Abstract
The circadian rhythm of pineal melatonin requires the nocturnal increment of serotonin N-acetyltransferase (arylalkylamine N-acetyltransferase [AANAT]) protein. To date, only limited information is available in the critical issue of how AANAT protein expression is up-regulated exclusively at night regardless of its species-specific mRNA profiles. Here we show that the circadian timing of AANAT protein expression is regulated by rhythmic translation of AANAT mRNA. This rhythmic control is mediated by both a highly conserved IRES (internal ribosome entry site) element within the AANAT 5′ untranslated region and its partner hnRNP Q (heterogeneous nuclear ribonucleoprotein Q) with a peak in the middle of the night. Consistent with the enhancing role of hnRNP Q in AANAT IRES activities, knockdown of the hnRNP Q level elicited a dramatic decrease of peak amplitude in the AANAT protein profile parallel to reduced melatonin production in pinealocytes. This translational regulation of AANAT mRNA provides a novel aspect for achieving the circadian rhythmicity of vertebrate melatonin.
Keywords: AANAT, translation, IRES, hnRNP Q, melatonin, circadian rhythm
Melatonin is the primary marker of circadian rhythm in vertebrates. The nocturnal increase in circulating melatonin is mirrored by the expression of arylalkylamine N-acetyltransferase (AANAT) protein, which is the rate-limiting enzyme in the melatonin biosynthetic pathway (Coon et al. 1995; Reppert and Weaver 1995; Ganguly et al. 2001). Interestingly, there are species-specific discrepancies between AANAT protein expression and its mRNA expression profiles during the circadian cycle (Klein et al. 1997; Ganguly et al. 2002). In rodents, there is a lag between a nocturnal increase in AANAT mRNA levels and an increase in AANAT protein levels (Fig. 1E, middle panel; Klein et al. 1997; Ganguly et al. 2002; Kim et al. 2005). In contrast, in primates and ungulates, AANAT protein levels increase exclusively nocturnally, despite steady-state, high levels of AANAT mRNAs (Stehle et al. 2001; Ganguly et al. 2002; Ackermann et al. 2006). Thus, AANAT protein expression is out of phase with its mRNA expression profile. Previous studies of nocturnal expression of AANAT protein have described transcriptional (feedback loop) (Baler et al. 1997; Foulkes et al. 1997), post-transcriptional (mRNA degradation) (Kim et al. 2005), and post-translational (proteolysis) (Gastel et al. 1998; Obsil et al. 2001) regulatory mechanisms. However, little work has been done to understand the role of translational (protein synthesis) control in the circadian rhythmicity of AANAT protein expression. Cued by the observations of species-specific differences in the circadian timing of AANAT mRNA and AANAT protein expression, we hypothesized that circadian phase-specific translational regulation of AANAT mRNA might be a novel and efficient mechanism for controlling melatonin production at night.
Figure 1.
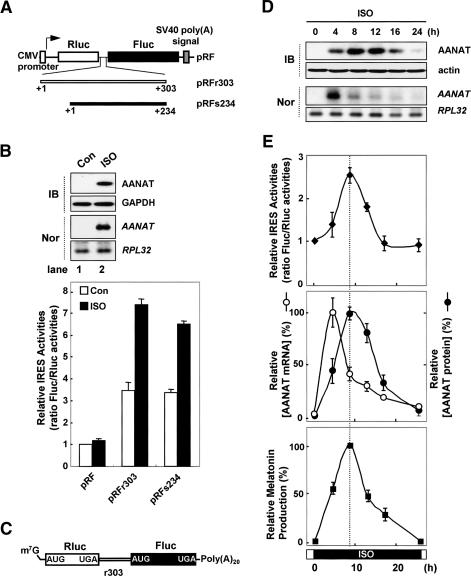
Nocturnal signaling activates IRES-mediated AANAT translation in rat pinealocytes. (A) Schematic diagrams of bicistronic reporter plasmids containing the full-length rat (r303) or sheep (s234) AANAT 5′UTR. (pRF) Bicistronic reporter plasmid; (Rluc) Renilla luciferase; (Fluc) firefly luciferase. (B) Phase-dependent AANAT IRES activity. Rat pinealocyte cells were transfected with bicistronic reporter plasmids and incubated for 48 h before treatment with (ISO, closed bar) or without (Con, open bar) ISO, and incubated for a further 12 h before harvesting. Cell lysates were prepared and subjected to immunoblotting (IB), Northern blotting (Nor), and luciferase assays. The ratio for the empty vector pRF was set to 1. (C) Schematic diagram of bicistronic reporter mRNA RFr303. (m7G) 7-Methylguanosine; [Poly(A)20] 20-nt-long poly(A) tail. (D) Pinealocytes were treated with ISO and transiently transfected with the capped bicistronic reporter mRNA RFr303 at intervals and incubated for 2 h. The medium was then exchanged with complete medium, and the cells were incubated for a further 4 h before harvesting at the indicated times. Cell lysates were then prepared and subjected to luciferase assays, Northern blotting, immunoblotting, and melatonin assays. (E) The ratio of Fluc/Rluc activities at zero time was set to 1. Intensities of AANAT mRNA and its protein levels were quantitated with a densitometer, normalized to ribosomal protein large subunit 32 (RPL32) mRNA and the actin signals, respectively, and plotted as a percentage of the maximum. Corresponding melatonin values are expressed as a percentage of the maximum of net synthesis using a melatonin ELISA kit. All results are expressed as the mean ± SD of three different experiments.
One mechanism of translational regulation is an internal ribosome-entry site (IRES), which allows ribosomes to bind mRNA in a cap-independent manner (Hellen and Sarnow 2001; Vagner et al. 2001; Gebauer and Hentze 2004). Since the discovery of viral IRESes (Jang et al. 1988; Pelletier and Sonenberg 1988), various cellular mRNAs have been shown to contain IRESes (http://www.rangueil.inserm.fr/iresdatabase). IRES-mediated translation, distinct from the canonical cap-dependent scanning model, is widely used by specific cellular mRNAs to regulate protein synthesis under subtle physiological circumstances (Vagner et al. 2001; Stoneley and Willis 2004), such as apoptosis, cell cycling, development, and differentiation. Here, we elucidate the molecular mechanisms for circadian rhythm-dependent AANAT translation, and provide a novel aspect for framing the circadian rhythmicity of vertebrate melatonin.
Results
Circadian rhythm-dependent AANAT IRES activity
Translational regulation frequently involves 5′ untranslated regions (UTRs) (Hellen and Sarnow 2001; Vagner et al. 2001; Gebauer and Hentze 2004). To investigate the translational regulation of AANAT mRNA, we determined two types of 5′UTRs from rat (Supplementary Fig. S1) and sheep (Supplementary Fig. S2) AANAT mRNAs that show quite discrepant mRNA profiles (Coon et al. 1996; Klein et al. 1997; Ganguly et al. 2002; Ackermann et al. 2006) during the circadian cycle (see Supplemental Material for details). The newly identified nucleotide sequences of rat and sheep AANAT 5′UTRs have been deposited in GenBank with the accession numbers DQ075321 and DQ839412, respectively.
To determine whether the AANAT 5′UTR shows IRES activity in pinealocytes, we generated bicistronic reporters (Fig. 1A; Jang et al. 1988; Pelletier and Sonenberg 1988; Vagner et al. 2001; Kim et al. 2003). In these constructs, the translation of Renilla luciferase (RLuc) from the first cistron is directed by cap-dependent ribosome scanning, but induction of firefly luciferase (FLuc) translation is directed by the IRES element in the intercistronic region. Both rat and sheep AANAT 5′UTRs (r303 and s234, respectively) enhanced Fluc translation by approximately fourfold, compared with those produced without the 5′UTR (pRF) (Fig. 1B, open bar in bottom panel). These results suggest that the AANAT mRNA contains a potential IRES element within its 5′UTR. To verify that the AANAT IRES functions under physiological conditions, we used isoproterenol (ISO) (Gastel et al. 1998; Kim et al. 2005). Treatment of primary rat pinealocytes with ISO elicits activation of β1 adrenergic receptor-mediated cyclic AMP signaling, which mediates AANAT gene expression and melatonin biosynthesis in a mode similar to that seen with nocturnal pineal glands (Fig. 1B, lane 2 in top panel; Gastel et al. 1998). The night-time environment dramatically increased AANAT IRES activity in reporters containing either rat or sheep 5′UTRs by approximately twofold, but did not influence a reporter that lacked a 5′UTR (Fig. 1B, closed bar in bottom panel; Supplementary Fig. S3). To confirm that phase-dependent AANAT translation occurs regardless of its mRNA fluctuation, we generated a bicistronic mRNA reporter (Cho et al. 2005, 2007), the capped RFr303 mRNA (Fig. 1C). Pinealocytes were treated with ISO and transiently transfected with the capped bicistronic reporter mRNA RFr303 at intervals (Fig. 1D). While translation of Rluc marginally increased under nocturnal conditions (data not shown), AANAT IRES-mediated translation was dramatically enhanced with a peak at 8 h after ISO treatment and declined gradually thereafter (Fig. 1E, top panel). Surprisingly, the rhythmicity of AANAT IRES activity was coupled with endogenous AANAT protein levels (Fig. 1E, middle panel) parallel to the profile of melatonin production (Fig. 1E, bottom panel). These results suggest the closed relationship between the rhythmic IRES-mediated translation of AANAT mRNA and the circadian melatonin production.
Insensitivity of AANAT translation to rapamycin
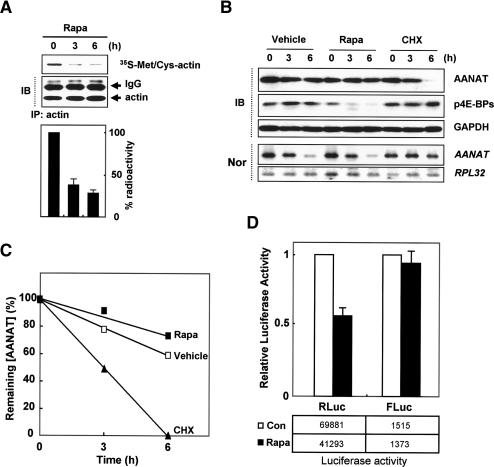
To gain insight into the mechanism by which rat AANAT mRNA is translated at night, we inhibited the mTOR (mammalian target of rapamycin) pathway in pinealocytes under nocturnal conditions induced by ISO (Gastel et al. 1998; Kim et al. 2005). Rapamycin causes inhibition of cap-dependent translation by inducing hypophosphorylation of eIF4E-binding proteins (4E-BPs) (Pyronnet et al. 2000; Gingras et al. 2004; Hay and Sonenberg 2004). General inhibition of protein synthesis (Kullmann et al. 2002) by rapamycin was confirmed in our study by metabolic labeling under nocturnal conditions; for example, the de novo synthesis rate of actin was reduced to ∼35% of control level (Fig. 2A). However, rapamycin had no effect on the AANAT protein kinetics or mRNA levels, and even marginally up-regulated AANAT protein expression (Fig. 2B,C). To exclude the possibility that the apparent insensitivity of AANAT kinetics to rapamycin was due to AANAT stabilization via β-adrenergic signaling (Gastel et al. 1998), we treated rat pinealocytes with the general translation inhibitor cycloheximide. Cycloheximide induced a dramatic decrease in AANAT protein levels despite the ISO-mediated stabilization of AANAT protein (Gastel et al. 1998) and increased AANAT mRNA stability (Fig. 2B,C; Bernard et al. 1997), which suggests that ongoing AANAT translation is required for the maintenance of its nocturnal protein kinetics. Moreover, rapamycin had no effect on the overall expression profile of AANAT protein in this assay, but rather slightly increased AANAT protein levels despite the hypophosphorylation of 4E-BP (Supplementary Fig. S4). Importantly, these findings can be recapitulated with the expression of bicistronic reporter containing the AANAT 5′UTR for IRES activity. Rapamycin treatment reduced expression of the upstream Rluc reporter, but had no effect on the induction of downstream Fluc expression in the presence of the rat AANAT 5′UTR (pRFr303) (Fig. 2D). Taken together, these data demonstrate that de novo synthesis of AANAT protein is mediated by a cap-independent translation mechanism via an IRES element within its 5′UTR.
