Abstract
To ligate exons in pre-messenger RNA (pre-mRNA) splicing, the spliceosome must reposition the substrate after cleaving the 5′ splice site. Because spliceosomal small nuclear RNAs (snRNAs) bind the substrate, snRNA structures may rearrange to reposition the substrate. However, such rearrangements have remained undefined. Although U2 stem IIc inhibits binding of U2 snRNP to pre-mRNA during assembly, we found that weakening U2 stem IIc suppressed a mutation in prp16, a DExD/H box ATPase that promotes splicing after 5′ splice site cleavage. The prp16 mutation was also suppressed by mutations flanking stem IIc, suggesting that Prp16p facilitates a switch from stem IIc to the mutually exclusive U2 stem IIa, which activates binding of U2 to pre-mRNA during assembly. Providing evidence that stem IIa switches back to stem IIc before exon ligation, disrupting stem IIa suppressed 3′ splice site mutations, and disrupting stem IIc impaired exon ligation. Disrupting stem IIc also exacerbated the 5′ splice site cleavage defects of certain substrate mutations, suggesting a parallel role for stem IIc at both catalytic stages. We propose that U2, much like the ribosome, toggles between two conformations—a closed stem IIc conformation that promotes catalysis and an open stem IIa conformation that promotes substrate binding and release.
Keywords: U2, snRNA, Prp16, spliceosome, DExD/H box, pre-mRNA splicing
Introns are excised from pre-messenger RNA (pre-mRNA) by the spliceosome, a large ribonucleoprotein machine composed of >100 proteins and five small nuclear RNAs (snRNAs) (for reviews, see Jurica and Moore 2003; Will and Lührmann 2006). The spliceosome excises introns in two sequential transesterification reactions. In the first chemical step, termed 5′ splice site cleavage, the 2′ hydroxyl of an intronic adenosine, termed the branch point, attacks the 5′ splice site, forming a lariat intermediate and a liberated 5′ exon with a free 3′ hydroxyl. In the second chemical step, termed exon ligation, the 3′ hydroxyl of the 5′ exon attacks the 3′ splice site, excising the lariat intron and ligating the exons to form mRNA. Because the leaving group of the first reaction becomes the attacking group for the second reaction, this two-step reaction presents a significant biochemical challenge to the spliceosome, which must consequently rearrange the substrate after 5′ splice site cleavage (for review, see Staley and Guthrie 1998). Specifically, because the spliceosome is thought to catalyze the two similar reactions in one active site, and the second reaction is essentially the reverse of the first (Steitz and Steitz 1993), the spliceosome likely removes the branched product of 5′ splice site cleavage from the active site and replaces the branch with the 3′ splice site. The mechanism by which the spliceosome rearranges the substrate is understood poorly.
The substrate is defined by intronic consensus sequences at the 5′ splice site, the branch site, and the 3′ splice site (for review, see Burge et al. 1999). These sequences are recognized dynamically by the spliceosome (for review, see Staley and Guthrie 1998). The 5′ splice site consensus sequence is first recognized by U1 and then by U6, which defines the site of 5′ splice site cleavage. Similarly, the branch site consensus sequence is first recognized by the branch-point-binding protein and then by U2, which defines the branch site. Consistent with the roles of U2 and U6 in defining the reactive sites for 5′ splice site cleavage, both U2 and U6 are necessary at the catalytic stages of splicing (for review, see Valadkhan 2005). Significantly, U2 and U6 interact through base-pairing (Datta and Weiner 1991; Madhani and Guthrie 1992; Sun and Manley 1995; Hilliker and Staley 2004) and are sufficient, in the absence of protein, to promote a splicing-related reaction, suggesting that U2 and U6 promote catalysis directly (Valadkhan and Manley 2001, 2003). Because U2 and U6 define the reactive sites for 5′ splice site cleavage and because the substrate must remodel after 5′ splice site cleavage, U2 and U6 are strong candidates for factors that rearrange after 5′ splice site cleavage to effect substrate remodeling.
Validating a requirement for substrate remodeling, the spliceosome alters its interactions with the substrate after 5′ splice site cleavage. In particular, the spliceosome dissociates the 5′ splice site from U6 (Konarska et al. 2006) and binds the 3′ splice site through unknown elements (Schwer and Guthrie 1992). Although the spliceosome likely removes the branched product from the active site, the spliceosome nonetheless interacts with the branched structure at the chemical stage of exon ligation to proofread splicing and presumably to restrict the branch site from the active site (Mayas et al. 2006).
Exon ligation requires two steps, an initial ATP-dependent step and a subsequent ATP-independent step (for review, see Umen and Guthrie 1995). These two steps likely promote substrate remodeling after 5′ splice site cleavage. The ATP-dependent step is required for 3′ splice site binding and depends on the activity of the DExD/H box ATPase Prp16p (Schwer and Guthrie 1992). Because Prp16p unwinds RNA duplexes in vitro (Wang et al. 1998), Prp16p has been hypothesized to destabilize RNA:RNA and/or RNA:protein structures. Consistent with this hypothesis, a mutation in PRP8 (Query and Konarska 2004), deletion of ISY1 (Villa and Guthrie 2005), or deletion of nucleotides upstream of the 5′-splice-site-binding sequence in U6 (Madhani and Guthrie 1994a) each suppress prp16 mutants, suggesting that structures involving Isy1p, Prp8p, and/or U6 snRNA are disrupted during the Prp16p-dependent step; however, specific interactions that are targeted for disruption have not yet been defined. After the ATP-dependent step, Prp22p as well as Slu7p promote 3′ splice site binding in an ATP-independent manner (Brys and Schwer 1996; Schwer and Gross 1998).
While interactions between the spliceosome and the substrate are clearly dynamic after 5′ splice site cleavage, it has not yet been clear whether structures within the snRNAs are similarly dynamic at this stage. One candidate structure is the stem II region of U2 (Fig. 1A), a region that is dynamic in the free U2 snRNP (Zavanelli and Ares 1991; Zavanelli et al. 1994). Intriguingly, exon ligation is impeded by one of several 2′-O-methyl oligonucleotides complimentary to the stem II region of U2, suggesting that this region of U2 may rearrange after 5′ splice site cleavage (Barabino et al. 1992). Stem II lies just downstream from the branch-site-binding region and can regulate binding of the free U2 snRNP to the branch site by alternating between two mutually exclusive structures, stem–loop IIa and stem IIc; stem IIc comprises an interaction between the loop sequence of stem–loop IIa and conserved, downstream sequences (Fig. 1A; Zavanelli and Ares 1991; Zavanelli et al. 1994). Stem–loop IIa, which is required for 5′ splice site cleavage (Ares and Igel 1990), promotes prespliceosome formation (Zavanelli and Ares 1991; Zavanelli et al. 1994), during which the U2 snRNP binds the branch site. In contrast, stem IIc inhibits prespliceosome formation (Zavanelli and Ares 1991; Zavanelli et al. 1994). The stem–loop IIa conformation of U2 is favored by the DExD/H box ATPase Prp5p, thereby accounting for the ATP-dependence of prespliceosome formation (Perriman and Ares 2007).
Figure 1.
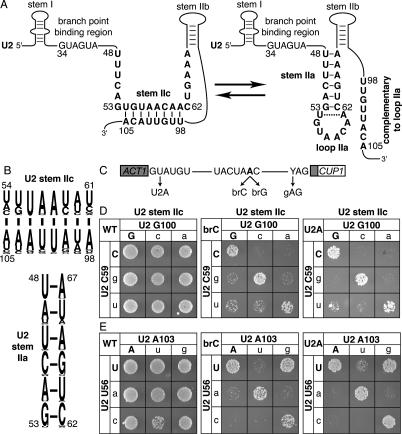
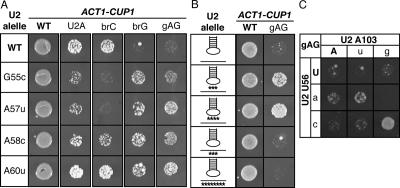
Disrupting U2 stem IIc exacerbates the splicing defect of a substrate mutated at the branch site or the 5′ splice site. (A) Structure of U2 stem IIc and stem–loop IIa. Nucleotides involved in stem IIc, stem IIa, and loop IIa are bold. The nucleotides of U2 that bind the intronic branch site consensus sequence are shown. Nucleotide sequence and numbers correspond to the Saccharomyces cerevisiae sequence. The dashed line marks a nonconserved base pair that can extend stem IIa in S. cerevisiae. (B) A pictogram of stem IIc and IIa showing the consensus sequence for organisms ranging from budding yeast to humans; numbering refers to the budding yeast sequence. The height of the letter is proportional to the frequency of the nucleotide in the alignment. (C) Schematic diagram of the ACT1-CUP1 splicing reporter, which when spliced confers copper resistance to budding yeast (Lesser and Guthrie 1993). The intron consensus sequences for budding yeast are shown along with point mutations used in this study. The branch site adenosine is bold. (D,E) Mutations that disrupt base-pairing in U2 stem IIc enhance the growth defect conferred by the 5′ splice site mutation U2A and the branch site mutation brC. Compensatory analysis by copper resistance of U2 stem IIc base pairs C59/G100 (D) and U56/A103 (E) is shown. The matrices show the copper resistance of the wild-type (WT; left), brC (middle), or U2A (right) ACT1-CUP1 splicing reporters in yJPS1035 expressing U2 variants having single or double mutations in the stem IIc base pairs. Cells were grown for 3–4 d at 30°C on solid media containing 0.05 mM copper sulfate. Wild-type residues are capitalized.
