya S., Butchbach M.E., Sahenk Z., Wang H., Saji M., Carathers M., Ringel M.D., Skipworth R.J., Fearon K.C., Hollingsworth M.A., Butchbach M.E., Sahenk Z., Wang H., Saji M., Carathers M., Ringel M.D., Skipworth R.J., Fearon K.C., Hollingsworth M.A., Sahenk Z., Wang H., Saji M., Carathers M., Ringel M.D., Skipworth R.J., Fearon K.C., Hollingsworth M.A., Wang H., Saji M., Carathers M., Ringel M.D., Skipworth R.J., Fearon K.C., Hollingsworth M.A., Saji M., Carathers M., Ringel M.D., Skipworth R.J., Fearon K.C., Hollingsworth M.A., Carathers M., Ringel M.D., Skipworth R.J., Fearon K.C., Hollingsworth M.A., Ringel M.D., Skipworth R.J., Fearon K.C., Hollingsworth M.A., Skipworth R.J., Fearon K.C., Hollingsworth M.A., Fearon K.C., Hollingsworth M.A., Hollingsworth M.A., et al. Dystrophin glycoprotein complex dysfunction: A regulatory link between muscular dystrophy and cancer cachexia. Cancer Cell. 2005;8:421–432. doi: 10.1016/j.ccr.2005.10.004. [DOI] [PubMed] [Google Scholar]
- Angus L.M., Chakkalakal J.V., Mejat A., Eibl J.K., Belanger G., Megeney L.A., Chin E.R., Schaeffer L., Michel R.N., Jasmin B.J., Chakkalakal J.V., Mejat A., Eibl J.K., Belanger G., Megeney L.A., Chin E.R., Schaeffer L., Michel R.N., Jasmin B.J., Mejat A., Eibl J.K., Belanger G., Megeney L.A., Chin E.R., Schaeffer L., Michel R.N., Jasmin B.J., Eibl J.K., Belanger G., Megeney L.A., Chin E.R., Schaeffer L., Michel R.N., Jasmin B.J., Belanger G., Megeney L.A., Chin E.R., Schaeffer L., Michel R.N., Jasmin B.J., Megeney L.A., Chin E.R., Schaeffer L., Michel R.N., Jasmin B.J., Chin E.R., Schaeffer L., Michel R.N., Jasmin B.J., Schaeffer L., Michel R.N., Jasmin B.J., Michel R.N., Jasmin B.J., Jasmin B.J. Calcineurin–NFAT signaling, together with GABP and peroxisome PGC-1α, drives utrophin gene expression at the neuromuscular junction. Am. J. Physiol. Cell Physiol. 2005;289:C908–C917. doi: 10.1152/ajpcell.00196.2005. [DOI] [PubMed] [Google Scholar]
- Baar K. Involvement of PPAR γ co-activator-1, nuclear respiratory factors 1 and 2, and PPAR α in the adaptive response to endurance exercise. Proc. Nutr. Soc. 2004;63:269–273. doi: 10.1079/PNS2004334. [DOI] [PubMed] [Google Scholar]
- Bezakova G., Ruegg M.A., Ruegg M.A. New insights into the roles of agrin. Nat. Rev. Mol. Cell Biol. 2003;4:295–308. doi: 10.1038/nrm1074. [DOI] [PubMed] [Google Scholar]
- Bogdanovich S., Perkins K.J., Krag T.O., Khurana T.S., Perkins K.J., Krag T.O., Khurana T.S., Krag T.O., Khurana T.S., Khurana T.S. Therapeutics for Duchenne muscular dystrophy: Current approaches and future directions. J. Mol. Med. 2004;82:102–115. doi: 10.1007/s00109-003-0484-1. [DOI] [PubMed] [Google Scholar]
- Briguet A., Ruegg M.A., Ruegg M.A. The Ets transcription factor GABP is required for postsynaptic differentiation in vivo. J. Neurosci. 2000;20:5989–5996. doi: 10.1523/JNEUROSCI.20-16-05989.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briguet A., Courdier-Fruh I., Foster M., Meier T., Magyar J.P., Courdier-Fruh I., Foster M., Meier T., Magyar J.P., Foster M., Meier T., Magyar J.P., Meier T., Magyar J.P., Magyar J.P. Histological parameters for the quantitative assessment of muscular dystrophy in the mdx-mouse. Neuromuscul. Disord. 2004;14:675–682. doi: 10.1016/j.nmd.2004.06.008. [DOI] [PubMed] [Google Scholar]