Figure 2.
An IRES element of the AANAT 5′UTR confers resistance to cap-dependent translation repression. (A) Primary pinealocytes were treated with rapamycin (Rapa), cycloheximide (CHX), and vehicle after 8 h of ISO treatment (0 h) and then harvested at the indicated times. Down-regulated de novo synthesis of actin under Rapa treatment was seen in radiograms (top panel) as well as radioactivity of 35S-Met/Cys-actin in ISO-treated rat pinealocytes (bottom panel). (IB) Immunoblotting; (IP) immunoprecipitation. (B) Pinealocytes were subjected to Northern blotting using the AANAT coding region and RPL 32 cDNA as probes, and subjected to immunoblotting with specific antibodies as depicted on the right. (C) For kinetic analysis of AANAT protein, AANAT intensities were quantitated with a densitometer, normalized to the GAPDH signals, and plotted against the duration of drug treatment. (p4E-BPs) Phosphorylated eIF4E-binding proteins; (Nor) Northern blotting. (D) Pinealocytes were transfected with bicistronic reporter plasmid pRFr303 and treated with (Rapa) or without (Con) rapamycin after 4 h of ISO treatment, and further incubated for 12 h before harvesting. The cell lysates were prepared and subjected to luciferase assays. The activities of firefly and Renilla luciferases are depicted in boxes below the graph. The results are expressed as the mean ± SD of two different experiments.
AANAT mRNA contains an IRES element within its 5′UTR
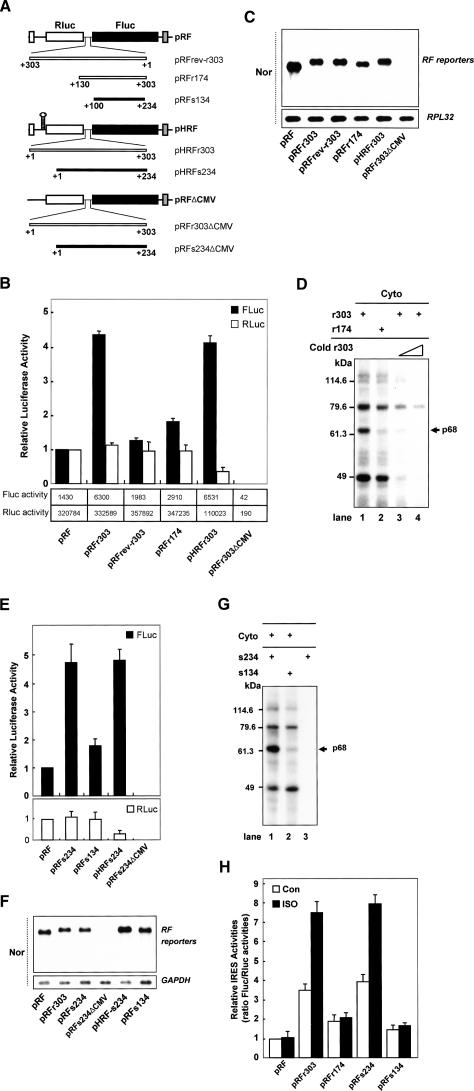
The induction of Fluc translation by AANAT 5′UTRs (Figs. 1B [bottom panel], 3B,E) was shown by Northern blotting not to be caused by altered mRNA stability, transcription, or the presence of cryptic promoter activity (Fig. 3C,F). Moreover, Fluc activity was not detected when promoter-lacking constructs (Fig. 3A, r303ΔCMV and s234ΔCMV) were used (Fig. 3B,E), confirming that the AANAT 5′UTR does not contain any cryptic promoters. To exclude alternative translational mechanisms, such as reinitiation, a plasmid (pHRF) was used in which a palindrome was inserted upstream of the Rluc gene (Fig. 3A, pHRFr303 and pHRFs234; Cho et al. 2005). The hairpin reduced expression of the upstream Rluc cistron, but had no effect on the induction of Fluc expression in the presence of the AANAT 5′UTRs (Fig. 3B,E, open bar). This argues against reinitiation as a possible mechanism. Moreover, an inverted sequence of the rat AANAT 5′UTR (rev-r303) failed to enhance FLuc activity, confirming lack of IRES activity in random sequences inserted into the intercistronic region of the bicistronic messenger (Fig. 3B, pRFrev-r303).
Figure 3.
AANAT mRNAs contain a highly conserved IRES element within their 5′UTRs among species. (A) Schematic diagram of bicistronic reporter plasmids. The AANAT 5′UTRs and their deletions were inserted into the intercistronic region of pRF, pHRF, and promoterless pRFΔCMV vectors. (B,E) AANAT 5′UTRs contain an IRES element. Pinealocytes were transfected with the pCMV·SPORT-β-gal control vector and bicistronic plasmids containing the rat or sheep AANAT 5′UTRs and their derivatives. Both Fluc (closed bar) and Rluc (open bar) activities were normalized to β-galactosidase and expressed relative to the values obtained from pRF, which was set to 1. The activities of firefly and Renilla luciferases are depicted in boxes below the graph in B. (C,F) Northern blot analysis of bicistronic mRNA expression. HEK-293T cells were transfected with β-gal as a control and various bicistronic plasmids containing the rat or sheep AANAT 5′UTRs and their derivatives, and incubated for 48 h before harvesting. Cell lysates were prepared and subjected to Northern blotting (Nor). Purified mRNA (2 μg) was hybridized with a specific probe for the Fluc coding region and loading controls GAPDH or RPL32. (D,G) The 68-kDa protein (p68) specifically binds to the AANAT IRES element. Radiolabeled riboprobes and cytoplasmic lysates (Cyto) extracted from HEK-293T cells were incubated with or without cold competitor, and subjected to UV cross-linking. Molecular mass markers are indicated on the left in kilodaltons. The arrow on the right indicates the position of p68. (H) Essential role of the AANAT IRES element in the nocturnal enhancement of the IRES activity. Transfection of bicistronic reporter plasmids into pinealocytes was performed, and the cells were incubated for 48 h and then treated with (ISO, closed bar) or without (Con, open bar) ISO and incubated for a further 12 h before harvesting. Cell lysates were prepared and subjected to luciferase assay. The ratio of Fluc/Rluc activities was expressed relative the values obtained from pRF in “Con,” which was set at 1. Each value represents the mean ± SD of five independent experiments.
Next, we generated reporter constructs containing deletions of either rat or sheep AANAT 5′UTRs (Fig. 3A, pRFr174 and pRFs134) in order to identify the functional element of the AANAT 5′UTR that is responsible for IRES activity. Pinealocytes transfected with both pRFr174 and pRFs134 exhibited Fluc activity similarly to those transfected with pRF (Fig. 3B,E). Interestingly, the 5′ end of the AANAT 5′UTR contains a purine-rich sequence and is highly conserved among vertebrate species (data not shown). Both human and bovine AANAT 5′UTRs also contain the homologous regions for IRES activity. IRES activity in the human and bovine is similar in structure and function to IRES activity in the rat or sheep (T.D. Kim and K.T. Kim, unpubl.). These results suggest that the 5′ end of the AANAT 5′UTR is essential for the AANAT IRES activity and that IRES-mediated AANAT translation is a common mechanism of regulation among vertebrate species.
Importantly, IRES-mediated translation usually requires IRES trans-acting factors (ITAFs) that regulate ribosome recruitment (Vagner et al. 2001; Stoneley and Willis 2004). To analyze the relationship between AANAT IRES function and the binding patterns of cellular proteins to the AANAT 5′UTR, we performed UV cross-linking assays. Both rat and sheep AANAT 5′UTRs specifically bound to four major cellular proteins in a similar manner (Fig. 3D [lanes 1,3,4], F [lane 1]). Among them, a 68-kDa protein (p68) showed strong binding to full-length r303 and s234, but not to the deletion constructs r174 and s134 (Fig. 3D,G, lane 2), which showed weak IRES activities. Consistent with AANAT IRES activity, UV cross-linking assays showed that p68 specifically bound to the 5′ end of the AANAT 5′UTR essential for the IRES activity. Moreover, the r174 and s134 constructs showed little induction of Fluc translation in spite of the nocturnal conditions in pinealocytes (Fig. 3H, closed bar). This suggests that p68 plays a major role in the nocturnal AANAT translation via the IRES element as an ITAF despite the possibility that additional factors are involved in the process. It was therefore of interest to identify p68.
Identification of p68 as heterogeneous nuclear ribonucleoprotein Q (hnRNP Q)
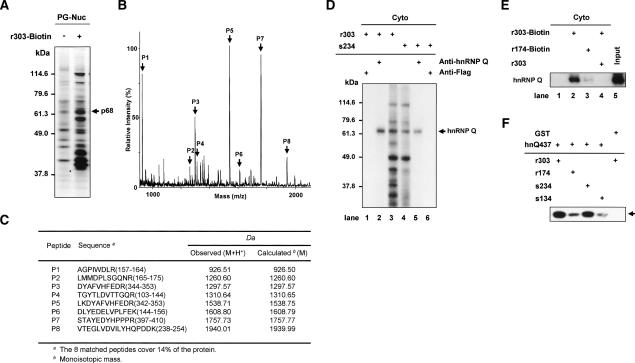
To isolate p68 interacting with the AANAT 5′UTR, we generated a biotinylated rat AANAT 5′UTR (r303-Biotin) by in vitro transcription and performed RNA affinity chromatography with nuclear extracts from rat pineal glands and the biotinylated RNA. After precipitation and washing of the RNA–protein complex, RNA-bound proteins were analyzed by SDS-PAGE (Fig. 4A). The p68 was excised from the gel and analyzed by peptide mass fingerprinting via MALDI-TOF. Figure 4B shows the MALDI mass spectrum of the peptides generated by trypsinization of the p68. The obtained masses were compared with those of proteins in the SWISS-PROT database by use of the MS-Fit peptide mass search program. As shown in Figure 4C, the peptides exhibited molecular masses that were in good agreement with the theoretically predicted tryptic peptides of hnRNP Q. The analyzed peptides covered 14% of the mouse hnRNP Q sequence. On the basis of these results, we concluded that the 68-kDa protein is hnRNP Q.
Figure 4.