We have found evidence that U2 stem II does indeed rearrange in the active spliceosome after 5′ splice site cleavage. Mutations that destabilize U2 stem IIc exacerbated the 5′ splice site cleavage defect of a branch site A-to-C (brC) mutation, which is limited at the 5′ splice site cleavage stage (Lesser and Guthrie 1993; Query and Konarska 2004), suggesting that stem IIc promotes 5′ splice site cleavage, despite inhibiting prespliceosome formation (Zavanelli and Ares 1991; Zavanelli et al. 1994). Supporting a role for stem IIc in 5′ splice site cleavage, mutations that destabilize U2 stem IIc suppressed a prp16 mutant. This suppression further suggests that Prp16p promotes disruption of stem IIc, directly or indirectly. The prp16 mutant was also suppressed by mutations that destabilized structures physically mutually exclusive with stem IIa, suggesting that Prp16p disrupts these structures to promote formation of stem–loop IIa. Providing evidence that stem–loop IIa does indeed form but then must unwind in turn to allow for exon ligation, mutations that disrupt the stem or loop of stem–loop IIa suppressed 3′ splice site mutations. Indicating that stem–loop IIa unfolds to allow reformation of stem IIc, mutations that destabilize stem IIc compromised exon ligation. Thus, our data provide evidence that U2 stem II toggles between two structures in the active spliceosome and that one of these structures, stem IIc, promotes both catalytic steps and the other, stem–loop IIa, promotes the transition from the 5′ splice site cleavage conformation to the exon ligation conformation of the spliceosome. We propose that this toggling reflects a larger conformational change that switches the spliceosome between a closed conformation that promotes catalysis and an open conformation that promotes substrate binding, rearrangement, and dissociation.
Results
U2 stem IIc promotes 5′ splice site cleavage
Because the stem II region (Fig. 1A) of U2 is dynamic in the free U2 snRNP (Zavanelli and Ares 1991; Zavanelli et al. 1994) and is sensitive to oligonucleotide hybridization in the spliceosome after 5′ splice site cleavage (Barabino et al. 1992), we hypothesized that this region is dynamic in the active spliceosome. To determine if the stem II region rearranges after 5′ splice site cleavage, we first assessed whether stem IIa or stem IIc functions at the stage of 5′ splice site cleavage and we found evidence that 5′ splice site cleavage is promoted by stem IIc. Because U2 stem IIc, although highly conserved (Fig. 1B), is not essential for growth (Ares and Igel 1990) or 5′ splice site cleavage (see below), we tested whether single U2 mutations that disrupt stem IIc enhanced pre-mRNA mutations that impair 5′ splice site cleavage. Specifically, we assayed for exacerbation of (1) an adenosine to cytidine mutation at the branch point (brC), a mutation that impairs 5′ splice site cleavage by destabilizing the 5′ splice site cleavage conformation of the spliceosome (Query and Konarska 2004), and (2) a uridine to adenosine mutation at position two of the 5′ splice site (U2A), a mutation that also decreases the efficiency of 5′ splice site cleavage (Ruis et al. 1994; Collins and Guthrie 1999). To assay for exacerbation in vivo in budding yeast, we cotransformed a cup1Δ strain with U2 mutants that destabilized stem IIc at either base pair C59/G100 or U56/A103 and with an ACT1-CUP1 splicing reporter having either the brC or U2A substrate mutation (Fig. 1C). The ACT1-CUP1 reporter confers resistance to copper in a splicing-dependent manner, allowing an indirect measure of splicing efficiency (Lesser and Guthrie 1993). Whereas a wild-type reporter supports growth up to 2.0 mM copper (data not shown), the brC and U2A mutations compromise growth, even at 0.05 mM copper (see below; Lesser and Guthrie 1993; Ruis et al. 1994). At all copper concentrations tested, single mutations that destabilized base pairs in U2 stem IIc did not compromise the copper resistance of strains expressing a wild-type reporter (Fig. 1D,E, left grids; data not shown). In contrast, at 0.05 mM copper nearly all single mutations that destabilized U2 stem IIc did compromise the copper resistance of strains expressing either the mutated brC reporter (Fig. 1D,E, middle grids) or the mutated U2A reporter (Fig. 1D,E, right grids). The sole exception was U2–A103G, which could maintain pairing through a wobble with U2–U56. Compensatory mutations that repaired stem IIc restored the copper resistance of the brC and U2A reporters; in contrast, mutations that failed to restore base-pairing failed to restore the copper resistance of the reporters (Fig. 1D,E), indicating that the single U2 mutations exacerbated the defects conferred by the U2A and brC reporters by destabilizing stem IIc. These results suggest that stem IIc cooperates with the 5′ splice site and the branch site, despite clear data indicating that stem IIc antagonizes binding of U2 to the branch site consensus sequence (Zavanelli and Ares 1991; Zavanelli et al. 1994).
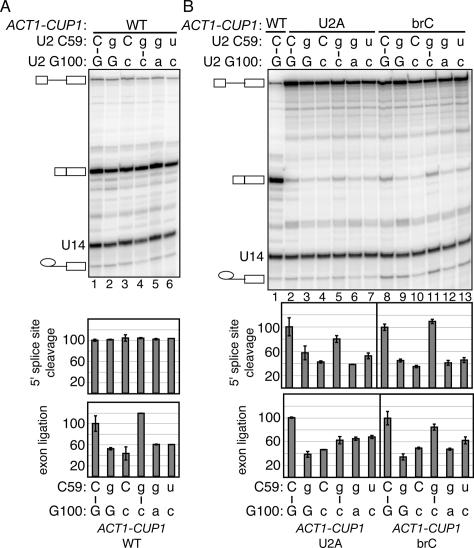
To assess whether U2 stem IIc cooperates with the 5′ splice site and the branch site to promote 5′ splice site cleavage, we analyzed splicing of the U2A and brC ACT1-CUP1 reporters in the stem IIc mutants directly by primer extension. We found evidence that stem IIc does promote 5′ splice site cleavage (Fig. 2). In a strain wild-type for U2, substrates mutated at the 5′ splice site (U2A) or the branch site (brC) compromised both 5′ splice site cleavage and exon ligation, as reflected by an increased level of pre-mRNA and an increased ratio of lariat intermediate to mRNA (Fig. 2B, lanes 1,2,8), as expected (Lesser and Guthrie 1993; Ruis et al. 1994; Collins and Guthrie 1999; Query and Konarska 2004). With a wild-type substrate, the single mutations C59G and G100C, which disrupt U2 stem IIc, did not compromise the efficiency of 5′ splice site cleavage (Fig. 2A). In contrast, with the mutated U2A and brC substrates, these single mutations did compromise the efficiency of 5′ splice site cleavage, further reducing the efficiency by 40%–60% (Fig. 2B, lanes 3,4,9,10). A double mutation that combines C59G and G100C and repairs stem IIc restored the efficiency of 5′ splice site cleavage to within 0%–20% of the wild-type U2 control (Fig. 2B, cf. lanes 2,5 and 8,11); in contrast, double mutations that did not repair stem IIc failed to restore the efficiency of 5′ splice site cleavage (Fig. 2B, cf. lanes 5–7,11–13). These results indicate that stem IIc promotes 5′ splice site cleavage. Consistent with these results, hyperstabilizing stem IIc suppresses the 5′ splice site cleavage defect of a mutated brC substrate (Perriman and Ares 2007). As U2 stem IIc impedes prespliceosome formation (Zavanelli and Ares 1991; Zavanelli et al. 1994), stem IIc likely promotes 5′ splice site cleavage at a later stage. Given that brC is limiting at the catalytic stage of 5′ splice site cleavage (Query and Konarska 2004), stem IIc likely promotes splicing at the catalytic stage of 5′ splice site cleavage. Additional evidence supports a role for stem IIc in exon ligation. The single mutations C59G and G100C, which disrupt stem IIc, exacerbated the exon ligation defect of the brC substrate (Fig. 2B); the C59G/G100C double mutation that repairs stem IIc suppressed the exon ligation defect, while double mutations that did not repair stem IIc failed to suppress. Further, disrupting stem IIc compromised exon ligation of a wild-type substrate (Fig. 2A). Additionally, relative to double mutations that maintain stem IIc, double mutations that disrupt stem IIc compromised a 3′ splice site mutation that impairs exon ligation (described below). Together, our data suggest that stem IIc promotes both 5′ splice site cleavage and exon ligation.
Figure 2.