- Brussee V., Tardif F., Tremblay J.P., Tardif F., Tremblay J.P., Tremblay J.P. Muscle fibers of mdx mice are more vulnerable to exercise than those of normal mice. Neuromuscul. Disord. 1997;7:487–492. doi: 10.1016/s0960-8966(97)00115-6. [DOI] [PubMed] [Google Scholar]
- Bulfield G., Siller W.G., Wight P.A., Moore K.J., Siller W.G., Wight P.A., Moore K.J., Wight P.A., Moore K.J., Moore K.J. X chromosome-linked muscular dystrophy (mdx) in the mouse. Proc. Natl. Acad. Sci. 1984;81:1189–1192. doi: 10.1073/pnas.81.4.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakkalakal J.V., Michel S.A., Chin E.R., Michel R.N., Jasmin B.J., Michel S.A., Chin E.R., Michel R.N., Jasmin B.J., Chin E.R., Michel R.N., Jasmin B.J., Michel R.N., Jasmin B.J., Jasmin B.J. Targeted inhibition of Ca2+/calmodulin signaling exacerbates the dystrophic phenotype in mdx mouse muscle. Hum. Mol. Genet. 2006;15:1423–1435. doi: 10.1093/hmg/ddl065. [DOI] [PubMed] [Google Scholar]
- Chen Y.W., Zhao P., Borup R., Hoffman E.P., Zhao P., Borup R., Hoffman E.P., Borup R., Hoffman E.P., Hoffman E.P. Expression profiling in the muscular dystrophies: Identification of novel aspects of molecular pathophysiology. J. Cell Biol. 2000;151:1321–1336. doi: 10.1083/jcb.151.6.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Disatnik M.H., Dhawan J., Yu Y., Beal M.F., Whirl M.M., Franco A.A., Rando T.A., Dhawan J., Yu Y., Beal M.F., Whirl M.M., Franco A.A., Rando T.A., Yu Y., Beal M.F., Whirl M.M., Franco A.A., Rando T.A., Beal M.F., Whirl M.M., Franco A.A., Rando T.A., Whirl M.M., Franco A.A., Rando T.A., Franco A.A., Rando T.A., Rando T.A. Evidence of oxidative stress in mdx mouse muscle: Studies of the pre-necrotic state. J. Neurol. Sci. 1998;161:77–84. doi: 10.1016/s0022-510x(98)00258-5. [DOI] [PubMed] [Google Scholar]
- Duclert A., Savatier N., Schaeffer L., Changeux J.P., Savatier N., Schaeffer L., Changeux J.P., Schaeffer L., Changeux J.P., Changeux J.P. Identification of an element crucial for the sub-synaptic expression of the acetylcholine receptor ε-subunit gene. J. Biol. Chem. 1996;271:17433–17438. doi: 10.1074/jbc.271.29.17433. [DOI] [PubMed] [Google Scholar]
- Escher P., Lacazette E., Courtet M., Blindenbacher A., Landmann L., Bezakova G., Lloyd K.C., Mueller U., Brenner H.R., Lacazette E., Courtet M., Blindenbacher A., Landmann L., Bezakova G., Lloyd K.C., Mueller U., Brenner H.R., Courtet M., Blindenbacher A., Landmann L., Bezakova G., Lloyd K.C., Mueller U., Brenner H.R., Blindenbacher A., Landmann L., Bezakova G., Lloyd K.C., Mueller U., Brenner H.R., Landmann L., Bezakova G., Lloyd K.C., Mueller U., Brenner H.R., Bezakova G., Lloyd K.C., Mueller U., Brenner H.R., Lloyd K.C., Mueller U., Brenner H.R., Mueller U., Brenner H.R., Brenner H.R. Synapses form in skeletal muscles lacking neuregulin receptors. Science. 2005;308:1920–1923. doi: 10.1126/science.1108258. [DOI] [PubMed] [Google Scholar]