Identification of p68 as hnRNP Q. (A–C) Nuclear extracts from pinealocytes (PG-Nuc) were incubated with or without a biotinylated probe (r303-Biotin), and then the resin-bound proteins were fractionated by SDS-PAGE, and silver stained in A. Peptide mass fingerprinting via MALDI-TOF was performed. (B) The peaks labeled with arrows match the estimated tryptic peptide masses of hnRNP Q with an error of <50 ppm. (C) Oligopeptide sequences from mass fingerprinting analysis. (D) Cytoplasmic extracts labeled by UV cross-linking with radiolabeled AANAT 5′UTRs were subjected to immunoprecipitation with antibodies against hnRNP Q or Flag as a control, separated by SDS-PAGE, and autoradiographed. (E) RNA pull-down experiments were performed with cytoplasmic extracts of rat pineal glands and biotinylated RNA (r303-Biotin and r174-Biotin) in the presence or absence of competitor r303. Purified proteins were immunoblotted using anti-hnRNP Q antibody. Cytoplasmic extract (Cyto) was loaded onto the “Input” lane. (F) The radiolabeled riboprobes (r303, r174, s234, and s134) were incubated with purified hnRNP Q (hnQ437) or GST, subjected to UV cross-linking analysis, and autoradiographed.
The identity of hnRNP Q was confirmed by immunoprecipitation of UV cross-linked proteins with radio-labeled r303 and s234 (Fig. 4D, lanes 3,4, respectively) using anti-hnRNP Q antibody. HnRNP Q was detected by the antibody, suggesting that the protein interacts directly with the AANAT 5′UTR (Fig. 4D, lanes 2,5). No bands were detected when an anti-Flag antibody was used as a negative control (Fig. 4D, lanes 1,6). RNA pull-down experiments were performed by using cytoplasmic extracts of rat pineal glands and biotinylated AANAT 5′UTRs. Immunoblot analyses of purified proteins showed that hnRNP Q is specifically associated with the 5′UTR (Fig. 4E, lanes 2,4), whereas the truncated r174 is weakly associated with the protein (Fig. 4E, lane 3). Moreover, direct binding between hnRNP Q and the AANAT 5′UTR was assessed by UV cross-linking with a purified hnRNP Q (hnQ437) and 32P-labeled riboprobes. Purified hnRNP Q interacted strongly with r303 and s234 RNAs but only weakly interacted with r174 or s134 RNA (Fig. 4F), consistent with the deletion analyses (Fig. 3D,G). Taken together, these results demonstrate that hnRNP Q directly interacts with the 5′ end of the AANAT IRES.
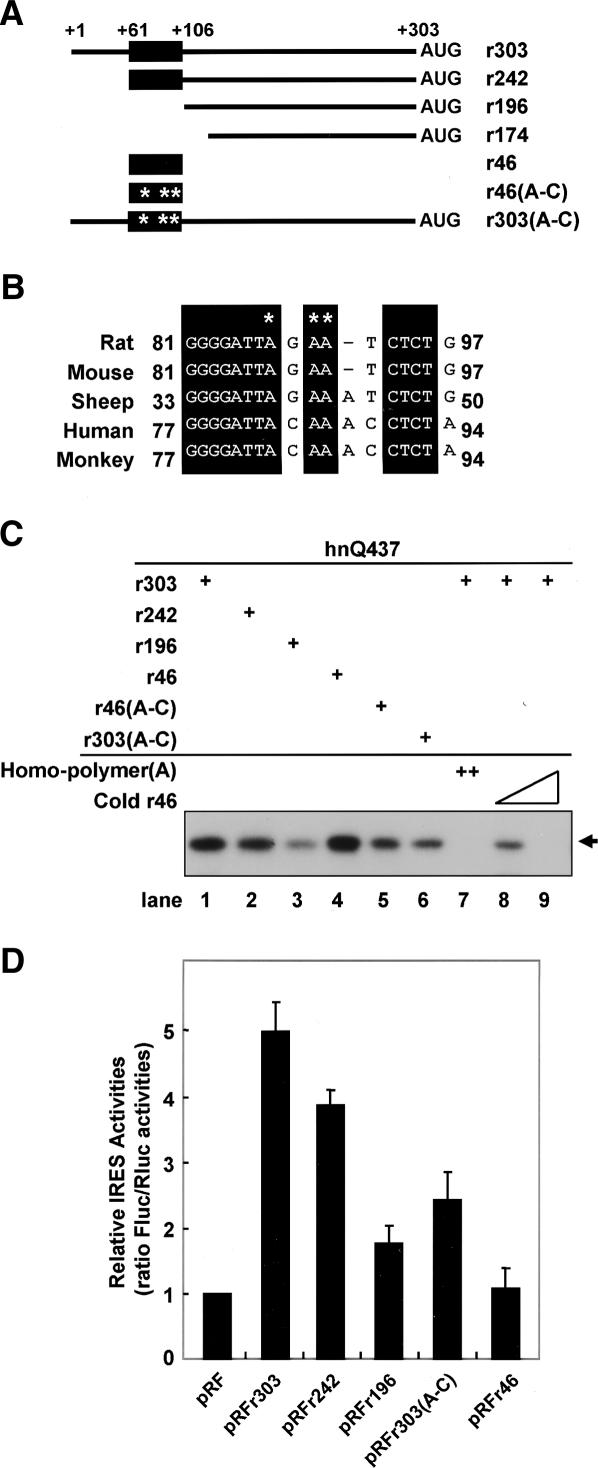
HnRNP Q specifically interacts with the 46-nucleotide (nt)-long element within the rat AANAT 5′UTR
In order to identify the region of the AANAT IRES that interacts with hnRNP Q, we generated serially deleted RNAs (Fig. 5A) and then performed UV cross-linking experiments. As shown in Figure 5C, purified hnRNP Q strongly bound to riboprobes containing the 46-nt-long region corresponding to nucleotides 61–106 of the rat AANAT IRES (Fig. 5C, lanes 1,2,4). In contrast, hnRNP Q bound only weakly to the probe r196 lacking the 46-nt-long region (Fig. 5C, lane 3). These results were further confirmed by competition assays. The interaction between hnRNP Q and probe r303 was strongly inhibited by poly(A) and unlabeled RNA probe r46 (Fig. 5C, lanes 7–9). The binding pattern for purified hnRNP Q was in good agreement with the binding patterns of the 68-kDa protein to the AANAT IRES observed by UV cross-linking with cytoplasmic extract from HEK-293T cells (Supplementary Fig. S5), suggesting that hnRNP Q directly interacts with the AANAT IRES and that a specific RNA–protein interaction can occur in cells in which numerous proteins exist together with hnRNP Q. The RNA-binding data demonstrate that the AANAT RNA fragment spanning nucleotides 61–106 is necessary and sufficient for the interaction with hnRNP Q. Moreover, homopolymeric RNA competition results and a previous report showed that hnRNP Q has a strong affinity for adenosine-rich RNAs (Mizutani et al. 2000; Mourelatos et al. 2001; Bannai et al. 2004; Kim et al. 2004). Interestingly, the 46-nt-long element contains highly conserved sequences involving an adenosine-rich region among species (Fig. 5B). In order to confirm that the 46-nt-long region participates in direct interaction with hnRNP Q, we generated a mutant construct in which adenosines of the element were replaced with cytosines (Fig. 5B, asterisks). The effect of the site-directed mutagenesis on hnRNP Q binding was measured by a UV cross-linking experiment. In these experiments, the binding affinity of the mutated RNA was dramatically reduced to a similar level as shown with the AANAT RNAs lacking the 46-nt-long element (Fig. 5C, lanes 3–6). Taken together, these data indicate that some or all of the adenosine residues in the 46-nt-long element play a key role in the interaction of the AANAT IRES with hnRNP Q.
Figure 5.
hnRNP Q specifically interacts with an adenosine-rich region in the 5′ end of the AANAT IRES. (A) Schematic diagram of the full-length rat AANAT IRES and its various mutant RNAs used for UV cross-linking experiments and monitoring efficiency of IRES-dependent translation in vivo. The conserved hnRNP Q-binding region among vertebrate species is represented as a closed box. Their locations in the AANAT 5′UTR are indicated as numbers from the 5′ end. (B) Three adenosine residues, within the conserved hnRNP Q-binding element, were replaced with cytosine residues. The asterisk denotes the nucleotide changes in the hnRNP R-binding region. Accession numbers for the AANAT 5′UTRs shown are as follows: rat, DQ075321; mouse, NM_009591 and U83462; human, U40347 and U40391; monkey, XM_523725; and sheep, DQ839412. (C) Radiolabeled riboprobes and purified hnRNP Q (hnQ437) were incubated with or without competitor RNAs [homopolymer(A) or cold r46], and subjected to UV cross-linking. (D) Rat pinealocyte cells were transfected with bicistronic reporter plasmids and incubated for 48 h before harvest. Cell lysates were prepared and subjected to luciferase assays. The ratio for the empty vector pRF was set to 1.
To further characterize the IRES element and to investigate the effect of the hnRNP Q-binding site on the AANAT IRES activity, we analyzed the effect of deletions in the 5′UTR of AANAT mRNA on translation in primary pinealocytes. Truncation of the IRES element from nucleotides 61–106 (pRFr196) dramatically diminished IRES activity, indicating that the 46-nt element is required for maximal IRES activity. Moreover, the mutation in the AANAT IRES [pRFr303(A–C)] showed a decreased IRES activity to <50% (Fig. 5D). These results were correlated with the binding affinity of the AANAT IRES to hnRNP Q as shown in Figure 5C. Interestingly, the 46-nt-long region containing the hnRNP Q-binding domain had only minimal IRES activity (Fig. 5D, pRFr46), indicating that additional factors, not identified yet, are necessary to function as cofactors in the AANAT IRES activation. Taken together, we concluded that the characterized hnRNP Q-binding region is essential for IRES activity of the AANAT 5′UTR.
Rhythmic control of AANAT IRES activity by hnRNP Q
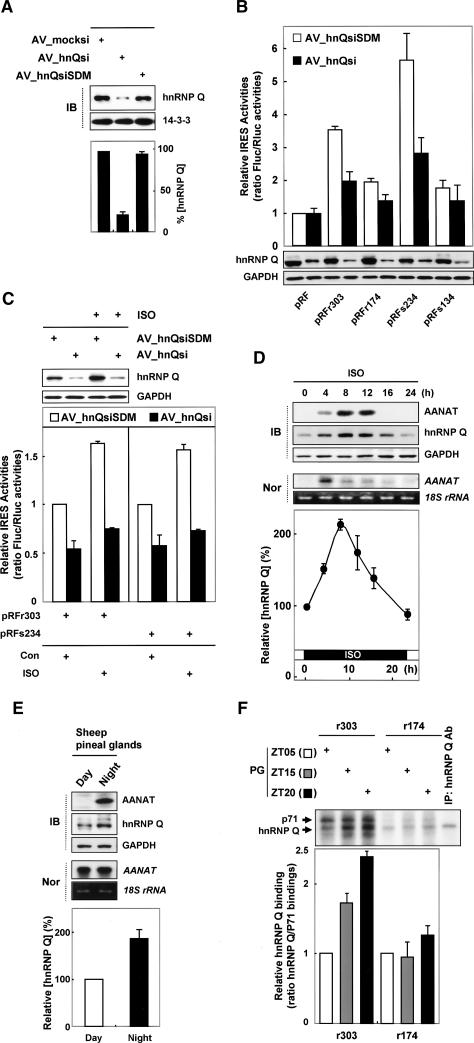
HnRNP Q is involved in various aspects of mRNA metabolism, such as pre-mRNA splicing (Mourelatos et al. 2001), degradation of AANAT mRNA (Kim et al. 2005) and c-fos mRNA (Grosset et al. 2000), and HCV IRES-mediated translational enhancement (Kim et al. 2004). Here, the role of hnRNP Q in AANAT mRNA translation was determined by RNA interference (RNAi) using the adenoviral RNAi system (Kuninger et al. 2004) in the rat pinealocytes. To create the adenoviral RNAi system, we generated adenoviral particles expressing small interfering RNA (siRNA) against hnRNP Q (AV_hnQsi) and mutated (AV_hnQsiSDM) or no (AV_mocksi) siRNA sequence (see Materials and Methods for details). Pinealocytes transiently transduced with AV_hnQsi showed a dramatic decrease in hnRNP Q level compared with those transduced with AV_hnQsiSDM and AV_mocksi (Fig. 6A). Knockdown of the hnRNP Q level dramatically decreased IRES activity of the AANAT IRES-element-containing mRNAs (r303 and s234) (Fig. 6B, closed bar). Importantly, pinealocytes lacking hnRNP Q protein showed little enhancement of the AANAT IRES activities despite nocturnal conditions (Fig. 6C, closed bar). In contrast, adenoviruses containing the mutated siRNA of hnRNP Q (AV_hnQsiSDM) had no effect on AANAT IRES activity (Fig. 6B,C, open bar).