Disrupting U2 stem IIc exacerbates the 5′ splice site cleavage defect of a substrate mutated at the branch site or the 5′ splice site and impairs exon ligation of a wild-type substrate. (A,B) Compensatory analysis by splicing in vivo of the U2 stem IIc base pair C59/G100 is shown. Primer extension analysis shows the in vivo splicing phenotype of wild-type (WT, A) or mutated U2A and brC (B) ACT1-CUP1 splicing reporters in yJPS1035 having single or double mutations in the stem IIc base pair. The pre-mRNA, mRNA, and lariat intermediate of the ACT1-CUP1 reporter and U14, which serves as an internal control, are highlighted. Quantitation of the primer extensions is shown below. The identities of the bases are indicated above the gel and below the graph; wild-type residues are capitalized. The apparent efficiency of 5′ splice site cleavage is calculated as (mRNA + lariat intermediate)/(pre-mRNA + lariat intermediate + mRNA). The apparent efficiency of exon ligation is calculated as (mRNA/lariat intermediate). Values are normalized to the strain expressing wild-type U2. The histograms show the mean of duplicate samples; the error bars indicate the range of values. While the trends of the exon ligation efficiencies in A repeated in independent experiments, the magnitude of the differences varied.
Evidence that Prp16p destabilizes U2 stem IIc, directly or indirectly
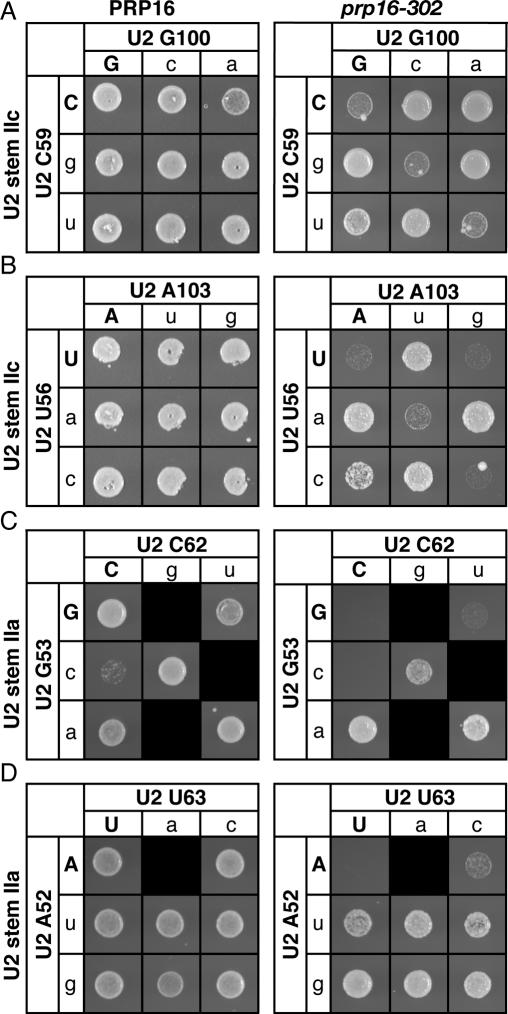
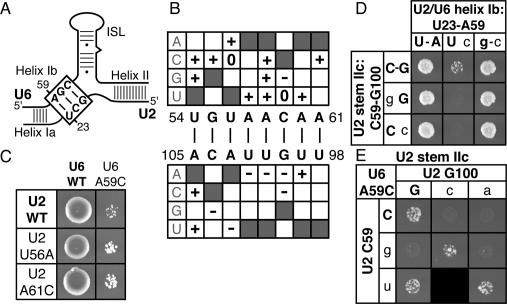
Because of the evidence suggesting that the dynamic stem II region of U2 (Zavanelli and Ares 1991; Zavanelli et al. 1994) rearranges after 5′ splice site cleavage (Barabino et al. 1992), we hypothesized that stem IIc is disrupted after 5′ splice site cleavage and that this disruption is promoted by Prp16p, the DEAH box ATPase that promotes splicing after 5′ splice site cleavage (Schwer and Guthrie 1991, 1992). To evaluate this hypothesis, we tested the prediction that the cold-sensitive growth defect of the prp16-302 mutant (Madhani and Guthrie 1994a) would be suppressed by mutations that disrupted U2 stem IIc, thereby relieving the requirement for PRP16. Indeed, 12 of 13 point mutants tested in either the upstream or downstream strands of stem IIc strongly suppressed the cold-sensitive defect of prp16-302 (Fig. 3A,B; data not shown). For example, all point mutations in either base pair C59/G100 (Fig. 3A, right) or base pair U56/A103 (Fig. 3B, right) suppressed prp16-302, with the exception of U2–A103G, which could maintain pairing through a wobble with U2–U56, as suggested by the data in Figure 1E. Importantly, compensatory mutations that repaired stem IIc abolished the suppression of prp16-302 in every case; in contrast, noncompensatory mutations that failed to repair stem IIc failed to abolish suppression (Fig. 3A,B). Consistent with these results, Perriman and Ares (2007) found that deleting the downstream strand of stem IIc also suppressed the growth defect of prp16-302. Our results suggest that Prp16p promotes, directly or indirectly, destabilization of stem IIc.
Figure 3.
Disrupting U2 stem IIc or flanking structures mutually exclusive with stem IIa suppresses a mutation in PRP16, a DExD/H box ATPase. (A,B) Disruption of U2 stem IIc suppresses the mutation prp16-302. Compensatory analysis by growth of the U2 stem IIc base pairs C59/G100 (A) and U56/A103 (B) in a wild-type PRP16 (left) or a mutant prp16-302 (right) strain is shown. The cells were grown for 3 d (left) or 6 d (right) at 20°C on solid media containing 5-FOA. (C,D) Disruption of structures mutually exclusive with stem IIa suppresses prp16-302. Compensatory analysis by growth of the U2 stem IIa base pairs G53/C62 (C) and A52/U63 (D) in a wild-type PRP16 (left) or a mutant prp16-302 (right) strain. Cells were grown for 3 d (D, left), 4 d (C, left) or 6 d (C,D, right) at 20°C on solid media containing 5-FOA. Black boxes indicate that the mutation is lethal and could not be tested. Matrices are labeled as in Figure 1.
Evidence that Prp16p destabilizes interactions that are physically mutually exclusive with stem IIa
Because stem IIc antagonizes stem IIa in the free U2 snRNP (Zavanelli and Ares 1991; Zavanelli et al. 1994), we hypothesized that Prp16p promotes the formation of stem–loop IIa both by destabilizing stem IIc, which is functionally mutually exclusive with stem IIa (Zavanelli and Ares 1991; Zavanelli et al. 1994), and also by destabilizing structures physically mutually exclusive with stem IIa. Indeed, seven out of eight point mutations tested in the strands of U2 stem IIa suppressed prp16-302; these mutations included G53A, A52U, and U63C (Fig. 3C,D; data not shown). To test whether these mutations suppressed prp16-302 by destabilizing U2 stem IIa or a mutually exclusive structure, we tested whether or not suppression of prp16-302 was abolished by compensatory mutations that repaired stem IIa. Importantly, compensatory mutations that repaired stem IIa failed to abolish the suppression prp16-302; of five compensatory mutations tested at three different positions in stem IIa, none abolished suppression (Fig. 3C,D; data not shown). These results indicate that point mutations in the strands of stem IIa do not suppress prp16-302 by disrupting stem IIa but rather by disrupting an interaction that is physically mutually exclusive with stem IIa, as stem IIa mutants suppress regardless of their base-pairing potential. Thus, our data suggest that Prp16p not only promotes destabilization of stem IIc, which is functionally mutually exclusive with stem IIa, but also promotes destabilization of structures physically mutually exclusive with stem IIa. By promoting destabilization of these structures, Prp16p may promote formation of stem IIa after 5′ splice site cleavage (see below).
Disrupting stem IIa suppresses a 3′ splice site mutation
To determine whether stem IIc unwinds to allow stem–loop IIa to form before stem IIc functions in exon ligation (Fig. 2; see below), we tested whether mutations that disrupt the stem of stem–loop IIa suppressed a mutation at the 3′ splice site that destabilizes the exon ligation conformation of the spliceosome (Query and Konarska 2004). Indeed, mutations that disrupt stem IIa suppressed the 3′ splice site mutation (Fig. 4).
Figure 4.