- Flory E., Hoffmeyer A., Smola U., Rapp U.R., Bruder J.T., Hoffmeyer A., Smola U., Rapp U.R., Bruder J.T., Smola U., Rapp U.R., Bruder J.T., Rapp U.R., Bruder J.T., Bruder J.T. Raf-1 kinase targets GA-binding protein in transcriptional regulation of the human immunodeficiency virus type 1 promoter. J. Virol. 1996;70:2260–2268. doi: 10.1128/jvi.70.4.2260-2268.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromm L., Burden S.J., Burden S.J. Neuregulin-1-stimulated phosphorylation of GABP in skeletal muscle cells. Biochemistry. 2001;40:5306–5312. doi: 10.1021/bi002649m. [DOI] [PubMed] [Google Scholar]
- Gramolini A.O., Burton E.A., Tinsley J.M., Ferns M.J., Cartaud A., Cartaud J., Davies K.E., Lunde J.A., Jasmin B.J., Burton E.A., Tinsley J.M., Ferns M.J., Cartaud A., Cartaud J., Davies K.E., Lunde J.A., Jasmin B.J., Tinsley J.M., Ferns M.J., Cartaud A., Cartaud J., Davies K.E., Lunde J.A., Jasmin B.J., Ferns M.J., Cartaud A., Cartaud J., Davies K.E., Lunde J.A., Jasmin B.J., Cartaud A., Cartaud J., Davies K.E., Lunde J.A., Jasmin B.J., Cartaud J., Davies K.E., Lunde J.A., Jasmin B.J., Davies K.E., Lunde J.A., Jasmin B.J., Lunde J.A., Jasmin B.J., Jasmin B.J. Muscle and neural isoforms of agrin increase utrophin expression in cultured myotubes via a transcriptional regulatory mechanism. J. Biol. Chem. 1998;273:736–743. doi: 10.1074/jbc.273.2.736. [DOI] [PubMed] [Google Scholar]
- Handschin C., Spiegelman B.M., Spiegelman B.M. PGC-1 coactivators, energy homeostasis, and metabolism. Endocr. Rev. 2006;27:728–735. doi: 10.1210/er.2006-0037. [DOI] [PubMed] [Google Scholar]
- Handschin C., Rhee J., Lin J., Tarr P.T., Spiegelman B.M., Rhee J., Lin J., Tarr P.T., Spiegelman B.M., Lin J., Tarr P.T., Spiegelman B.M., Tarr P.T., Spiegelman B.M., Spiegelman B.M. An autoregulatory loop controls peroxisome proliferator-activated receptor γ coactivator 1α expression in muscle. Proc. Natl. Acad. Sci. 2003;100:7111–7116. doi: 10.1073/pnas.1232352100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handschin C., Lin J., Rhee J., Peyer A.K., Chin S., Wu P.H., Meyer U.A., Spiegelman B.M., Lin J., Rhee J., Peyer A.K., Chin S., Wu P.H., Meyer U.A., Spiegelman B.M., Rhee J., Peyer A.K., Chin S., Wu P.H., Meyer U.A., Spiegelman B.M., Peyer A.K., Chin S., Wu P.H., Meyer U.A., Spiegelman B.M., Chin S., Wu P.H., Meyer U.A., Spiegelman B.M., Wu P.H., Meyer U.A., Spiegelman B.M., Meyer U.A., Spiegelman B.M., Spiegelman B.M. Nutritional regulation of hepatic heme biosynthesis and porphyria through PGC-1α. Cell. 2005;122:505–515. doi: 10.1016/j.cell.2005.06.040. [DOI] [PubMed] [Google Scholar]
- Hughes B.W., Kusner L.L., Kaminski H.J., Kusner L.L., Kaminski H.J., Kaminski H.J. Molecular architecture of the neuromuscular junction. Muscle Nerve. 2006;33:445–461. doi: 10.1002/mus.20440. [DOI] [PubMed] [Google Scholar]
- Jaworski A., Burden S.J., Burden S.J. Neuromuscular synapse formation in mice lacking motor neuron- and skeletal muscle-derived Neuregulin-1. J. Neurosci. 2006;26:655–661. doi: 10.1523/JNEUROSCI.4506-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandarian S.C., Jackman R.W., Jackman R.W. Intracellular signaling during skeletal muscle atrophy. Muscle Nerve. 2006;33:155–165. doi: 10.1002/mus.20442. [DOI] [PubMed] [Google Scholar]