Figure 6.
hnRNP Q mediates rhythmic AANAT translation via the IRES element. (A) Pinealocytes were transduced by adding adenoviral particles expressing siRNA against hnRNP Q (AV_hnQsi) and mutated (AV_hnQsiSDM) or no (AV_mocksi) siRNA sequence, and the cells were incubated for 48 h before harvesting. Cell lysates were prepared and subjected to immunoblotting. Intensities of hnRNP Q level were quantitated with a densitometer, normalized to the 14–3–3 signals, and plotted as a percentage. (B,C) Transfections of pinealocytes with bicistronic reporter plasmids were performed by overlaying the cells with a precipitate of Metafectene and reporter DNAs for 20 min before adding adenoviral particles expressing siRNA against hnRNP Q (AV_hnQsi) or mutated siRNA sequence (AV_hnQsiSDM). The cells were incubated for 48 h and treated with or without ISO for a further 12 h before harvesting. Cell lysates were prepared and subjected to immunoblotting using antibodies against hnRNP Q and GAPDH. The ratio of Fluc/Rluc activities was expressed relative to the values obtained from pRF in “Con,” which was set at 1. The values represent the mean ± SD of four transfections. (D) Rat pinealocytes were treated with ISO and then harvested at the indicated times. Cells lysates were subjected to Northern blotting and immunoblotting. For kinetic analysis of hnRNP Q protein, hnRNP Q intensities were normalized to the actin signals, and plotted as a percentage. The value at zero time was set to 100%. (E) Sheep were field-maintained (11 h light, 13 h dark). The day sheep (Day) in this group were sacrificed at noon, and the night sheep (Night) at midnight. Cell lysates extracted from sheep pineal glands were subjected to Northern blotting and immunoblotting. hnRNP Q intensities were normalized to the GAPDH signals, and plotted as a percentage. The value at the daytime (Day) was set to 100%. (F) Representative UV cross-linking using cell lysates extracted from rat pineal glands (PG) isolated at different time points as indicated (ZT05, ZT15, and ZT20) and radiolabeled riboprobes (r303 and r174). The UV cross-linked complexes were immunoprecipitated with anti-hnRNP Q antibody (lane “IP”). The samples were fractionated by SDS-PAGE and autoradiographed. The arrows on the left indicate the 71-kDa protein (p71) and hnRNP Q. hnRNP Q intensities were normalized to the p71 signals, and plotted as the ratio of hnRNP Q/p71 bindings. The value at ZT05 was set to 1. Each result represents the mean ± SD of two independent experiments.
To further analyze the role of hnRNP Q in AANAT IRES-dependent translation, we additionally used an overexpression approach in HEK293T cells. HnRNP Q overexpression augmented the IRES activity of r303containing mRNA more than twofold, but did not influence that of rev-r303-containing mRNA (Supplementary Fig. S6A,B). The activity of the control, polioviral IRES (polio), was not affected by hnRNP Q, as has been described elsewhere (Supplementary Fig. S6B,D, pRFpolio; Kim et al. 2004; Cho et al. 2005, 2007). The integrity and abundance of the reporter mRNAs under knockdown of hnRNP Q level were confirmed by Northern blots (Supplementary Fig. S6D, bottom panels). Taken together, these results suggest that hnRNP Q plays an essential role in the activation of the AANAT IRES.
We next investigated the physiological relevance of rhythmic IRES-mediated AANAT translation to circadian expression of hnRNP Q. In vivo hnRNP Q is expressed in both rat and sheep pineal glands in a circadian rhythm-dependent manner (Fig. 6D,E, respectively; Kim et al. 2005). HnRNP Q displayed relatively significant basal levels at all times, which reach a maximum at ∼8 h after ISO treatment, to about two times that observed at zero time, and they gradually decline to basal levels in rat pinealocytes (Fig. 6D). Moreover, the night-time hnRNP Q level in sheep pineal glands increases in a mode similar to that seen with rat pineal glands (Fig. 6E). Thus, it is expressed concurrently with AANAT protein under nocturnal conditions (Fig. 6D,E). Interestingly, interactions between r303 and hnRNP Q were nocturnally enhanced in parallel with the hnRNP Q level. In contrast, truncated r174 showed no changes in its binding to hnRNP Q (Fig. 6F). These results indicate that the nocturnal enhancement of AANAT IRES activity is mediated by the rhythmic expression of hnRNP Q, with a peak in the middle of night.
Essential role of rhythmic AANAT translation in robust melatonin production
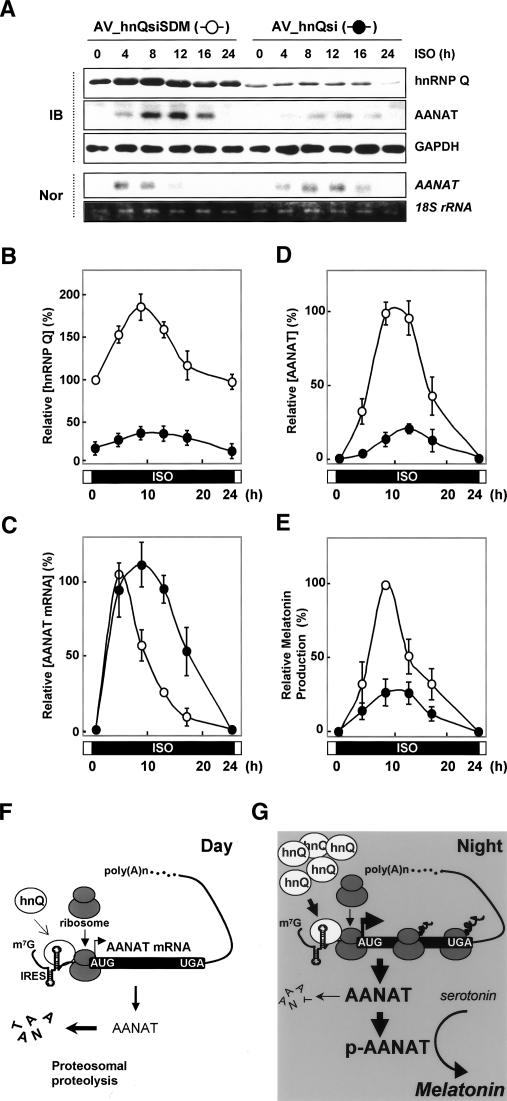
To verify that the circadian rhythmicity of the AANAT protein level is due to hnRNP Q-mediated AANAT IRES activities, we performed a knockdown experiment of hnRNP Q in pinealocytes. Adenoviral particles expressing siRNA against hnRNP Q (AV_hnQsi) efficiently reduced the hnRNP Q level. Time course experiments of hnRNP Q in AV_hnQsiSDM-transduced pinealocytes showed that its level reaches a maximum after 8 h of ISO stimulation and declines gradually thereafter (Fig. 7A,B, open circle). Similarly, the reduced hnRNP Q level in AV_hnQsi-transduced cells marginally fluctuated in a circadian rhythm-dependent manner (Fig. 7A,B, closed circle). The reduced hnRNP Q level elicited an increase of peak amplitude and a delay of peak time in the AANAT mRNA profile as previously reported (Kim et al. 2005; Fig. 7C, closed circle). Surprisingly, the AANAT protein levels showed a dramatic decrease of peak amplitude despite sustained high levels of AANAT mRNA in hnRNP Q-reduced cells (Fig. 7D, closed circle). Moreover, the melatonin production profile in hnRNP Q-attenuated pinealocytes was concomitant with the reduced level of AANAT protein (Fig. 7E, closed circle). Importantly, these results provide the first evidence that the circadian timing of AANAT protein expression by hnRNP Q is essential for the robustness of melatonin rhythmicity.
Figure 7.
Essential role of rhythmic AANAT translation in robust melatonin production. (A–D) Pinealocytes were transduced with adenoviral particles expressing either siRNA against hnRNP Q (AV_hnQsi, closed circle) or mutated siRNA sequence (AV_hnQsiSDM, open cirlce), incubated for 48 h, and treated with ISO before harvesting at the indicated times. (A) Cell lysates were extracted and subjected to immunoblotting (IB) and Northern blotting (Nor). The intensities of proteins (hnRNP Q in B and AANAT in D) and AANAT mRNA in C were quantitated with a densitometer, normalized to the GAPDH and 18S rRNA signals, respectively, and plotted as a percentage. The maximum value in the AV_hnQsiSDM-transduced pinealocytes was set to 100%. (E) Corresponding melatonin values are expressed as a percentage. The maximum value in the AV_hnQsiSDM-transduced pinealocytes was set to 100%. The values represent the mean ± SD of four independent experiments. (F,G) Proposed model for rhythmic AANAT translation as a key regulatory mechanism in nocturnal melatonin production. The circadian timing of AANAT protein expression can be explained by two cooperative nocturnal processes: (1) IRES-mediated AANAT synthesis up-regulated by rhythmic hnRNP Q, which peaks in the middle of the night; and (2) post-translational AANAT phosphorylation mediated by β-adrenergic signaling, which is required for its stability and enzymatic activity in melatonin biosynthesis. (hnQ) hnRNP Q; (p-AANAT) phosphorylated AANAT.
Discussion
The net amount of protein is determined by the balance between protein synthesis (translation) and protein degradation (post-translational regulation) (Gebauer and Hentze 2004; Shu and Hong-Hui 2004). As translation of mRNA into protein represents the final step in the gene expression pathway, its regulation is an important mechanism that enables spatiotemporal modulations of protein levels (Gebauer and Hentze 2004). The molecular mechanism that governs melatonin rhythmicity is based on the nocturnal increment of AANAT protein (Klein et al. 1997; Ganguly et al. 2002). In this study, we demonstrate that the rhythmic control of AANAT IRES activity by hnRNP Q is an indispensable mechanism for the robustness of circadian melatonin rhythmicity.