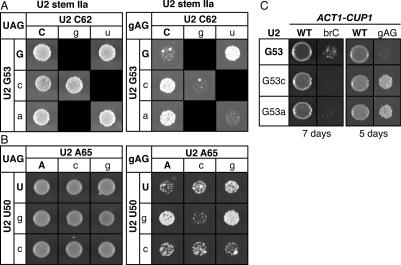
Destabilization of stem IIa suppresses the 3′ splice site mutant gAG. (A,B) Disruption of base-pairing in U2 stem IIa suppresses a 3′ splice site mutation. Compensatory analysis by copper resistance of the U2 stem IIa base pair G53/C62 (A) or U2–U50/A65 (B) is shown. The matrices show the copper resistance of a wild-type (left) or mutated 3′ splice site mutant (gAG; right) ACT1-CUP1 splicing reporter in yJPS1035 expressing a U2 variant having single or double mutations in the stem IIa base pairs. Cells were grown for 3 d (left) or 5 d (right) at 30°C on solid media containing 0.05 mM CuSO4. Black boxes represent lethal U2 alleles that could not be included in the analysis. (C) Disrupting U2 stem IIa does not suppress the mutated branch site substrate brC. For comparison, suppression of the gAG mutated 3′ splice site substrate is shown. Mutations were tested in strain yJPS1035. The identity of U2 is indicated to the left and the identity of the reporter is indicated at the top. Cells were grown for 5 or 7 d, as indicated below the data, at 30°C on 0.1 mM CuSO4.
To test in vivo for genetic interactions between stem IIa and the 3′ splice site, we utilized an ACT1-CUP1 splicing reporter containing a 3′ splice site mutation (gAG) in the antepenultimate position of the consensus, a mutation that decreases the ratio of mRNA to lariat intermediate and decreases growth in the presence of copper (Umen and Guthrie 1996). We found that this defect in copper resistance is suppressed strongly by the U2 point mutations U50G, G53C, G53A, C62U, U63C, and G64A and moderately by the U2 point mutations U50C, A65C, and A65G (Fig. 4A,B; data not shown). Although we have been unable to recapitulate the suppression by primer extension analysis of RNA from cells grown in liquid culture (data not shown), the suppression conferred by the stem IIa mutations G53A and G53C, for example, is specific to a 3′ splice site mutation, because these mutations exacerbated the branch site mutation brC (Fig. 4C; data not shown); note that the exacerbation of the brC mutation was not abolished upon repair of stem IIa (data not shown), consistent with a role for stem IIc rather than stem IIa at the 5′ splice site cleavage stage. The lack of observable suppression by primer extension analysis as compared with the suppression by copper resistance may reflect the significant differences in the growth conditions of these two assays—growth in liquid culture in the absence of selection for copper resistance as compared with growth on solid media selecting for copper resistance, respectively. Alternatively, the negative results may reflect the limits of the primer extension assay, with regard to its inability to explicitly assay some stages of splicing, such as mRNA release from the spliceosome. The suppression of the 3′ splice site mutation conferred by the stem IIa mutations was abolished by stem IIa compensatory mutations that restored base-pairing (Fig. 4A,B). In contrast, at least for base pair U2–U50/A65, noncompensatory mutations that do not restore base-pairing failed to abolish suppression (Fig. 4B). Note that we were unable to test for suppression by noncompensatory mutations that failed to repair the G53/C62 base pair, because these double mutations were lethal. Together, these data suggest that the exon ligation defect conferred by the gAG 3′ splice site mutation is suppressed by destabilizing stem IIa, perhaps because the destabilizing effect of stem IIa offsets the destabilizing effect of the 3′ splice site mutation on the exon ligation conformation (Query and Konarska 2004). Thus, these data suggest both that stem IIa competes with the second catalytic conformation of the spliceosome and that stem IIa must unwind to permit the catalysis of exon ligation. Because stem IIc promotes the first catalytic conformation, these observations suggest that stem IIa stabilizes an intermediate in the transition from the first catalytic conformation of the spliceosome to the second.
Mutating the loop of stem–loop IIa also suppresses substrates defective in exon ligation
Just as mutations in the stem of stem–loop IIa suppressed a 3′ splice site mutation, mutations in the loop of stem–loop IIa also suppressed mutated substrates defective for exon ligation, further supporting a role for stem–loop IIa after 5′ splice site cleavage and before exon ligation. To test in vivo for genetic interactions between loop mutations and substrate mutations that impair exon ligation, we utilized an ACT1-CUP1 splicing reporter having a mutation of the 3′ splice site (gAG) or a mutation of the branch site adenosine to guanosine (brG). Like the gAG mutation, the brG mutation limits splicing at the stage of exon ligation, as revealed by a decrease in the ratio of mRNA to lariat intermediate and a decrease in growth of budding yeast in the presence of copper (Umen and Guthrie 1996; Query and Konarska 2004). We found that point mutations throughout the loop of stem–loop IIa, such as U2–G55C and U2–A60U, suppressed the copper resistance phenotypes of the gAG and brG substrates (Fig. 5A,C). Mutations that alter multiple nucleotides in the loop, such as UAA56–58AUU and GUAA55–58CAUU, suppressed the exon ligation mutations brG and gAG more robustly (Fig. 5B; data not shown); these multiple U2 mutations also suppressed substrate mutations at the second (UAG to UgG) and third (UAG to UAc) positions of the 3′ splice site (data not shown). As for the suppression by stem IIa mutations, we were unable to recapitulate the suppression by loop IIa mutations by primer extension analysis of RNA from cells grown in liquid culture (data not shown), thereby complicating a determination of the stage of suppression. Unlike mutations in the upstream strand of stem IIc—i.e., loop IIa—mutations in the downstream strand of stem IIc failed to suppress 3′ splice site mutations (Fig. 5B,C; data not shown).
Figure 5.
Mutations in U2 loop IIa suppress the 3′ splice site mutation gAG and the branch site mutation brG, independently of disrupting stem IIc. (A–C) Copper resistance of U2 mutants in yJPS1035 expressing the wild-type, U2A, brC, brG, or gAG ACT1-CUP1 reporters. Cells were grown at 30°C on solid media containing 0.2 mM (A,C) or 0.1 mM (B) CuSO4. (A) Point mutants in loop IIa suppress the 3′ splice site mutation gAG and the branch site mutation brG but exacerbate or fail to suppress the U2A and brC mutations. Cells were grown for 2 d (WT; U2A), 3 d (gAG), 4 d (brC), or 5 d (brG). For each strain, the identity of U2 is indicated to the left (cf. Fig. 1A) and the identity of the ACT1-CUP1 reporter is shown at the top. (B) Mutations that severely alter loop IIa suppress gAG strongly, but mutations that severely alter the downstream strand of stem IIc do not. The position of the mutations in loop IIa or the downstream strand of stem IIc is indicated by asterisks below the affected strand. The U2 alleles are (from top row to bottom) wild type, UAA56–58AUU, GUAA57–58CAUU, UUA101–103AAU, and UUGUUACA98–105AACAAUGU. Cells were grown for 3 d (WT) or 6 d (gAG) at 30°C on solid media containing 0.2 mM CuSO4. (C) Restoration of stem IIc does not abolish suppression but rather improves suppression. A compensatory analysis by copper resistance of the U2 stem IIc base pair U56/A103 is shown. The matrix shows the copper resistance of the gAG mutated 3′ splice site reporter in yJP1035 expressing U2 variants having single or double mutations in the stem IIc base pair. Cells were grown for 6 d at 0.1 mM CuSO4.
The asymmetry between the two strands of stem IIc in suppressing the gAG 3′ splice site mutation suggests strongly that the mutations in the upstream strand of stem IIc—i.e., mutations in loop IIa—suppressed independently of destabilizing stem IIc. Indeed, the weak suppression conferred by the loop IIa mutation U2–U56A was not abolished by the compensatory mutation U2–A103U, which repaired base-pairing in stem IIc; rather, the compensatory mutation improved suppression (Fig. 5C). Furthermore, whereas the single mutants U56C or A103G did not suppress the 3′ splice site mutation, the double mutation U56C/A103G, in which stem IIc was repaired, did suppress (Fig. 5C). In contrast, double mutants, such as U56A/A103G, which fail to restore base-pairing in stem IIc, failed to suppress or to increase suppression of the 3′ splice site mutation. The requirement for stem IIc in strongly suppressing a 3′ splice site mutation is consistent with the requirement for stem IIc in exon ligation (Fig. 2; see below). Our data suggest that loop IIa mutations suppress exon ligation mutations indirectly by disrupting an unidentified interaction mutually exclusive with stem IIc. Thus, these data suggest (1) that the loop of stem–loop IIa cooperates with the stem of stem–loop IIa in competing with the second catalytic conformation of the spliceosome, and (2) that, as for stem IIa, an unknown interaction involving loop IIa must dissociate to permit the catalysis of exon ligation. Further, these observations suggest that the loop of stem–loop IIa cooperates with the stem of stem–loop IIa in stabilizing an intermediate in the transition from the first catalytic conformation of the spliceosome to the second catalytic conformation.
Evidence that stem IIc promotes exon ligation
Just as mutations in the loop of stem–loop IIa suppressed the defect conferred by the 3′ splice site mutation gAG (Fig. 5), mutations in the loop suppressed the growth defect of U6–A59C, a mutation in the conserved AGC triad of U6 snRNA (Fig. 6A). The U6–A59C mutation permits 5′ splice site cleavage but impedes exon ligation both in vitro (Fabrizio and Abelson 1990) and in vivo (Hilliker and Staley 2004). The exon ligation defect results from disrupting the conserved U2/U6 helix Ib (Fig. 6A; Hilliker and Staley 2004), but it has been unclear whether helix Ib stabilizes an intermediate after 5′ splice site cleavage, promotes the conformation of the spliceosome that catalyzes exon ligation, or both.