- Kapsa R., Kornberg A.J., Byrne E., Kornberg A.J., Byrne E., Byrne E. Novel therapies for Duchenne muscular dystrophy. Lancet Neurol. 2003;2:299–310. doi: 10.1016/s1474-4422(03)00382-x. [DOI] [PubMed] [Google Scholar]
- Koike S., Schaeffer L., Changeux J.P., Schaeffer L., Changeux J.P., Changeux J.P. Identification of a DNA element determining synaptic expression of the mouse acetylcholine receptor δ-subunit gene. Proc. Natl. Acad. Sci. 1995;92:10624–10628. doi: 10.1073/pnas.92.23.10624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krag T.O., Bogdanovich S., Jensen C.J., Fischer M.D., Hansen-Schwartz J., Javazon E.H., Flake A.W., Edvinsson L., Khurana T.S., Bogdanovich S., Jensen C.J., Fischer M.D., Hansen-Schwartz J., Javazon E.H., Flake A.W., Edvinsson L., Khurana T.S., Jensen C.J., Fischer M.D., Hansen-Schwartz J., Javazon E.H., Flake A.W., Edvinsson L., Khurana T.S., Fischer M.D., Hansen-Schwartz J., Javazon E.H., Flake A.W., Edvinsson L., Khurana T.S., Hansen-Schwartz J., Javazon E.H., Flake A.W., Edvinsson L., Khurana T.S., Javazon E.H., Flake A.W., Edvinsson L., Khurana T.S., Flake A.W., Edvinsson L., Khurana T.S., Edvinsson L., Khurana T.S., Khurana T.S. Heregulin ameliorates the dystrophic phenotype in mdx mice. Proc. Natl. Acad. Sci. 2004;101:13856–13860. doi: 10.1073/pnas.0405972101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kummer T.T., Misgeld T., Sanes J.R., Misgeld T., Sanes J.R., Sanes J.R. Assembly of the postsynaptic membrane at the neuromuscular junction: Paradigm lost. Curr. Opin. Neurobiol. 2006;16:74–82. doi: 10.1016/j.conb.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Kuznetsov A.V., Winkler K., Wiedemann F.R., von Bossanyi P., Dietzmann K., Kunz W.S., Winkler K., Wiedemann F.R., von Bossanyi P., Dietzmann K., Kunz W.S., Wiedemann F.R., von Bossanyi P., Dietzmann K., Kunz W.S., von Bossanyi P., Dietzmann K., Kunz W.S., Dietzmann K., Kunz W.S., Kunz W.S. Impaired mitochondrial oxidative phosphorylation in skeletal muscle of the dystrophin-deficient mdx mouse. Mol. Cell. Biochem. 1998;183:87–96. doi: 10.1023/a:1006868130002. [DOI] [PubMed] [Google Scholar]
- Leone T.C., Lehman J.J., Finck B.N., Schaeffer P.J., Wende A.R., Boudina S., Courtois M., Wozniak D.F., Sambandam N., Bernal-Mizrachi C., Lehman J.J., Finck B.N., Schaeffer P.J., Wende A.R., Boudina S., Courtois M., Wozniak D.F., Sambandam N., Bernal-Mizrachi C., Finck B.N., Schaeffer P.J., Wende A.R., Boudina S., Courtois M., Wozniak D.F., Sambandam N., Bernal-Mizrachi C., Schaeffer P.J., Wende A.R., Boudina S., Courtois M., Wozniak D.F., Sambandam N., Bernal-Mizrachi C., Wende A.R., Boudina S., Courtois M., Wozniak D.F., Sambandam N., Bernal-Mizrachi C., Boudina S., Courtois M., Wozniak D.F., Sambandam N., Bernal-Mizrachi C., Courtois M., Wozniak D.F., Sambandam N., Bernal-Mizrachi C., Wozniak D.F., Sambandam N., Bernal-Mizrachi C., Sambandam N., Bernal-Mizrachi C., Bernal-Mizrachi C., et al. PGC-1α deficiency causes multi-system energy metabolic derangements: Muscle dysfunction, abnormal weight control and hepatic steatosis. PLoS Biol. 2005;3:e101. doi: 