HnRNP Q shows multiple functions in the circadian rhythmicity of AANAT expression: It has been identified as a factor involved in the destabilization (Kim et al. 2005) and IRES-mediated translation of AANAT mRNA via its 3′UTR and 5′UTR, respectively. This raises an interesting question of whether specific functions of hnRNP Q are a consequence of its binding to distinct A-rich sequences in the 3′UTR or 5′UTR of AANAT mRNA and/or of additional binding proteins in rat pinealocytes. Most RNA-binding proteins show the broad range of RNA-binding specificity and exhibit diverse biological roles that are general characteristics of hnRNPs (Dreyfuss et al. 1993; Wilkinson and Shyu 2001; Dreyfuss et al. 2002), as an example shown by the functions of hnRNP E1/E2 in translational inhibition and stabilization of specific mRNAs in hematopoiesis (Weiss and Liebhaber 1994; Ostareck et al. 1997; Ostareck-Lederer et al. 1998; Ostareck-Lederer and Ostareck 2004). The binding region of hnRNP Q in the AANAT 5′UTR is the 46-nt-long region that contains adenosine-rich sequence, but shows no sequence homology with the rat AANAT 3′UTR. Since hnRNP Q can bind the rat AANAT 3′UTR, it would be highly informative if the 3′UTR were evaluated for IRES activity. Importantly, rat AANAT 3′UTR within the bicistronic context showed no IRES activity in pinealocytes (Supplementary Fig. S7). We previously showed that hnRNP R, hnRNP Q, and hnRNP L are associated as a complex with the rat AANAT 3′UTR to mediate the AANAT mRNA degradation (Kim et al. 2005). However, overexpression of hnRNP L showed little effect on AANAT IRES activity (data not shown). Taken together, these results suggest that two distinct functions of hnRNP Q in the translation and degradation of AANAT mRNA are due to different binding partners corresponding to distinct binding sequences. Therefore, it cannot be excluded that additional factors, not identified yet, function as cofactors in the IRES-dependent translation of AANAT mRNA.
Although it is not clear how hnRNP Q augments translation of AANAT mRNA, we can speculate on several modes of action. First, hnRNP Q may conformationally change the IRES into a structure that enables it to interact with the translational machinery such as the 40S ribosomal subunit. That is, a putative hnRNP Q–ribosome interaction may assist in binding and/or guiding ribosomes to the IRES. Second, hnRNP Q may enhance IRES-mediated AANAT translation through interactions with other unknown ITAFs. Third, it is possible that hnRNP Q recruits a canonical translation factor through a putative protein–protein interaction.
Next, we had a question regarding the functional relevance of hnRNP Q to the circadian AANAT protein profile. There is no evidence of a regulatory mechanism that coordinates the two different actions through hnRNP Q in AANAT gene expression. However, AANAT mRNA degradation would be dependent on the AANAT translation, as shown in stabilization of AANAT mRNA by CHX treatment (Fig. 2B; Bernard et al. 1997). AANAT protein expression is apparently out of phase with its mRNA level after midnight. During this phase, the IRES-mediated translational enhancement could compromise the decline of AANAT mRNA levels to maintain the nocturnally high level of AANAT protein. Interestingly, the reduced hnRNP Q level in AV_hnQsi-transduced pinealocytes showed a tendency to induce a putative lag of peak time in AANAT protein level (Fig. 7A,D, closed circle). This suggests the possibility that the delayed shift of the AANAT protein profile is due to translation of sustained high levels of AANAT mRNA with a delayed peak time in hnRNP Q-reduced cells. In summary, the nocturnal condition causes an increase of hnRNP Q in rat pinealocytes. Up-regulated hnRNP Q concentration-dependently binds to the 3′UTR or 5′UTR of AANAT mRNA according to the specific binding sequence with presumably additional binding partners. Distinct complexes regulate the stability and translation of AANAT mRNA via its 3′UTR and 5′UTR, respectively. HnRNP Q causes the remarkable differences in AANAT mRNA profiles via their species-specific 3′UTRs (Kim et al. 2005), but it induces convergence of nocturnal AANAT protein elevation via its highly conserved 5′UTR among species. Thus, hnRNP Q plays a key role in controlling the circadian rhythmicity of AANAT expression at the level of post-transcriptional regulation. Now, the nocturnal increment of AANAT protein can be explained by two cooperative nocturnal processes (Fig. 7F,G): (1) translational regulation—IRES-mediated AANAT synthesis regulated by rhythmic hnRNP Q, which peaks in the middle of the night; and (2) post-translational regulation—AANAT phosphorylation mediated by mainly adrenergic signaling, which is required for its stability and enzymatic activity in melatonin biosynthesis (Gastel et al. 1998; Ganguly et al. 2001; Obsil et al. 2001; Zheng et al. 2003; Choi et al. 2004).
Circadian rhythms are fundamental biological phenomena in most living organisms (Dunlap 1999; Harms et al. 2004; Bell-Pedersen et al. 2005). The molecular machinery that governs the rhythmicity is based on circadian phase-specific expressions of clock proteins (Harms et al. 2004; Shu and Hong-Hui 2004). Importantly, there is still an open question in clock gene expression—the regulatory mechanisms mediating the dissociation between mRNA and protein level. The present study reveals a yet unappreciated level of regulation within the core mechanism of circadian melatonin production: rhythmic IRES-mediated AANAT synthesis. Similar circadian rhythm-dependent translation could possibly function as rhythmic switches in other clock genes that have discrepancies between their mRNA and protein oscillation profiles, as the 5′UTRs of various clock genes also appear to have IRES activities (K.H. Lee, D.Y. Kim, T.D. Kim, and K.T. Kim, unpubl.). Thus, this example of rhythmic AANAT translation defines a new paradigm for circadian phase-dependent protein oscillation in the biological clock system.
Materials and methods
Rats and analyses of primary pinealocyte cells
Rats were maintained in a controlled environment (12 h light, 12 h darkness; lights on Zeitgeber time, ZT, ZT0–12). Dissociated primary pinealocyte cells were prepared and maintained as previously described (Han et al. 2005; Kim et al. 2005). To determine the de novo protein synthesis rate under rapamycin treatment, metabolic radiolabeling experiments were performed as described previously (Cho et al. 2005). Treatment of rat pinealocytes with norepinephrine or the β-adrenergic agonist ISO induces AANAT expression and melatonin biosynthesis in a mode similar to that seen with nocturnal pineal glands (Gastel et al. 1998; Han et al. 2005; Kim et al. 2005). In brief, pinealocytes were treated with 5 μM ISO for 8 h before adding 5 μg/mL actinomycin D (Act D) and 20 nM rapamycin. Cells were washed twice in methionine- and cysteine-free Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen), incubated for 60 min, and further incubated for 90 min following supplementation with 35S-labeled methionine (35S-Met) and cysteine (35S-Cys) (500 μCi/mL; NEN Life Science Products). Cells were then harvested at the times indicated relative to addition of rapamycin (0, 3, and 6 h). Cell lysates were prepared and subjected to immunoprecipitation and immunoblotting with a monoclonal antibody against actin (Cell Signaling). Immunoprecipitates were subjected to SDS-PAGE and transferred to a nitrocellulose membrane. After autoradiography, the band corresponding to actin was excised, and radioactivity was measured using a liquid scintillation counter. Incorporation of radioactivity into actin was represented by values relative to that measured at time zero, to which a value of 100% was assigned. Due to the lack of the capacity of AANAT antibody to be immunoprecipitated, the kinetics of AANAT protein were determined under rapamycin or cycloheximide treatment as described above. Pinealocytes were treated with ISO for 8 h before Act D was added in the absence or presence of rapamycin or 50 μg/mL cycloheximide, and then harvested at the indicated times (0, 3, and 6 h). Cell lysates were subjected to immunoblotting and Northern blot analyses with specific antibodies and 32P-labeled probes, respectively, as depicted in Figure 2B. For kinetic analysis of AANAT protein, its intensities were quantitated with a densitometer, normalized to the GAPDH signals, and represented by values relative to that measured at time zero, to which a value of 100% was assigned.
Transfections of pinealocytes with bicistronic reporter DNAs and pCMV · SPORT-β-gal were performed by overlaying the cells with a precipitate of 1.5 μL of Metafectene (Biontex) and 0.2 μg of DNA for 20 min before adding ∼4 × 105 adenovirus shuttle particles as previously described (Kim et al. 2005), and the cells were incubated for 48 h and then treated with or without ISO and incubated for a further 12 h before harvesting at the indicated times. Cell lysates were prepared and subjected to immunoblotting, Northern blotting, and luciferase assay. The ratio (Fluc/Rluc activities) between firefly and Renilla luciferase activities was calculated. The ratio for the empty vector pRF was set to 1. Adenovirus-mediated RNA transfection was performed as described above (Kim et al. 2005). In brief, pinealocytes were treated with ISO and transiently transfected with 0.8 μg of the capped bicistronic reporter mRNA RFr303 at intervals and incubated for 2 h; the medium was then exchanged with complete medium, and the cells were incubated for a further 4 h before harvesting at the indicated times. Cell lysates were then prepared and subjected to luciferase assays, immunoblotting, Northern blotting, and melatonin assay (Pfeffer et al. 1999). The ratio of Fluc/Rluc activities at zero time was set to 1. Intensities of AANAT mRNA and its protein levels were quantitated with a densitometer, normalized to RPL32 mRNA and the actin signals, respectively, and plotted as a percentage of the maximum. Corresponding melatonin values are expressed as a percentage of the maximum of net synthesis using a melatonin ELISA kit. Pinealocytes were transduced with adenoviral particles expressing siRNA against hnRNP Q (AV_hnQsi) or mutated siRNA sequence (AV_hnQsiSDM) and incubated for 48 h, and the cells were treated with ISO before harvesting at the indicated times. Cell lysates were extracted and subjected to immunoblotting and Northern blotting. Intensities of proteins (hnRNP Q and AANAT) and AANAT mRNA were quantitated with a densitometer, normalized to the GAPDH and the 18S rRNA signals, respectively, and plotted as a percentage. The maximum value in the AV_hnQsiSDM-transduced pinealocytes was set to 100%.