Figure 6.
Disrupting U2 stem IIc exacerbates disruption of U2/U6 helix Ib, which includes the U6–AGC triad. (A) Schematic of U2/U6 base-pairing; U2/U6 helix Ib is boxed. (B–E) Mutations in the 5′ side of stem IIc largely suppress U6–A59C, while mutations in the 3′ side largely exacerbate. Genetic interactions between U2 stem IIc point or double mutations and U6 variants in strain XYU96 (Xu et al. 1998) are shown. (B) Summary of genetic interactions between U2 stem IIc point mutations and U6–A59C, a mutation in the AGC triad that disrupts U2/U6 helix Ib. Weak suppressors (+), enhancers (−), and those that do not interact (0) are indicated. Shaded boxes mark wild-type nucleotides; white boxes mark mutations that were not tested. Note: For all mutations tested, repair of stem IIc failed to abolish suppression (data not shown) but did abolish enhancement (see E; data not shown). (C) Representative mutations in loop IIa that weakly suppress the growth defect of U6–A59C. The identity of U2 is indicated to the right and the identity of U6 is indicated on top. Cells were grown for 6 d at 30°C on solid media containing 5-FOA. (D) Mutations in U2 stem IIc that exacerbate the growth defect of U6–A59C do so by enhancing disruption of U2/U6 helix Ib. The matrix shows the growth of cells expressing U2 variants that disrupt the U2 stem IIc base pair C59/G100 (rows) and U2 and/or U6 mutations that alter base-pairing in U2/U6 helix Ib (columns). Cells were grown for 5 d at 30°C on solid media containing 5-FOA. (E) Mutations in U2 stem IIc that exacerbate the growth defect of U6–A59C do so by disrupting stem IIc. A compensatory analysis by growth is shown for the U2 stem IIc base pair C59/G100. All strains expressed U6–A59C. Growth conditions were the same as in D.
To gain further insight into the function of U2/U6 helix Ib and the dynamics of the U2 stem II region in exon ligation, we assayed for genetic interactions between U6–A59C and mutations in stem II, starting with mutations in the loop of stem–loop IIa. Ten of 13 mutations tested in the loop weakly suppressed the growth defect of U6–A59C (Fig. 6B,C). Similar to the loop IIa suppressors of the 3′ splice site mutation gAG (Fig. 5C), the loop IIa suppressors of U6–A59C suppressed independently of disrupting stem IIc (data not shown), indicating that the loop IIa mutations disrupted a mutually exclusive interaction. Given that many different loop IIa mutations suppressed U6–A59C, these mutations likely suppressed U6–A59C indirectly by destabilizing a conformation of the spliceosome that competes with an alternative conformation stabilized by U2/U6 helix Ib. Given the parallel suppression of U6–A59C and 3′ splice site mutations by loop IIa mutations, these data suggest a role for U2/U6 helix Ib at the stage of 3′ splice site recognition and consequently at the stage of catalyzing exon ligation.
In contrast to the loop IIa mutations, which reside in the upstream strand of stem IIc, mutations in the downstream strand of stem IIc exacerbated the growth defect of U6–A59C; further, these mutations exacerbated the growth defect of U6–A59C by disrupting stem IIc. Specifically, six of nine downstream mutations were synthetically lethal with U6–A59C (Fig. 6B,D,E; data not shown). Compensatory mutations that restored base-pairing abolished the synthetic lethality, whereas noncompensatory mutations that did not restore base-pairing failed to abolish the synthetic lethality (Fig. 6E). These data indicate that stem IIc cooperates with a structure that includes U6–A59.
U6–A59 participates in at least four structures, including U2/U6 helix Ib (Madhani and Guthrie 1992; Hilliker and Staley 2004), the central stem of U6 (Fortner et al. 1994), U4/U6 stem I (Brow and Guthrie 1988), and an extension of the intramolecular stem–loop (ISL) of U6 (Sun and Manley 1995; Sashital et al. 2004). To determine whether the synthetic lethality between stem IIc mutations and U6–A59C resulted from disruption of one of these structures, we repaired each structure individually and reassessed the growth phenotype. Repair of U2/U6 helix Ib abolished the synthetic lethality (Fig. 6D) whereas repair of the other structures failed to abolish the synthetic lethality (data not shown), indicating that stem IIc cooperates with U2/U6 helix Ib. Given the implication of a role for helix Ib in the catalytic conformation of exon ligation (Fig. 6B,C; Hilliker and Staley 2004), these data suggest a role for stem IIc at the catalytic stage of exon ligation, consistent with independent observations described above (Figs. 2, 5C). Thus, our data implicate not only a requirement for dissociating stem–loop IIa for exon ligation but also a requirement for reforming stem IIc for exon ligation.
Discussion
To catalyze exon ligation, the spliceosome must reposition the substrate after 5′ splice site cleavage. Suggesting that rearrangements in U2 may help reposition the substrate, we have found evidence that after 5′ splice site cleavage the stem II region of U2 snRNA toggles between two mutually exclusive structures, stem IIc and stem–loop IIa. First, destabilizing U2 stem IIc exacerbated the 5′ splice site cleavage defect of two substrates mutated at either the 5′ splice site or the branch site (Figs. 1, 2), demonstrating that stem IIc promotes 5′ splice site cleavage despite repressing prespliceosome formation (Zavanelli and Ares 1991; Zavanelli et al. 1994). Second, disrupting stem IIc suppressed the cold-sensitive growth defect of prp16-302 (Fig. 3), which (1) supports a role for stem IIc at the catalytic stage of 5′ splice site cleavage, (2) indicates that stem IIc must be disrupted after 5′ splice site cleavage, and (3) suggests that Prp16p promotes destabilization of stem IIc. Because U2 stem IIc is functionally mutually exclusive with stem IIa (Zavanelli and Ares 1991; Zavanelli et al. 1994), Prp16p may promote disruption of stem IIc to promote formation of stem–loop IIa. Consistent with this possibility, disrupting structures that are physically mutually exclusive with stem IIa also suppressed the prp16-302 mutant (Fig. 3). Third, destabilizing the stem or loop of stem–loop IIa suppressed a 3′ splice site mutation defective for exon ligation (Figs. 4, 5), suggesting that stem–loop IIa does form after 5′ splice site cleavage but then must unwind before exon ligation. Fourth, disrupting stem IIc (1) impaired exon ligation of a wild-type and a mutated brC substrate (Fig. 2), (2) exacerbated a 3′ splice site mutation (Fig. 5C), and (3) exacerbated U2/U6 helix Ib disruption (Fig. 6), which impairs exon ligation (Hilliker and Staley 2004), suggesting a role for stem IIc in exon ligation and supporting a requirement to unwind stem–loop IIa before exon ligation. Thus, our data suggest that during the catalytic phase of splicing the stem II region of U2 toggles from stem IIc to stem–loop IIa and then back again to stem IIc (Fig. 7, iii–v). Further, our data suggest that stem IIc promotes the catalytic states of the spliceosome and that stem–loop IIa promotes a transient intermediate that both facilitates the transition from the first catalytic state to the second and contributes to the fidelity of exon ligation (Figs. 4, 5).
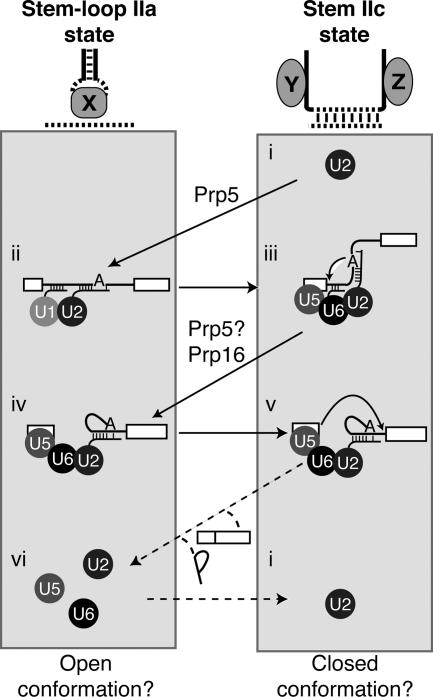
Figure 7.