10.1371/journal.pbio.0030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Czubryt M.P., McAnally J., Bassel-Duby R., Richardson J.A., Wiebel F.F., Nordheim A., Olson E.N., Czubryt M.P., McAnally J., Bassel-Duby R., Richardson J.A., Wiebel F.F., Nordheim A., Olson E.N., McAnally J., Bassel-Duby R., Richardson J.A., Wiebel F.F., Nordheim A., Olson E.N., Bassel-Duby R., Richardson J.A., Wiebel F.F., Nordheim A., Olson E.N., Richardson J.A., Wiebel F.F., Nordheim A., Olson E.N., Wiebel F.F., Nordheim A., Olson E.N., Nordheim A., Olson E.N., Olson E.N. Requirement for serum response factor for skeletal muscle growth and maturation revealed by tissue-specific gene deletion in mice. Proc. Natl. Acad. Sci. 2005;102:1082–1087. doi: 10.1073/pnas.0409103102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J., Puigserver P., Donovan J., Tarr P., Spiegelman B.M., Puigserver P., Donovan J., Tarr P., Spiegelman B.M., Donovan J., Tarr P., Spiegelman B.M., Tarr P., Spiegelman B.M., Spiegelman B.M. Peroxisome proliferator-activated receptor γ coactivator 1β (PGC-1β), a novel PGC-1-related transcription coactivator associated with host cell factor. J. Biol. Chem. 2002a;277:1645–1648. doi: 10.1074/jbc.C100631200. [DOI] [PubMed] [Google Scholar]
- Lin J., Wu H., Tarr P.T., Zhang C.Y., Wu Z., Boss O., Michael L.F., Puigserver P., Isotani E., Olson E.N., Wu H., Tarr P.T., Zhang C.Y., Wu Z., Boss O., Michael L.F., Puigserver P., Isotani E., Olson E.N., Tarr P.T., Zhang C.Y., Wu Z., Boss O., Michael L.F., Puigserver P., Isotani E., Olson E.N., Zhang C.Y., Wu Z., Boss O., Michael L.F., Puigserver P., Isotani E., Olson E.N., Wu Z., Boss O., Michael L.F., Puigserver P., Isotani E., Olson E.N., Boss O., Michael L.F., Puigserver P., Isotani E., Olson E.N., Michael L.F., Puigserver P., Isotani E., Olson E.N., Puigserver P., Isotani E., Olson E.N., Isotani E., Olson E.N., Olson E.N., et al. Transcriptional co-activator PGC-1 α drives the formation of slow-twitch muscle fibres. Nature. 2002b;418:797–801. doi: 10.1038/nature00904. [DOI] [PubMed] [Google Scholar]
- Lin J., Wu P.H., Tarr P.T., Lindenberg K.S., St-Pierre J., Zhang C.Y., Mootha V.K., Jager S., Vianna C.R., Reznick R.M., Wu P.H., Tarr P.T., Lindenberg K.S., St-Pierre J., Zhang C.Y., Mootha V.K., Jager S., Vianna C.R., Reznick R.M., Tarr P.T., Lindenberg K.S., St-Pierre J., Zhang C.Y., Mootha V.K., Jager S., Vianna C.R., Reznick R.M., Lindenberg K.S., St-Pierre J., Zhang C.Y., Mootha V.K., Jager S., Vianna C.R., Reznick R.M., St-Pierre J., Zhang C.Y., Mootha V.K., Jager S., Vianna C.R., Reznick R.M., Zhang C.Y., Mootha V.K., Jager S., Vianna C.R., Reznick R.M., Mootha V.K., Jager S., Vianna C.R., Reznick R.M., Jager S., Vianna C.R., Reznick R.M., Vianna C.R., Reznick R.M., Reznick R.M., et al. Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1α null mice. Cell. 2004;119:121–135. doi: 10.1016/j.cell.2004.09.013. [DOI] [PubMed] [Google Scholar]
- Lyons P.R., Slater C.R., Slater C.R. Structure and function of the neuromuscular junction in young adult mdx mice. J. Neurocytol. 1991;20:969–981. doi: 10.1007/BF01187915. [DOI] [PubMed] [Google Scholar]