Plasmid constructions
In the absence of convenient restriction sites for cloning, standard PCR techniques were used to amplify the desired sequences. Amplification of cDNA was performed with Pfu polymerase (SolGent) and confirmed by sequencing. To construct chimeric reporter plasmids, the rat and sheep AANAT 5′UTRs were amplified from full-length rat AANAT cDNA (accession no. DQ075321) and sheep AANAT cDNA (accession no. DQ839412), respectively. The resulting products were digested with SalI and SmaI and then cloned into the SalI/SmaI site of the intercistronic region of a pRF bicistronic vector containing Renilla (Rluc) as the first cistron and firefly luciferase (Fluc) as the second cistron (Kim et al. 2003; Cho et al. 2005), yielding pRFr303 and pRFs234. To create pHRFr303 and pHRFs234 reporters, a palindrome was inserted at the NheI site upstream of the pRFr303 and pRFs234, respectively (Cho et al. 2005). To generate the deletion constructs as shown in Figures 3A and 5A and the inverse construct pRFrev-r303, the rat and sheep AANAT 5′UTR fragments were amplified from pRFr303 and pRFs234, respectively, and then inserted into the SalI/SmaI site of the mock vector pRF. For the construction of the promoter-lacking reporter plasmids pRFr303ΔCMV and pRFs234ΔCMV, the CMV promoter region was removed from pRFr303 and pRFs234 via BglII/NheI digestion, and then the DNA fragment was self-ligated. To create the construct of point mutations [pRFr303(A–C)] as shown in Figure 5, A and B, two-step PCR amplification was performed with the following primers (Kwak et al. 2006): former fragment (5′-AAGTCGACTCAGCAGGAT TGGGTCAG-3′ and 5′-GTGATGGCTCAGAGAGGCGAAT CCC-3′) and latter fragment (5′-GCAGGGCCAGGGGGGATT CGCCTCTCTGAGC-3′ and 5′-GGGCATGGGTATCTGGC CACTGACC-3′) (the flanking sequences for cloning purposes are underlined, and mutated sequences are italic). The second PCR fragment was digested and cloned in the SalI/SmaI site of the mock vector pRF. For the construction of pRFr46(A–C), PCR was performed with pRFr303(A–C) as a template and the oligomers (5′-AAGTCGACATGGGAGGCAGGGCCA-3′ and 5′-GGGTGATGGCTCAGAGAGGCGAATC-3′) (the flanking sequences for cloning purposes are underlined, and mutated sequences are italic). Then the amplified fragments were digested and inserted into the SalI/SmaI site of the mock vector pRF. To perform in vitro binding assays using AANAT 5′UTRs and cell extracts, fragments of the rat and sheep AANAT 5′UTRs were amplified as described above. The PCR products were digested with EcoRI and XbaI and then subcloned into the pSK′ vector (Kim et al. 2005) to generate pSK′-r303, pSK′-r303(A–C), pSK′-r242, pSK′-r196, pSK′-r174, pSK′-r46, pSK′-r46(A–C), pSK′-s234, and pSK′-s134. Sense strands were transcribed with T7 RNA polymerase (Roche). Plasmids pEGFP-hnRNP Q and pRFpolio have been described elsewhere (Kim et al. 2004; Cho et al. 2005, 2007). For the generation of pKJ-Tev-hnQ437 for purification of hnRNP Q (amino acids 1–437), PCR was performed with plasmid pEGFP-hnRNP Q as a template and hnRNP Q-specific primers (5′ primer, 5′-GCGCGCATATGGTGAAGATGGCG GCGGCG-3′; and 3′ primer, 5′-CCCTCGAGCTAATAATAG TAGTAATCATC-3′) (the flanking sequences for cloning purposes are underlined). The amplified DNA fragment was digested with NdeI and XhoI and cloned into pKJ-Tev (Kwon et al. 2005). For the generation of the bicistronic mRNA reporter using in vitro transcription, pCY2-RFr303 was constructed as follows: A DNA fragment composed of tandem arrays of the Renilla luciferase gene and rat AANAT 5′UTR as the intercistronic region and the firefly luciferase gene was amplified with plasmid pRFr303 as a template and primers 5′-AAAGCTTGC TAGCCACCATGACTTCG-3′ and 5′-AACTCGAGTCTAGC TAGAATTACACG-3′; this was then inserted into the HindIII/XhoI site of pCY2 (Chen et al. 2001). Sense strands were transcribed with SP6 RNA polymerase (Roche).
Adenoviral RNAi system
To create the modified adenoviral RNAi system (Kuninger et al. 2004; Cho et al. 2007), vector backbone was made by PCR amplification with two primers specific for the pShuttle-IRES-hrGFP vector (Stratagene) (5′ primer, 5′-AACTCGAGGGG TGGGAAAGAATATATA-3′; and 3′ primer, 5′-AAAGGCCT TACGCGCTATGAGTAAGTG-3′) (the flanking sequences for cloning purposes are underlined). The amplified DNA fragment was digested with XhoI and StuI. The H1 promoter fragment from the pSuper RNAi system (Oligoengine) was digested with EcoR1-T4 DNA polymerase followed by XhoI and cloned into the XhoI/StuI site of pShuttle backbone, and designated pShuttle-RNAi vector. The sequence of siRNA against hnRNP Q (hnQsi) is 5′-ACTGGAACGAGTGAAGAAG-3′ (accession no. AY034483). To generate short hairpin RNA (shRNA) against hnRNP Q, the sequences of synthesized oligomers were as follows: sense, 5′-GATCTCCACTGGAACGAGTGAAGAAGT TCAAGAGACTTCTTCACTCGTTCCAGTTTTTTGGAAA-3′; antisense, 5′-AGGTGACCTTGCTCACTTCTTCAAGTTCTCT GAAGAAGTGAGCAAGGTCAAAAAACCTTTTCGA-3′ (the flanking sequences for cloning purposes are underlined; hnQsi sequences are italicized). Two oligomers were annealed and cloned into the BglII/HindIII site of the pShuttle-RNAi vector to generate pShuttle-hnQsi. For constructing mutated shRNA against hnRNP Q, the hnQsi sequence was mutated by site-directed mutagenesis (SDM) as follows: 5′-AGTCGAACGA GTCAACAAC-3′ (mutated sequences are bold). Two oligomers were annealed and cloned into the BglII/HindIII site of the pShuttle-RNAi vector, and named pShuttle-hnQsiSDM. The next steps (Generating AdEasy recombinants of pShuttle-mocksi, pShuttle-hnQsi, and pShuttle-hnQsiSDM, and amplification of adenoviral particles containing recombinants) were performed according to the manufacturer’s recommendations (Stratagene) to generate three types of adenoviral RNAi systems, and abbreviated AV_mocksi, AV_hnQsi, and AV_hnQsiSDM, respectively.
Melatonin assay
Melatonin content in pinealocytes was measured with a melatonin ELISA kit (IBL). Homogenized samples in phosphate buffer (pH 6.8) were diluted and extracted according to the manufacturer’s instructions. The detection limit for melatonin in this assay is 1.6 pg/mL. Melatonin content was calculated against standard values provided by the manufacturer and normalized to protein content.
In vitro RNA synthesis and in vitro binding assay
For in vitro binding assays, [32P]UTP-labeled RNAs, unlabeled competitor RNA, and biotinylated RNAs were transcribed in XbaI-linearized recombinant pSK′ vectors with T7 RNA polymerase (Roche) as described elsewhere (Kim et al. 2005). For mRNA transfection, the bicistronic construct pCY2-RFr303 was linearized with EcoRI. This plasmid contains a 20-nt-long poly(A) stretch between XhoI and EcoRI restriction sites. Reporter mRNA was generated in vitro from the linearized plasmid with SP6 RNA polymerase (Roche) in the presence of the cap analog m7G(5′)ppp(5′) (Roche). UV cross-linking experiments, biotinylated RNA-affinity purification using rat pineal extracts, peptide sequencing using MALDI-TOF mass spectrometry, and immunoprecipitation of UV cross-linked proteins were performed as previously described (Kim et al. 2003, 2005; Cho et al. 2005, 2007), except that radiolabeled RNAs corresponding to AANAT IRES elements and purified hnRNP Q were used as interacting RNA and protein, respectively. Rhythmic binding of hnRNP Q to the AANAT 5′UTR was confirmed by UV cross-linking analyses between [32P]UTP-labeled RNAs and cell lysates extracted from rat pineal glands isolated at different Zeitgeber times, and calculated as the relative value to p71 binding to AANAT 5′UTRs. The ratio for ZT05 was set at 1.
Protein preparation and immunoblot analysis
Fractionation of pineal glands and HEK-293T cells into cytoplasmic and nuclear extracts was performed as previously described (Kim et al. 2005). Immunoblot analyses were performed with polyclonal anti-hnRNP Q (anti-SYNCRIP-N antibody), polyclonal anti-phospho-4EBP (Cell Signaling), monoclonal anti-EGFP (Santa Cruz Biotechnology), polyclonal anti-AANAT, monoclonal anti-actin (Cell Signaling), and monoclonal anti-GAPDH (ICN) as primary antibodies. The secondary antibodies were visualized using a SUPEX Kit (Neuronex) (Kim et al. 2005), according to the manufacturer’s instructions.
Acknowledgments
We thank Dr. D.C. Klein and Dr. S.L. Coon for critical comments, anti-AANAT antibody, sheep pineal glands, and unpublished sequence information of the sheep AANAT 5′UTR; Dr. H. Okamura for critical comments; Dr. K. Mikoshiba for anti-SYNCRIP-N antibody; Dr. C.R. Astell for plasmid pRSETC9-15; Dr. K.Y. Lee for the pCY2 vector; Dr. K.J. Kim for hnRNP Q437 purification; and Mr. Y.S. Park for adenovirus preparation. This work was supported by the Brain Neurobiology Research Program and Systems-Biodynamics NCRC sponsored by the Korean Ministry of Science and Technology and the Basic Science Research Fund from KOSEF. This work was also supported by the Brain Korea 21 program of the Korean Ministry of Education.