U2 stem–loop IIa and stem IIc toggle during both the assembly phase and the catalytic phase of splicing. Our data suggest a model in which U2 stem II toggles between stem–loop IIa and stem IIc throughout splicing; this toggling may reflect larger conformational rearrangements in the spliceosome. We propose that the stem IIc state stabilizes a closed state of the spliceosome that promotes catalysis (right conformation states) and that stem–loop IIa stabilizes an open state of the spliceosome that promotes substrate sampling, rearrangement, and release (left conformational states). First, the free U2 snRNP in the stem IIc state (i) is converted by the DExD/H box ATPase Prp5p to the stem–loop IIa state to promote binding of U2 to the pre-mRNA and formation of the prespliceosome (Perriman and Ares 2007). Next, U2 must toggle from the stem–loop IIa state back to the stem IIc state, as the stem IIc state promotes 5′ splice site cleavage (iii; Figs. 1, 2). After 5′ splice site cleavage, stem IIc is destabilized by the DExD/H box ATPase Prp16p (Fig. 3), directly or indirectly, promoting formation of stem–loop IIa in the inferred intermediate (iv), a reaction that may also be promoted by Prp5p (Perriman and Ares 2007). Stem–loop IIa, in turn, is destabilized (Figs. 4, 5) to promote reformation of stem IIc and exon ligation (v; Figs. 2, 5C, 6). After exon ligation, toggling of stem IIc back to stem–loop IIa (vi) could reconfigure the spliceosome to promote product release and/or spliceosome disassembly. The free U2 snRNP may then switch back to the stem IIc state (i). The connectivity or changing connectivity of the substrate is shown for each conformation, and the snRNPs are shown as circles. Our data suggest that loop IIa interacts with an unknown factor, shown here as “X,” in the intermediate (iv; Figs. 5, 6) and that the strands of stem IIa interact with unknown factors, shown here as “Y” and “Z,” in the 5′ splice site cleavage conformation (ii; Fig. 3C,D), precluding stem IIa formation at this stage.
Unexpectedly, U2 stem IIc promotes the catalytic stages of splicing
Our observations support a role for U2 stem IIc at both catalytic stages of splicing. Given that stem IIc inhibits prespliceosome formation (Zavanelli and Ares 1991; Zavanelli et al. 1994), a role for stem IIc during 5′ splice site cleavage is surprising. In addition, as stem–loop IIa promotes prespliceosome formation and therefore the binding of U2 snRNP to the branch site (Zavanelli and Ares 1991; Zavanelli et al. 1994), a role for stem IIc during 5′ splice site cleavage implicates a switch from the stem–loop IIa state to the stem IIc state at some point during spliceosome assembly or activation (Fig. 7, cf. ii,iii). The proximity of the stem II region to the branch-site-binding region and the role of the stem II region in regulating the interaction between the free U2 snRNP and pre-mRNA suggest several possible roles for stem IIc during 5′ splice site cleavage, roles in which stem IIc could permit the branch site to access the catalytic core of the spliceosome. First, since stem IIc antagonizes binding of the free U2 snRNP to the branch site (Zavanelli and Ares 1991; Zavanelli et al. 1994), reformation of stem IIc during spliceosome assembly could release the branch site from U2 to allow the branch site to interact with another partner. However, there is no evidence that the U2–branch point helix is disrupted during splicing. Second, upon stem IIc formation, the U2–branch point interaction may persist but dissociate from the U2 snRNP protein factors. Third, on forming stem IIc within the active spliceosome, U2 may bind the substrate even more stably than in the stem–loop IIa state. In this model, although stem IIc would not permit initial binding of U2 to the branch site, this block would represent only an insurmountable kinetic barrier and not a thermodynamic constraint. In this way, the stem–loop IIa state could function as a “snRNP loader” state, allowing U2 to engage the substrate, after which rearrangement to the stem IIc state could allow U2 to clamp down on the substrate. In each case, reformation of stem–loop IIa after 5′ splice site cleavage could allow removal of the branch site from the catalytic core after 5′ splice site cleavage and thereby account for its role in the inferred intermediate (see below). In group II introns, RNA recognition of the branch site may persist through both catalytic steps of splicing (Hamill and Pyle 2006). As the branch site is monitored at the exon ligation stage (Mayas et al. 2006) and our results suggest that stem IIc reforms to promote exon ligation, stem IIc could similarly promote branch site recognition during both 5′ splice site cleavage and exon ligation.
Intriguingly, paralleling the two conformations of stem II, the U2 snRNP factor, SF3b155p, a member of the dynamic HEAT-repeat protein family, assumes two conformations as visualized by cryoelectron microscopy (Golas et al. 2005). Within the purified SF3b complex, SF3b155p wraps around p14, a protein that cross-links to the branch point adenosine (Query et al. 1996; Will et al. 2001), sterically precluding the interaction between p14 and the branch site (Golas et al. 2003). In contrast, within the U11/U12 di-snRNP of the minor metazoan spliceosome, a snRNP that corresponds to the U1 and U2 snRNPs of the major spliceosome (for review, see Patel and Steitz 2003), SF3b155p opens up into a conformation that would allow p14 to interact with the branch site (Golas et al. 2005). Given this parallel between the positive and negative roles of stem II and SF3b155p, it is tempting to speculate that the stem IIc state of U2 correlates with the closed state of SF3b155p and the stem–loop IIa state correlates with the open state of SF3b155p. Further, because the SF3b complex both cross-links to the branch site region (Champion-Arnaud and Reed 1994; MacMillan et al. 1994; Gozani et al. 1996, 1998) and protects the stem II region of U2 from micrococcal nuclease (Kramer et al. 1999), the SF3b complex may couple stem II rearrangements with branch site binding.
Although stem IIc is not essential for growth (Figs. 1C, 5B; data not shown; Ares and Igel 1990), it is highly conserved from budding yeast to humans (Fig. 1B), implicating stem IIc in a fundamental role in splicing. We propose that stem IIc functions redundantly with other interactions to compete with stem–loop IIa. For example, stem IIc may cooperate with SF3b, as discussed above. In addition, stem IIc may cooperate with the interactions implicated by mutations in the strands of stem IIa that suppress prp16-302, interactions that are necessarily mutually exclusive with stem IIa (Figs. 3, 7 [interactions with factors Y and Z]). Consistent with a redundant role for stem IIc, stem IIc is not conserved in the U12 snRNP of the minor spliceosome, whereas stem IIa is conserved (Tarn et al. 1995). Paralleling this difference between the major and minor spliceosomes, the SF3a complex does not interact with the minor spliceosome, whereas SF3b does (Will et al. 1999). Given genetic data suggesting that SF3a cooperates with stem–loop IIa (Wells and Ares 1994), the absence of SF3a may offset the absence of stem IIc, especially if the two antagonize one another in the major spliceosome.
A parallel between assembling and rearranging the spliceosome
At the stage of prespliceosome formation, splicing requires that stem IIc unwinds and that stem–loop IIa forms (Zavanelli and Ares 1991; Zavanelli et al. 1994). Our data suggest that after stem IIc promotes 5′ splice site cleavage, splicing again requires that stem IIc unwinds and that stem–loop IIa forms (Figs. 3, 4, 5). Thus, toggling from the stem IIc state to the stem–loop IIa state promotes both assembly of the spliceosome and rearrangement of the spliceosome. Because the stem II region lies just downstream from the branch-site-binding region and because the toggling in the free U2 snRNP regulates substrate binding (Zavanelli and Ares 1991; Zavanelli et al. 1994), we propose that toggling in the active spliceosome after 5′ splice site cleavage similarly regulates interactions with the substrate. The toggling from stem IIc to stem IIa may promote rearrangements required for exon ligation, such as removal of the branched product from the catalytic core to permit engagement of the 3′ splice site with the 5′ exon. Switching from stem IIc to the stem–loop IIa state likely reverses rearrangements, such as those considered above, that occur during stem IIc formation. For example, if stem IIc formation results in the dissociation of U2 snRNA from the branch site, then stem–loop IIa formation may result in recapture of the branch site by U2. Alternatively, if stem IIc formation results in dissociation of the U2–branch site helix from the U2 snRNP proteins, then stem–loop IIa formation may result in recapture of the helix by the U2 snRNP proteins. Finally, if stem IIa formation acts like a snRNP loader and stem IIc formation allows the U2 snRNP to clamp down on the U2–branch site interaction, then stem–loop IIa formation may allow the U2 snRNP to open and to allow movement of the U2–branch site interaction. Just as stem–loop IIa forms anew after the spliceosome cleaves the 5′ splice site, the loop of domain VI, which includes the branch site helix, forms a new interaction with domain II after the Sc.cox1/5 group II intron cleaves the 5′ splice site; this conformational change may help remove the branch product from the catalytic core of group II introns (Chanfreau and Jacquier 1996). Thus, our work extends the analogy between the spliceosome and group II introns.
A role for the DEAH box ATPase Prp16p in the rearrangement of the U2 stem II region
We found evidence that Prp16p promotes disruption of stem IIc. Specifically, mutations that disrupt stem IIc suppressed prp16-302 (Fig. 3; Perriman and Ares 2007), indicating that stem IIc unwinding cooperates with Prp16p function. Although stem IIc unwinding could conceivably occur upstream of Prp16p to activate it, given that Prp16p destabilizes RNA structures in vitro (Wang et al. 1998), the simplest model is that stem IIc unwinding occurs downstream from Prp16p, as a direct or indirect consequence of Prp16p activity. Previously, suppressors of prp16 mutants have been identified in U6 snRNA (Madhani and Guthrie 1994a), PRP8 (Query and Konarska 2004), and ISY1 (Villa and Guthrie 2005), implicating additional interactions that may be destabilized by Prp16p; however, the interacting partners for these suppressors remain to be identified. The multiplicity of genes that suppress prp16-302 suggests that Prp16p destabilizes multiple interactions in distinct spliceosomal components, implying either that Prp16p destabilizes multiple interactions directly or destabilizes one interaction directly and others indirectly.