- Miura P., Jasmin B.J., Jasmin B.J. Utrophin upregulation for treating Duchenne or Becker muscular dystrophy: How close are we? Trends Mol. Med. 2006;12:122–129. doi: 10.1016/j.molmed.2006.01.002. [DOI] [PubMed] [Google Scholar]
- Mootha V.K., Handschin C., Arlow D., Xie X., St Pierre J., Sihag S., Yang W., Altshuler D., Puigserver P., Patterson N., Handschin C., Arlow D., Xie X., St Pierre J., Sihag S., Yang W., Altshuler D., Puigserver P., Patterson N., Arlow D., Xie X., St Pierre J., Sihag S., Yang W., Altshuler D., Puigserver P., Patterson N., Xie X., St Pierre J., Sihag S., Yang W., Altshuler D., Puigserver P., Patterson N., St Pierre J., Sihag S., Yang W., Altshuler D., Puigserver P., Patterson N., Sihag S., Yang W., Altshuler D., Puigserver P., Patterson N., Yang W., Altshuler D., Puigserver P., Patterson N., Altshuler D., Puigserver P., Patterson N., Puigserver P., Patterson N., Patterson N., et al. Errα and Gabpa/b specify PGC-1α-dependent oxidative phosphorylation gene expression that is altered in diabetic muscle. Proc. Natl. Acad. Sci. 2004;101:6570–6575. doi: 10.1073/pnas.0401401101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilegaard H., Saltin B., Neufer P.D., Saltin B., Neufer P.D., Neufer P.D. Exercise induces transient transcriptional activation of the PGC-1α gene in human skeletal muscle. J. Physiol. 2003;546:851–858. doi: 10.1113/jphysiol.2002.034850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puigserver P., Rhee J., Lin J., Wu Z., Yoon J.C., Zhang C.Y., Krauss S., Mootha V.K., Lowell B.B., Spiegelman B.M., Rhee J., Lin J., Wu Z., Yoon J.C., Zhang C.Y., Krauss S., Mootha V.K., Lowell B.B., Spiegelman B.M., Lin J., Wu Z., Yoon J.C., Zhang C.Y., Krauss S., Mootha V.K., Lowell B.B., Spiegelman B.M., Wu Z., Yoon J.C., Zhang C.Y., Krauss S., Mootha V.K., Lowell B.B., Spiegelman B.M., Yoon J.C., Zhang C.Y., Krauss S., Mootha V.K., Lowell B.B., Spiegelman B.M., Zhang C.Y., Krauss S., Mootha V.K., Lowell B.B., Spiegelman B.M., Krauss S., Mootha V.K., Lowell B.B., Spiegelman B.M., Mootha V.K., Lowell B.B., Spiegelman B.M., Lowell B.B., Spiegelman B.M., Spiegelman B.M. Cytokine stimulation of energy expenditure through p38 MAP kinase activation of PPARγ coactivator-1. Mol. Cell. 2001;8:971–982. doi: 10.1016/s1097-2765(01)00390-2. [DOI] [PubMed] [Google Scholar]
- Rosenblatt J.D., Lunt A.I., Parry D.J., Partridge T.A., Lunt A.I., Parry D.J., Partridge T.A., Parry D.J., Partridge T.A., Partridge T.A. Culturing satellite cells from living single muscle fiber explants. In Vitro Cell. Dev. Biol. 1995;31:773–779. doi: 10.1007/BF02634119. [DOI] [PubMed] [Google Scholar]
- Ruegg M.A. Organization of synaptic myonuclei by Syne proteins and their role during the formation of the nerve-muscle synapse. Proc. Natl. Acad. Sci. 2005;102:5643–5644. doi: 10.1073/pnas.0501516102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell A.P., Feilchenfeldt J., Schreiber S., Praz M., Crettenand A., Gobelet C., Meier C.A., Bell D.R., Kralli A., Giacobino J.P., Feilchenfeldt J., Schreiber S., Praz M., Crettenand A., Gobelet C., Meier C.A., Bell D.R., Kralli A., Giacobino J.P., Schreiber S., Praz M., Crettenand A., Gobelet C., Meier C.A., Bell D.R., Kralli