Footnotes
Supplemental Material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1519507
References
- Ackermann K., Bux R., Rub U., Korf H.W., Kauert G., Stehle J.H., Bux R., Rub U., Korf H.W., Kauert G., Stehle J.H., Rub U., Korf H.W., Kauert G., Stehle J.H., Korf H.W., Kauert G., Stehle J.H., Kauert G., Stehle J.H., Stehle J.H. Characterization of human melatonin synthesis using autoptic pineal tissue. Endocrinology. 2006;147:3235–3242. doi: 10.1210/en.2006-0043. [DOI] [PubMed] [Google Scholar]
- Baler R., Covington S., Klein D.C., Covington S., Klein D.C., Klein D.C. The rat arylalkylamine N-acetyltransferase gene promoter. cAMP activation via a cAMP-responsive element–CCAAT complex. J. Biol. Chem. 1997;272:6979–6985. doi: 10.1074/jbc.272.11.6979. [DOI] [PubMed] [Google Scholar]
- Bannai H., Fukatsu K., Mizutani A., Natsume T., Iemura S., Ikegami T., Inoue T., Mikoshiba K., Fukatsu K., Mizutani A., Natsume T., Iemura S., Ikegami T., Inoue T., Mikoshiba K., Mizutani A., Natsume T., Iemura S., Ikegami T., Inoue T., Mikoshiba K., Natsume T., Iemura S., Ikegami T., Inoue T., Mikoshiba K., Iemura S., Ikegami T., Inoue T., Mikoshiba K., Ikegami T., Inoue T., Mikoshiba K., Inoue T., Mikoshiba K., Mikoshiba K. An RNA-interacting protein, SYNCRIP (heterogeneous nuclear ribonuclear protein Q1/NSAP1) is a component of mRNA granule transported with inositol 1,4,5-trisphosphate receptor type 1 mRNA in neuronal dendrites. J. Biol. Chem. 2004;279:53427–53434. doi: 10.1074/jbc.M409732200. [DOI] [PubMed] [Google Scholar]
- Bell-Pedersen D., Cassone V.M., Earnest D.J., Golden S.S., Hardin P.E., Thomas T.L., Zoran M.J., Cassone V.M., Earnest D.J., Golden S.S., Hardin P.E., Thomas T.L., Zoran M.J., Earnest D.J., Golden S.S., Hardin P.E., Thomas T.L., Zoran M.J., Golden S.S., Hardin P.E., Thomas T.L., Zoran M.J., Hardin P.E., Thomas T.L., Zoran M.J., Thomas T.L., Zoran M.J., Zoran M.J. Circadian rhythms from multiple oscillators: Lessons from diverse organisms. Nat. Rev. Genet. 2005;6:544–556. doi: 10.1038/nrg1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard M., Klein D.C., Zatz M., Klein D.C., Zatz M., Zatz M. Chick pineal clock regulates serotonin N-acetyltransferase mRNA rhythm in culture. Proc. Natl. Acad. Sci. 1997;94:304–309. doi: 10.1073/pnas.94.1.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.Y., Gherzi R., Ong S.E., Chan E.L., Raijmakers R., Pruijn G.J., Stoecklin G., Moroni C., Mann M., Karin M., Gherzi R., Ong S.E., Chan E.L., Raijmakers R., Pruijn G.J., Stoecklin G., Moroni C., Mann M., Karin M., Ong S.E., Chan E.L., Raijmakers R., Pruijn G.J., Stoecklin G., Moroni C., Mann M., Karin M., Chan E.L., Raijmakers R., Pruijn G.J., Stoecklin G., Moroni C., Mann M., Karin M., Raijmakers R., Pruijn G.J., Stoecklin G., Moroni C., Mann M., Karin M., Pruijn G.J., Stoecklin G., Moroni C., Mann M., Karin M., Stoecklin G., Moroni C., Mann M., Karin M., Moroni C., Mann M., Karin M., Mann M., Karin M., Karin M. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell. 2001;107:451–464. doi: 10.1016/s0092-8674(01)00578-5. [DOI] [PubMed] [Google Scholar]
- Cho S., Kim J.H., Back S.H., Jang S.K., Kim J.H., Back S.H., Jang S.K., Back S.H., Jang S.K., Jang S.K. Polypyrimidine tract-binding protein enhances the internal ribosomal entry site-dependent translation of p27Kip1 mRNA and modulates transition from G1 to S phase. Mol. Cell. Biol. 2005;25:1283–1297. doi: 10.1128/MCB.25.4.1283-1297.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S., Park S.M., Kim T.D., Kim J.H., Kim K.T., Jang S.K., Park S.M., Kim T.D., Kim J.H., Kim K.T., Jang S.K., Kim T.D., Kim J.H., Kim K.T., Jang S.K., Kim J.H., Kim K.T., Jang S.K., Kim K.T., Jang S.K., Jang S.K. BiP internal ribosomal entry site activity is controlled by heat-induced interaction of NSAP1. Mol. Cell. Biol. 2007;27:368–383. doi: 10.1128/MCB.00814-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi B.H., Chae H.D., Park T.J., Oh J., Lim J., Kang S.S., Ha H., Kim K.T., Chae H.D., Park T.J., Oh J., Lim J., Kang S.S., Ha H., Kim K.T., Park T.J., Oh J., Lim J., Kang S.S., Ha H., Kim K.T., Oh J., Lim J., Kang S.S., Ha H., Kim K.T., Lim J., Kang S.S., Ha H., Kim K.T., Kang S.S., Ha H., Kim K.T., Ha H., Kim K.T., Kim K.T. Protein kinase C regulates the activity and stability of serotonin N-acetyltransferase. J. Neurochem. 2004;90:442–454. doi: 10.1111/j.1471-4159.2004.02495.x. [DOI] [PubMed] [Google Scholar]
- Coon S.L., Roseboom P.H., Baler R., Weller J.L., Namboodiri M.A., Koonin E.V., Klein D.C., Roseboom P.H., Baler R., Weller J.L., Namboodiri M.A., Koonin E.V., Klein D.C., Baler R., Weller J.L., Namboodiri M.A., Koonin E.V., Klein D.C., Weller J.L., Namboodiri M.A., Koonin E.V., Klein D.C., Namboodiri M.A., Koonin E.V., Klein D.C., Koonin E.V., Klein D.C., Klein D.C. Pineal serotonin N-acetyltransferase: Expression cloning and molecular analysis. Science. 1995;270:1681–1683. doi: 10.1126/science.270.5242.1681. [DOI] [PubMed] [Google Scholar]
- Coon S.L., Mazuruk K., Bernard M., Roseboom P.H., Klein D.C., Rodriguez I.R., Mazuruk K., Bernard M., Roseboom P.H., Klein D.C., Rodriguez I.R., Bernard M., Roseboom P.H., Klein D.C., Rodriguez I.R., Roseboom P.H., Klein D.C., Rodriguez I.R., Klein D.C., Rodriguez I.R., Rodriguez I.R. The human serotonin N-acetyltransferase (EC 2.3.1.87) gene (AANAT): Structure, chromosomal localization, and tissue expression. Genomics. 1996;34:76–84. doi: 10.1006/geno.1996.0243. [DOI] [PubMed] [Google Scholar]
- Dreyfuss G., Matunis M.J., Pinol-Roma S., Burd C.G., Matunis M.J., Pinol-Roma S., Burd C.G., Pinol-Roma S., Burd C.G., Burd C.G. hnRNP proteins and the biogenesis of mRNA. Annu. Rev. Biochem. 1993;62:289–321. doi: 10.1146/annurev.bi.62.070193.001445. [DOI] [PubMed] [Google Scholar]
- Dreyfuss G., Kim V.N., Kataoka N., Kim V.N., Kataoka N., Kataoka N. Messenger-RNA-binding proteins and the messages they carry. Nat. Rev. Mol. Cell Biol. 2002;3:195–205. doi: 10.1038/nrm760. [DOI] [PubMed] [Google Scholar]
- Dunlap J.C. Molecular bases for circadian clocks. Cell. 1999;96:271–290. doi: 10.1016/s0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- Foulkes N.S., Borjigin J., Snyder S.H., Sassone-Corsi P., Borjigin J., Snyder S.H., Sassone-Corsi P., Snyder S.H., Sassone-Corsi P., Sassone-Corsi P. Rhythmic transcription: The molecular basis of circadian melatonin synthesis. Trends Neurosci. 1997;20:487–492. doi: 10.1016/s0166-2236(97)01109-0. [DOI] [PubMed] [Google Scholar]
- Ganguly S., Gastel J.A., Weller J.L., Schwartz C., Jaffe H., Namboodiri M.A., Coon S.L., Hickman A.B., Rollag M., Obsil T., Gastel J.A., Weller J.L., Schwartz C., Jaffe H., Namboodiri M.A., Coon S.L., Hickman A.B., Rollag M., Obsil T., Weller J.L., Schwartz C., Jaffe H., Namboodiri M.A., Coon S.L., Hickman A.B., Rollag M., Obsil T., Schwartz C., Jaffe H., Namboodiri M.A., Coon S.L., Hickman A.B., Rollag M., Obsil T., Jaffe H., Namboodiri M.A., Coon S.L., Hickman A.B., Rollag M., Obsil T., Namboodiri M.A., Coon S.L., Hickman A.B., Rollag M., Obsil T., Coon S.L., Hickman A.B., Rollag M., Obsil T., Hickman A.B., Rollag M., Obsil T., Rollag M., Obsil T., Obsil T., et al. Role of a pineal cAMP-operated arylalkylamine N-acetyltransferase/14–3–3-binding switch in melatonin synthesis. Proc. Natl. Acad. Sci. 2001;98:8083–8088. doi: 10.1073/pnas.141118798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguly S., Coon S.L., Klein D.C., Coon S.L., Klein D.C., Klein D.C. Control of melatonin synthesis in the mammalian pineal gland: The critical role of serotonin acetylation. Cell Tissue Res. 2002;309:127–137. doi: 10.1007/s00441-002-0579-y. [DOI] [PubMed] [Google Scholar]
- Gastel J.A., Roseboom P.H., Rinaldi P.A., Weller J.L., Klein D.C., Roseboom P.H., Rinaldi P.A., Weller J.L., Klein D.C., Rinaldi P.A., Weller J.L., Klein D.C., Weller J.L., Klein D.C., Klein D.C. Melatonin production: Proteasomal proteolysis in serotonin N-acetyltransferase regulation. Science. 1998;279:1358–1360. doi: 10.1126/science.279.5355.1358. [DOI] [PubMed] [Google Scholar]
- Gebauer F., Hentze M.W., Hentze M.W. Molecular mechanisms of translational control. Nat. Rev. Mol. Cell Biol. 2004;5:827–835. doi: 10.1038/nrm1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras A.C., Raught B., Sonenberg N., Raught B., Sonenberg N., Sonenberg N. mTOR signaling to translation. Curr. Top. Microbiol. Immunol. 2004;279:169–197. doi: 10.1007/978-3-642-18930-2_11. [DOI] [PubMed] [Google Scholar]
- Grosset C., Chen C.Y., Xu N., Sonenberg N., Jacquemin-Sablon H., Shyu A.B., Chen C.Y., Xu N., Sonenberg N., Jacquemin-Sablon H., Shyu A.B., Xu N., Sonenberg N., Jacquemin-Sablon H., Shyu A.B., Sonenberg N., Jacquemin-Sablon H., Shyu A.B., Jacquemin-Sablon H., Shyu A.B., Shyu A.B. A mechanism for translationally coupled mRNA turnover: Interaction between the poly(A) tail and a c-fos RNA coding determinant via a protein complex. Cell. 2000;103:29–40. doi: 10.1016/s0092-8674(00)00102-1. [DOI] [PubMed] [Google Scholar]
- Han S., Kim T.D., Ha D.C., Kim K.T., Kim T.D., Ha D.C., Kim K.T., Ha D.C., Kim K.T., Kim K.T. Rhythmic expression of adenylyl cyclase VI contributes to the differential regulation of serotonin N-acetyltransferase by bradykinin in rat pineal glands. J. Biol. Chem. 2005;280:38228–38234. doi: 10.1074/jbc.M508130200. [DOI] [PubMed] [Google Scholar]
- Harms E., Kivimae S., Young M.W., Saez L., Kivimae S., Young M.W., Saez L., Young M.W., Saez L., Saez L. Posttranscriptional and posttranslational regulation of clock genes. J. Biol. Rhythms. 2004;19:361–373. doi: 10.1177/0748730404268111. [DOI] [PubMed] [Google Scholar]
- Hay N., Sonenberg N., Sonenberg N. Upstream and downstream of mTOR. Genes & Dev. 2004;18:1926–1945. doi: 10.1101/gad.1212704. [DOI] [PubMed] [Google Scholar]
- Hellen C.U., Sarnow P., Sarnow P. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes & Dev. 2001;15:1593–1612. doi: 10.1101/gad.891101. [DOI] [PubMed] [Google Scholar]