We found evidence that Prp16p not only promotes destabilization of stem IIc, a structure functionally mutually exclusive with stem–loop IIa, but that Prp16p also promotes destabilization of structures that are physically mutually exclusive with stem–loop IIa (Fig. 7, interactions with factors Y and Z). Specifically, point mutations in either strand of stem IIa suppress prp16-302 (Fig. 3). Repairing stem IIa does not reverse suppression, suggesting strongly that suppression is not due to stem IIa disruption but rather to disruption of mutually exclusive interactions that flank stem IIc, interactions that may cooperate with stem IIc during 5′ splice site cleavage but that remain to be identified.
Although Prp16p promotes the transition from stem IIc to stem–loop IIa after 5′ splice site cleavage, Prp5p promotes this transition during prespliceosome formation (Perriman and Ares 2007). Prp16p may assume the role of promoting this transition after 5′ splice site cleavage, implying that a single rearrangement can be promoted by distinct DExD/H box ATPases. Alternatively, Prp16p may cooperate with Prp5p. In this case, Prp5p would function again after 5′ splice site cleavage to promote the switch of stem IIc to stem–loop IIa and Prp16p would destabilize distinct interactions, such as those implied by the suppressors in U6, PRP8, and ISY1 (Madhani and Guthrie 1994a; Query and Konarska 2004; Villa and Guthrie 2005). Consistent with a second role for Prp5p and the possibility of cooperation with Prp16p, Perriman and Ares (2007) have found that mutations in PRP5 suppress the 5′ splice site cleavage defect of the brC mutant substrate, just as mutations in PRP16 suppress. Given that Prp5p is antagonized by Cus2p at the prespliceosome stage (Perriman et al. 2003) and that Cus2p binds to the stem IIc state of the free U2 snRNP (Perriman and Ares 2007), Cus2p may also function again in the active spliceosome to bind the stem IIc state of U2 and/or to antagonize Prp5p.
Evidence for an intermediate after 5′ splice site cleavage and before exon ligation
Our data suggest that a spliceosomal intermediate forms after 5′ splice site cleavage, an intermediate that is characterized by the presence of U2 stem–loop IIa (Fig. 7, iv). Specifically, mutations that destabilize the stem (Fig. 4) or the loop (Fig. 5) of stem–loop IIa suppress the copper sensitivity of a 3′ splice site mutant, suggesting that stem IIa is formed and that loop IIa interacts with an unknown factor (Fig. 7, factor X) prior to exon ligation. It is conceivable that the stem–loop IIa-containing intermediate only forms when the exon ligation conformation of the spliceosome is destabilized by a substrate mutation. However, consistent with the more general formation of an intermediate, 5′ splice site cleavage is followed by and exon ligation is preceded by two steps (for review, see Umen and Guthrie 1995): an ATP-dependent step, requiring Prp16p (Schwer and Guthrie 1992), and a subsequent ATP-independent step that results in 3′-splice-site-binding and that requires Prp22p, Slu7p, and Prp18p (Ansari and Schwer 1995; Jones et al. 1995; Schwer and Gross 1998; Wagner et al. 1998). The intermediate may serve as a stage to release factors such as Prp16p and to bind factors such as Prp22p, Slu7p, and Prp18p. Moreover, as a conformational state structurally unique from the catalytic states, the intermediate state may promote rearrangement of the substrate by stabilizing an open conformation of the spliceosome.
Toggling of U2 stem II suggests a mechanism for rearranging the substrate in the activated spliceosome that is analogous to rearrangements in the ribosome
We propose that the toggling of the stem II region reflects a larger toggling of the spliceosome between a closed, catalytic conformation and an open, noncatalytic conformation. Specifically, we propose that stem IIc stabilizes a closed conformation of the spliceosome that binds the substrate tightly to promote chemistry and that stem–loop IIa stabilizes an open conformation of the spliceosome that binds the substrate loosely to promote binding, rearrangement, and release of the substrate and perhaps discard of incorrect substrates (Fig. 7); this open conformation corresponds to a stable version of the hypothetical “unlocked” state proposed by Konarska and Query (2005). This model is supported by our evidence that U2 stem IIc promotes both catalytic steps in splicing and that stem–loop IIa promotes an intermediate between the two catalytic steps (see also Perriman and Ares 2007). In addition, this model is supported by the role for stem–loop IIa in promoting binding of U2 snRNP to pre-mRNA (Zavanelli and Ares 1991; Zavanelli et al. 1994). No evidence yet supports a role for stem–loop IIa after exon ligation in promoting release of the mRNA and/or excised lariat intron product, but consistent with such a role, stem–loop IIa must reform after exon ligation (Fig. 7, v) in preparation for the next round of splicing (Fig. 7, ii). In the model, the spliceosome toggles between an open stem–loop IIa state and a closed stem IIc state to promote the two rounds of transesterification required in pre-mRNA splicing.
This model suggests an analogy with the ribosome, which similarly toggles between an open and closed conformation during each round of peptide bond formation (Ogle et al. 2002). In the open state, the ribosome recruits aminoacyl tRNA. In the closed state, the ribosome promotes peptide bond formation. Opening of the ribosome allows recruitment of the next aminoacyl tRNA and multiple rounds of peptide bond formation.
Previously, Query and Konarska (2004) discovered evidence that the spliceosome promotes fidelity in a manner similar to the ribosome. Studies of the ribosome suggest that the open and closed states of the ribosome can compete, thereby allowing the open state to serve as a sink for incorrect substrates that destabilize the closed, catalytic state (Ogle et al. 2003). Similarly, Query and Konarska (2004) found compelling evidence that the two catalytic conformations of the spliceosome can compete. Specifically, they found that substrate mutations that destabilized one catalytic conformation could be suppressed by spliceosomal mutations that destabilized the other conformation, indicating that one catalytic conformation can serve as a sink for incorrect substrates inappropriate for the other catalytic conformation. This fidelity mechanism for the spliceosome differs from the fidelity mechanism for the ribosome in that the splicing fidelity mechanism depends on competition between two catalytic and likely closed conformations, whereas the ribosome fidelity mechanism depends on competition between a closed catalytic conformation and an open noncatalytic conformation.
Our work suggests a related mechanism for establishing fidelity in splicing, a mechanism that implies a deeper, more direct analogy between the spliceosome and the ribosome. In the ribosome, destabilizing the closed state improves fidelity whereas destabilizing the open state compromises fidelity (Ogle et al. 2003). Similarly, in the spliceosome, destabilizing the closed stem IIc state improves fidelity whereas destabilizing the open stem–loop IIa state compromises fidelity. Specifically, we have found that destabilizing stem IIc in the putative closed state improves the fidelity of branch site selection; whereas destabilizing stem IIc has no effect on the efficiency of 5′ splice site cleavage for a wild-type substrate, destabilizing stem IIc compromises the efficiency of 5′ splice site cleavage for a mutated brC substrate, increasing the specificity for 5′ splice site cleavage (Figs. 1D,E, 2). Similarly, mutations that destabilize the first catalytic conformation specifically, such as U6–U57A (McPheeters 1996), comprise the efficiency of 5′ splice site cleavage for a mutated brC substrate without compromising a wild-type substrate, thereby increasing the specificity for 5′ splice site cleavage. In contrast to stem IIc destabilization, which improves fidelity, destabilizing the open stem–loop IIa state compromises fidelity. Specifically, as indicated by the ACT1-CUP1 splicing reporter, whereas destabilizing stem–loop IIa has no effect on the production of mRNA from a wild-type substrate, destabilizing stem–loop IIa improves production of mRNA from a substrate mutated at the 3′ splice site, thereby decreasing specificity (Figs. 4, 5). Given the broad range of 3′ splice site suppressors in stem–loop IIa, we interpret the suppression as indirect and resulting from a shift in an equilibrium from the intermediate stem–loop IIa state back to the exon ligation conformation of the spliceosome, which is effectively destabilized by a 3′ splice site mutation (Query and Konarska 2004). In this view, the intermediate state, like the 5′ splice site cleavage conformation (Query and Konarska 2004), serves as an important sink to sequester incorrect substrates from the exon ligation conformation of the spliceosome and perhaps promotes discard of aberrant substrates. Thus, the switch of stem IIc to stem–loop IIa not only promotes prespliceosome formation and rearrangement of the spliceosome but also the fidelity of splicing.
Materials and methods
Pictogram analysis
Thirty-seven U2 sequences from 35 organisms were chosen from the seed set of the Rfam database (Griffiths-Jones et al. 2005); duplicate U2 genes from the same organism that did not differ in the stem II region of U2 were discarded. The sequences were aligned using ClustalW (Thompson et al. 1994), and then the alignment was edited manually. The pictogram was generated from the alignment using the Pictrogram program created by C.B. Burge at http://genes.mit.edu/pictogram.html.