A., Giacobino J.P., Praz M., Crettenand A., Gobelet C., Meier C.A., Bell D.R., Kralli A., Giacobino J.P., Crettenand A., Gobelet C., Meier C.A., Bell D.R., Kralli A., Giacobino J.P., Gobelet C., Meier C.A., Bell D.R., Kralli A., Giacobino J.P., Meier C.A., Bell D.R., Kralli A., Giacobino J.P., Bell D.R., Kralli A., Giacobino J.P., Kralli A., Giacobino J.P., Giacobino J.P., et al. Endurance training in humans leads to fiber type-specific increases in levels of peroxisome proliferator-activated receptor-γ coactivator-1 and peroxisome proliferator-activated receptor-α in skeletal muscle. Diabetes. 2003;52:2874–2881. doi: 10.2337/diabetes.52.12.2874. [DOI] [PubMed] [Google Scholar]
- Sandri M., Lin J., Handschin C., Yang W., Arany Z.P., Lecker S.H., Goldberg A.L., Spiegelman B.M., Lin J., Handschin C., Yang W., Arany Z.P., Lecker S.H., Goldberg A.L., Spiegelman B.M., Handschin C., Yang W., Arany Z.P., Lecker S.H., Goldberg A.L., Spiegelman B.M., Yang W., Arany Z.P., Lecker S.H., Goldberg A.L., Spiegelman B.M., Arany Z.P., Lecker S.H., Goldberg A.L., Spiegelman B.M., Lecker S.H., Goldberg A.L., Spiegelman B.M., Goldberg A.L., Spiegelman B.M., Spiegelman B.M. PGC-1{α} protects skeletal muscle from atrophy by suppressing FoxO3 action and atrophy-specific gene transcription. Proc. Natl. Acad. Sci. 2006;103:16260–16265. doi: 10.1073/pnas.0607795103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpulla R.C. Nuclear activators and coactivators in mammalian mitochondrial biogenesis. Biochim. Biophys. Acta. 2002;1576:1–14. doi: 10.1016/s0167-4781(02)00343-3. [DOI] [PubMed] [Google Scholar]
- St-Pierre J., Drori S., Uldry M., Silvaggi J.M., Rhee J., Jager S., Handschin C., Zheng K., Lin J., Yang W., Drori S., Uldry M., Silvaggi J.M., Rhee J., Jager S., Handschin C., Zheng K., Lin J., Yang W., Uldry M., Silvaggi J.M., Rhee J., Jager S., Handschin C., Zheng K., Lin J., Yang W., Silvaggi J.M., Rhee J., Jager S., Handschin C., Zheng K., Lin J., Yang W., Rhee J., Jager S., Handschin C., Zheng K., Lin J., Yang W., Jager S., Handschin C., Zheng K., Lin J., Yang W., Handschin C., Zheng K., Lin J., Yang W., Zheng K., Lin J., Yang W., Lin J., Yang W., Yang W., et al. Suppression of reactive oxygen species and neurodegeneration by the PGC-1 transcriptional coactivators. Cell. 2006;127:397–408. doi: 10.1016/j.cell.2006.09.024. [DOI] [PubMed] [Google Scholar]
- Sunesen M., Changeux J.P., Changeux J.P. Transcription in neuromuscular junction formation: Who turns on whom? J Neurocytol. 2003;32:677–684. doi: 10.1023/B:NEUR.0000020616.53664.80. [DOI] [PubMed] [Google Scholar]
- Sunesen M., Huchet-Dymanus M., Christensen M.O., Changeux J.P., Huchet-Dymanus M., Christensen M.O., Changeux J.P., Christensen M.O., Changeux J.P., Changeux J.P. Phosphorylation-elicited quaternary changes of GA binding protein in transcriptional activation. Mol. Cell. Biol. 2003;23:8008–8018. doi: 10.1128/MCB.23.22.8008-8018.