- Jang S.K., Krausslich H.G., Nicklin M.J., Duke G.M., Palmenberg A.C., Wimmer E., Krausslich H.G., Nicklin M.J., Duke G.M., Palmenberg A.C., Wimmer E., Nicklin M.J., Duke G.M., Palmenberg A.C., Wimmer E., Duke G.M., Palmenberg A.C., Wimmer E., Palmenberg A.C., Wimmer E., Wimmer E. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol. 1988;62:2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.H., Paek K.Y., Choi K., Kim T.D., Hahm B., Kim K.T., Jang S.K., Paek K.Y., Choi K., Kim T.D., Hahm B., Kim K.T., Jang S.K., Choi K., Kim T.D., Hahm B., Kim K.T., Jang S.K., Kim T.D., Hahm B., Kim K.T., Jang S.K., Hahm B., Kim K.T., Jang S.K., Kim K.T., Jang S.K., Jang S.K. Heterogeneous nuclear ribonucleoprotein C modulates translation of c-myc mRNA in a cell cycle phase-dependent manner. Mol. Cell. Biol. 2003;23:708–720. doi: 10.1128/MCB.23.2.708-720.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.H., Paek K.Y., Ha S.H., Cho S., Choi K., Kim C.S., Ryu S.H., Jang S.K., Paek K.Y., Ha S.H., Cho S., Choi K., Kim C.S., Ryu S.H., Jang S.K., Ha S.H., Cho S., Choi K., Kim C.S., Ryu S.H., Jang S.K., Cho S., Choi K., Kim C.S., Ryu S.H., Jang S.K., Choi K., Kim C.S., Ryu S.H., Jang S.K., Kim C.S., Ryu S.H., Jang S.K., Ryu S.H., Jang S.K., Jang S.K. A cellular RNA-binding protein enhances internal ribosomal entry site-dependent translation through an interaction downstream of the hepatitis C virus polyprotein initiation codon. Mol. Cell. Biol. 2004;24:7878–7890. doi: 10.1128/MCB.24.18.7878-7890.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.D., Kim J.S., Kim J.H., Myung J., Chae H.D., Woo K.C., Jang S.K., Koh D.S., Kim K.T., Kim J.S., Kim J.H., Myung J., Chae H.D., Woo K.C., Jang S.K., Koh D.S., Kim K.T., Kim J.H., Myung J., Chae H.D., Woo K.C., Jang S.K., Koh D.S., Kim K.T., Myung J., Chae H.D., Woo K.C., Jang S.K., Koh D.S., Kim K.T., Chae H.D., Woo K.C., Jang S.K., Koh D.S., Kim K.T., Woo K.C., Jang S.K., Koh D.S., Kim K.T., Jang S.K., Koh D.S., Kim K.T., Koh D.S., Kim K.T., Kim K.T. Rhythmic serotonin N-acetyltransferase mRNA degradation is essential for the maintenance of its circadian oscillation. Mol. Cell. Biol. 2005;25:3232–3246. doi: 10.1128/MCB.25.8.3232-3246.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein D.C., Coon S.L., Roseboom P.H., Weller J.L., Bernard M., Gastel J.A., Zatz M., Iuvone P.M., Rodriguez I.R., Begay V., Coon S.L., Roseboom P.H., Weller J.L., Bernard M., Gastel J.A., Zatz M., Iuvone P.M., Rodriguez I.R., Begay V., Roseboom P.H., Weller J.L., Bernard M., Gastel J.A., Zatz M., Iuvone P.M., Rodriguez I.R., Begay V., Weller J.L., Bernard M., Gastel J.A., Zatz M., Iuvone P.M., Rodriguez I.R., Begay V., Bernard M., Gastel J.A., Zatz M., Iuvone P.M., Rodriguez I.R., Begay V., Gastel J.A., Zatz M., Iuvone P.M., Rodriguez I.R., Begay V., Zatz M., Iuvone P.M., Rodriguez I.R., Begay V., Iuvone P.M., Rodriguez I.R., Begay V., Rodriguez I.R., Begay V., Begay V., et al. 1997The melatonin rhythm-generating enzyme: Molecular regulation of serotonin N-acetyltransferase in the pineal gland Recent Prog. Horm. Res. 52307 –357. Discussion 357358 [PubMed] [Google Scholar]
- Kullmann M., Gopfert U., Siewe B., Hengst L., Gopfert U., Siewe B., Hengst L., Siewe B., Hengst L., Hengst L. ELAV/Hu proteins inhibit p27 translation via an IRES element in the p27 5′UTR. Genes & Dev. 2002;16:3087–3099. doi: 10.1101/gad.248902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuninger D., Stauffer D., Eftekhari S., Wilson E., Thayer M., Rotwein P., Stauffer D., Eftekhari S., Wilson E., Thayer M., Rotwein P., Eftekhari S., Wilson E., Thayer M., Rotwein P., Wilson E., Thayer M., Rotwein P., Thayer M., Rotwein P., Rotwein P. Gene disruption by regulated short interfering RNA expression, using a two-adenovirus system. Hum. Gene Ther. 2004;15:1287–1292. doi: 10.1089/hum.2004.15.1287. [DOI] [PubMed] [Google Scholar]
- Kwak E., Kim T.D., Kim K.T., Kim T.D., Kim K.T., Kim K.T. Essential role of 3′-untranslated region-mediated mRNA decay in circadian oscillations of mouse Period3 mRNA. J. Biol. Chem. 2006;281:19100–19106. doi: 10.1074/jbc.M511927200. [DOI] [PubMed] [Google Scholar]
- Kwon S.Y., Choi Y.J., Kang T.H., Lee K.H., Cha S.S., Kim G.H., Lee H.S., Kim K.T., Kim K.J., Choi Y.J., Kang T.H., Lee K.H., Cha S.S., Kim G.H., Lee H.S., Kim K.T., Kim K.J., Kang T.H., Lee K.H., Cha S.S., Kim G.H., Lee H.S., Kim K.T., Kim K.J., Lee K.H., Cha S.S., Kim G.H., Lee H.S., Kim K.T., Kim K.J., Cha S.S., Kim G.H., Lee H.S., Kim K.T., Kim K.J., Kim G.H., Lee H.S., Kim K.T., Kim K.J., Lee H.S., Kim K.T., Kim K.J., Kim K.T., Kim K.J., Kim K.J. Highly efficient protein expression and purification using bacterial hemoglobin fusion vector. Plasmid. 2005;53:274–282. doi: 10.1016/j.plasmid.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Mizutani A., Fukuda M., Ibata K., Shiraishi Y., Mikoshiba K., Fukuda M., Ibata K., Shiraishi Y., Mikoshiba K., Ibata K., Shiraishi Y., Mikoshiba K., Shiraishi Y., Mikoshiba K., Mikoshiba K. SYNCRIP, a cytoplasmic counterpart of heterogeneous nuclear ribonucleoprotein R, interacts with ubiquitous synaptotagmin isoforms. J. Biol. Chem. 2000;275:9823–9831. doi: 10.1074/jbc.275.13.9823. [DOI] [PubMed] [Google Scholar]
- Mourelatos Z., Abel L., Yong J., Kataoka N., Dreyfuss G., Abel L., Yong J., Kataoka N., Dreyfuss G., Yong J., Kataoka N., Dreyfuss G., Kataoka N., Dreyfuss G., Dreyfuss G. SMN interacts with a novel family of hnRNP and spliceosomal proteins. EMBO J. 2001;20:5443–5452. doi: 10.1093/emboj/20.19.5443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obsil T., Ghirlando R., Klein D.C., Ganguly S., Dyda F., Ghirlando R., Klein D.C., Ganguly S., Dyda F., Klein D.C., Ganguly S., Dyda F., Ganguly S., Dyda F., Dyda F. Crystal structure of the 14–3–3ζ:serotonin N-acetyltransferase complex. A role for scaffolding in enzyme regulation. Cell. 2001;105:257–267. doi: 10.1016/s0092-8674(01)00316-6. [DOI] [PubMed] [Google Scholar]
- Ostareck D.H., Ostareck-Lederer A., Wilm M., Thiele B.J., Mann M., Hentze M.W., Ostareck-Lederer A., Wilm M., Thiele B.J., Mann M., Hentze M.W., Wilm M., Thiele B.J., Mann M., Hentze M.W., Thiele B.J., Mann M., Hentze M.W., Mann M., Hentze M.W., Hentze M.W. mRNA silencing in erythroid differentiation: hnRNP K and hnRNP E1 regulate 15-lipoxygenase translation from the 3′ end. Cell. 1997;89:597–606. doi: 10.1016/s0092-8674(00)80241-x. [DOI] [PubMed] [Google Scholar]
- Ostareck-Lederer A., Ostareck D.H., Ostareck D.H. Control of mRNA translation and stability in haematopoietic cells: The function of hnRNPs K and E1/E2. Biol. Cell. 2004;96:407–411. doi: 10.1016/j.biolcel.2004.03.010. [DOI] [PubMed] [Google Scholar]
- Ostareck-Lederer A., Ostareck D.H., Hentze M.W., Ostareck D.H., Hentze M.W., Hentze M.W. Cytoplasmic regulatory functions of the KH-domain proteins hnRNPs K and E1/E2. Trends Biochem. Sci. 1998;23:409–411. doi: 10.1016/s0968-0004(98)01301-2. [DOI] [PubMed] [Google Scholar]
- Pelletier J., Sonenberg N., Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- Pfeffer M., Maronde E., Molina C.A., Korf H.W., Stehle J.H., Maronde E., Molina C.A., Korf H.W., Stehle J.H., Molina C.A., Korf H.W., Stehle J.H., Korf H.W., Stehle J.H., Stehle J.H. Inducible cyclic AMP early repressor protein in rat pinealocytes: A highly sensitive natural reporter for regulated gene transcription. Mol. Pharmacol. 1999;56:279–289. doi: 10.1124/mol.56.2.279. [DOI] [PubMed] [Google Scholar]
- Pyronnet S., Pradayrol L., Sonenberg N., Pradayrol L., Sonenberg N., Sonenberg N. A cell cycledependent internal ribosome entry site. Mol. Cell. 2000;5:607–616. doi: 10.1016/s1097-2765(00)80240-3. [DOI] [PubMed] [Google Scholar]
- Reppert S.M., Weaver D.R., Weaver D.R. Melatonin madness. Cell. 1995;83:1059–1062. doi: 10.1016/0092-8674(95)90131-0. [DOI] [PubMed] [Google Scholar]
- Shu Y., Hong-Hui L., Hong-Hui L. Transcription, translation, degradation, and circadian clock. Biochem. Biophys. Res. Commun. 2004;321:1–6. doi: 10.1016/j.bbrc.2004.06.093. [DOI] [PubMed] [Google Scholar]
- Stehle J.H., von Gall C., Schomerus C., Korf H.W., von Gall C., Schomerus C., Korf H.W., Schomerus C., Korf H.W., Korf H.W. Of rodents and ungulates and melatonin: Creating a uniform code for darkness by different signaling mechanisms. J. Biol. Rhythms. 2001;16:312–325. doi: 10.1177/074873001129002033. [DOI] [PubMed] [Google Scholar]
- Stoneley M., Willis A.E., Willis A.E. Cellular internal ribosome entry segments: Structures, trans-acting factors and regulation of gene expression. Oncogene. 2004;23:3200–3207. doi: 10.1038/sj.onc.1207551. [DOI] [PubMed] [Google Scholar]
- Vagner S., Galy B., Pyronnet S., Galy B., Pyronnet S., Pyronnet S. Irresistible IRES. Attracting the translation machinery to internal ribosome entry sites. EMBO Rep. 2001;2:893–898. doi: 10.1093/embo-reports/kve208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss I.M., Liebhaber S.A., Liebhaber S.A. Erythroid cell-specific determinants of α-globin mRNA stability. Mol. Cell. Biol. 1994;14:8123–8132. doi: 10.1128/mcb.14.12.8123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson M.F., Shyu A.B., Shyu A.B. Multifunctional regulatory proteins that control gene expression in both the nucleus and the cytoplasm. Bioessays. 2001;23:775–787. doi: 10.1002/bies.1113. [DOI] [PubMed] [Google Scholar]
- Zheng W., Zhang Z., Ganguly S., Weller J.L., Klein D.C., Cole P.A., Zhang Z., Ganguly S., Weller J.L., Klein D.C., Cole P.A., Ganguly S., Weller J.L., Klein D.C., Cole P.A., Weller J.L., Klein D.C., Cole P.A., Klein D.C., Cole P.A., Cole P.A. Cellular stabilization of the melatonin rhythm enzyme induced by nonhydrolyzable phosphonate incorporation. Nat. Struct. Biol. 2003;10:1054–1057. doi: 10.1038/nsb1005. [DOI] [PubMed] [Google Scholar]