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons J.A., Larsson O., Jansson E., Fischer H., Gustafsson T., Greenhaff P.L., Ridden J., Rachman J., Peyrard-Janvid M., Wahlestedt C., Larsson O., Jansson E., Fischer H., Gustafsson T., Greenhaff P.L., Ridden J., Rachman J., Peyrard-Janvid M., Wahlestedt C., Jansson E., Fischer H., Gustafsson T., Greenhaff P.L., Ridden J., Rachman J., Peyrard-Janvid M., Wahlestedt C., Fischer H., Gustafsson T., Greenhaff P.L., Ridden J., Rachman J., Peyrard-Janvid M., Wahlestedt C., Gustafsson T., Greenhaff P.L., Ridden J., Rachman J., Peyrard-Janvid M., Wahlestedt C., Greenhaff P.L., Ridden J., Rachman J., Peyrard-Janvid M., Wahlestedt C., Ridden J., Rachman J., Peyrard-Janvid M., Wahlestedt C., Rachman J., Peyrard-Janvid M., Wahlestedt C., Peyrard-Janvid M., Wahlestedt C., Wahlestedt C., et al. Human muscle gene expression responses to endurance training provide a novel perspective on Duchenne muscular dystrophy. FASEB J. 2005;19:750–760. doi: 10.1096/fj.04-1980com. [DOI] [PubMed] [Google Scholar]
- Tinsley J., Deconinck N., Fisher R., Kahn D., Phelps S., Gillis J.M., Davies K., Deconinck N., Fisher R., Kahn D., Phelps S., Gillis J.M., Davies K., Fisher R., Kahn D., Phelps S., Gillis J.M., Davies K., Kahn D., Phelps S., Gillis J.M., Davies K., Phelps S., Gillis J.M., Davies K., Gillis J.M., Davies K., Davies K. Expression of full-length utrophin prevents muscular dystrophy in mdx mice. Nat. Med. 1998;4:1441–1444. doi: 10.1038/4033. [DOI] [PubMed] [Google Scholar]
- Vogel J.L., Kristie T.M., Kristie T.M. The novel coactivator C1 (HCF) coordinates multiprotein enhancer formation and mediates transcription activation by GABP. EMBO J. 2000;19:683–690. doi: 10.1093/emboj/19.4.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson M.H., Deschenes M.R., Deschenes M.R. The neuromuscular junction: Anatomical features and adaptations to various forms of increased, or decreased neuromuscular activity. Int. J. Neurosci. 2005;115:803–828. doi: 10.1080/00207450590882172. [DOI] [PubMed] [Google Scholar]
- Wu Z., Puigserver P., Andersson U., Zhang C., Adelmant G., Mootha V., Troy A., Cinti S., Lowell B., Scarpulla R.C., Puigserver P., Andersson U., Zhang C., Adelmant G., Mootha V., Troy A., Cinti S., Lowell B., Scarpulla R.C., Andersson U., Zhang C., Adelmant G., Mootha V., Troy A., Cinti S., Lowell B., Scarpulla R.C., Zhang C., Adelmant G., Mootha V., Troy A., Cinti S., Lowell B., Scarpulla R.C., Adelmant G., Mootha V., Troy A., Cinti S., Lowell B., Scarpulla R.C., Mootha V., Troy A., Cinti S., Lowell B., Scarpulla R.C., Troy A., Cinti S., Lowell B., Scarpulla R.C., Cinti S., Lowell B., Scarpulla R.C., Lowell B., Scarpulla R.C., Scarpulla R.C., et al. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98:115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- Wu H., Kanatous S.B., Thurmond F.A., Gallardo T., Isotani E., Bassel-Duby R., Williams R.S., Kanatous S.B., Thurmond F.A., Gallardo T., Isotani E., Bassel-Duby R., Williams R.S., Thurmond F.A., Gallardo T., Isotani E., Bassel-Duby R., Williams R.S., Gallardo T., Isotani E., Bassel-Duby R., Williams R.S., Isotani E., Bassel-Duby R., Williams R.S., Bassel-Duby R., Williams R.S., Williams R.S. Regulation of mitochondrial biogenesis in skeletal muscle by CaMK. Science. 2002;296:349–352. doi: 10.1126/science.1071163. [DOI] [PubMed] [Google Scholar]