Plasmids
The shuffle plasmid pU2U6U was described previously (Madhani and Guthrie 1994b). The wild-type plasmids pJPS216 (Shuster and Guthrie 1988) and pJPS796 (Yan and Ares 1996), expressing full-length U2, and the wild-type plasmids pSX6 (Madhani and Guthrie 1992) and pJPS464 (Hilliker and Staley 2004), expressing U6 and U4, respectively, were described previously. A wild-type ACT1-CUP1 vector (Lesser and Guthrie 1993) marked with URA3 (pCG92, provided by C. Guthrie, University of California at San Francisco, San Francisco, CA) was mutagenized to introduce mutations U2A, brC (A259C), brG (A259G), and gAG (U301G/A304G). These same mutants, except U2A, were introduced into an ACT1-CUP1 vector marked with LEU2 (pCG157, provided by C. Guthrie) by subcloning the SacI–SalI fragment from the pCG92 series into pCG157; the wild-type and U2A LEU2 plasmids (pCG157 and pCG158, provided by C. Guthrie), relative to the subcloned mutants, had a 14-codon insertion in the ACT1 portion of the reporter that does not alter the sequence encoding Cup1p. All mutagenesis was performed by the QuikChange method (Stratagene).
Yeast strains
To make yJPS1035 (MATa ade2, his3, trp1, leu2, ura3, GAL+, cup1Δ∷ura3, snr20∷LYS, snr6∷KanMX4 [pU2U6U]), we replaced SNR6 with KanMX4 (Wach et al. 1994) in strain K3 already deleted for SNR20 and CUP1 (Lesser and Guthrie 1993). The prp16-302 strain YHM187 (Madhani and Guthrie 1994a) and the wild-type isogenic strain YHM118 (Madhani and Guthrie 1994b), as well as the strain XYU96, deleted for U2 and U6 (Xu et al. 1998), were described previously. Transformations were performed by the lithium acetate method (Ito et al. 1983). The strains YHM118, YHM187, and yJPS1035 were transformed with the pSX6 and pJPS216 variants, whereas the strain XYU96 was transformed with the pSX6 and pJPS796 variants.
ACT1-CUP1 splicing reporter assays
With the exception of the experiment shown in Figures 2 and 5C, to test U2 and/or U6 alleles in ACT1-CUP1 splicing assays, we cotransformed yJPS1035 with pJPS216, pSX6, and pCG157. Colonies were streaked onto 5-fluoroorotic acid media (5-FOA) (Sikorski and Boeke 1991) lacking leucine and then were colony-purified on media lacking leucine. For the experiment shown in Figures 2 and 5C, we cotransformed yJPS1035 initially with only pJPS796 and pSX6 variants. After streaking onto 5-FOA media and colony-purifying on rich media, the strains were transformed with pCG92 variants. For all experiments, to assess copper resistance, purified colonies were grown overnight in liquid media that selected for the reporter. Resulting saturated cultures were diluted to equivalent optical densities (OD600) of ∼1.2 and spotted onto media that selected for the ACT1-CUP1 reporter and that contained copper sulfate at a concentration of 0.013 mM, 0.05 mM, 0.1 mM, 0.15 mM, 0.2 mM, or 0.4 mM. Cells were grown for 3–10 d at 30°C.
Growth assays on 5-FOA
To test for genetic interactions between U2 alleles and prp16-302, we cotransformed YHM118 or YHM187 with pJPS216 variants and pSX6. To test for genetic interactions between U2 alleles and U6–A59C, we cotransformed XYU96 with pJPS796 and pSX6 variants. For growth assays, colonies were grown overnight in liquid media that selected for the transformed plasmids. Saturated cultures were then diluted in the same media and grown for two to three doublings at 30°C, and harvested in mid-log phase. Cultures were diluted to equivalent OD600 and spotted onto media containing 5-FOA. Growth was assayed for 3–7 d at 30°C or for 3–6 d at 20°C.
Primer extension analysis
To assay the RNA levels of the ACT1-CUP1 splicing reporter in the presence of U2 alleles, we cotransformed yJPS1035 with pJPS216 and pSX6. Colonies were streaked onto 5-FOA media and then colony-purified on rich media. The strains were then transformed with pCG92 variants. Transformants were grown overnight at 30°C in media lacking uracil. Saturated cultures were diluted to an OD600 of 0.3 and grown to an OD600 of 1.2. Total RNA was isolated and then assayed for specific RNA species by primer extension (Stevens and Abelson 2002) using the 32P-end-labeled primers oJPS233, complementary to the 3′ exon of ACT1-CUP1 (Mayas et al. 2006), and oJPS234 (5′-GTAC TAACGATGGGTTCGTAAGCGTACTCCTA-3′), complimentary to U14. Products were separated on a 6% polyacrylamide gel and visualized by PhosphorImager (Molecular Dynamics).
The efficiency of exon ligation was quantified using the ratio mRNA/lariat intermediate, rather than the ratio mRNA/(mRNA + lariat intermediate), because the former ratio is sensitive to changes in either the lariat intermediate or the mRNA, regardless of the relative levels of mRNA and lariat intermediate. Nonetheless, the trends of the exon ligation efficiencies shown in Figure 2 are the same with either ratio. Because the levels of lariat intermediate are affected by both steps of splicing, the efficiency of 5′ splice site cleavage was quantified using (mRNA + lariat intermediate)/(pre-mRNA + lariat intermediate + mRNA).
Acknowledgments
We thank Erik Sontheimer, Rhonda Perriman, and members of the Staley laboratory for critical reading of this manuscript; Rhonda Perriman and Manny Ares for sharing their unpublished results; Christine Guthrie and Manny Ares for strains and plasmids; and Channon Jordan, Martha Norman, and the Biology Department at St. Mary’s College of Maryland for technical assistance. This funding was supported by a grant from the U.S. National Institutes of Health (GM62264) to J.P.S.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1536107
References
- Ansari A., Schwer B., Schwer B. SLU7 and a novel activity, SSF1, act during the PRP16-dependent step of yeast pre-mRNA splicing. EMBO J. 1995;16:4001–4009. doi: 10.1002/j.1460-2075.1995.tb00071.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ares M., Jr., Igel A.H., Igel A.H. Lethal and temperature-sensitive mutations and their suppressors identify an essential structural element in U2 small nuclear RNA. Genes & Dev. 1990;12A:2132–2145. doi: 10.1101/gad.4.12a.2132. [DOI] [PubMed] [Google Scholar]
- Barabino S.M., Sproat B.S., Lamond A.I., Sproat B.S., Lamond A.I., Lamond A.I. Antisense probes targeted to an internal domain in U2 snRNP specifically inhibit the second step of pre-mRNA splicing. Nucleic Acids Res. 1992;17:4457–4464. doi: 10.1093/nar/20.17.4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brow D.A., Guthrie C., Guthrie C. Spliceosomal RNA U6 is remarkably conserved from yeast to mammals. Nature. 1988;6179:213–218. doi: 10.1038/334213a0. [DOI] [PubMed] [Google Scholar]
- Brys A., Schwer B., Schwer B. Requirement for SLU7 in yeast pre-mRNA splicing is dictated by the distance between the branchpoint and the 3′ splice site. RNA. 1996;7:707–717. [PMC free article] [PubMed] [Google Scholar]
- Burge C.B., Tuschl T., Sharp P.A., Tuschl T., Sharp P.A., Sharp P.A. Splicing of precursors to mRNAs by the spliceosome. In: Gesteland R.F., et al., editors. The RNA world. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1999. pp. 525–560. [Google Scholar]
- Champion-Arnaud P., Reed R., Reed R. The prespliceosome components SAP 49 and SAP 145 interact in a complex implicated in tethering U2 snRNP to the branch site. Genes & Dev. 1994;16:1974–1983. doi: 10.1101/gad.8.16.1974. [DOI] [PubMed] [Google Scholar]
- Chanfreau G., Jacquier A., Jacquier A. An RNA conformational change between the two chemical steps of group II self-splicing. EMBO J. 1996;13:3466–3476. [PMC free article] [PubMed] [Google Scholar]
- Collins C.A., Guthrie C., Guthrie C. Allele-specific genetic interactions between Prp8 and RNA active site residues suggest a function for Prp8 at the catalytic core of the spliceosome. Genes & Dev. 1999;15:1970–1982. doi: 10.1101/gad.13.15.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta B., Weiner A.M., Weiner A.M. Genetic evidence for base pairing between U2 and U6 snRNA in mammalian mRNA splicing. Nature. 1991;6338:821–824. doi: 10.1038/352821a0. [DOI] [PubMed] [Google Scholar]
- Fabrizio P., Abelson J., Abelson J. Two domains of yeast U6 small nuclear RNA required for both steps of nuclear precursor messenger RNA splicing. Science. 1990;4979:404–409. doi: 10.1126/science.2145630. [DOI] [PubMed] [Google Scholar]
- Fortner D.M., Troy R.G., Brow D.A., Troy R.G., Brow D.A., Brow D.A. A stem/loop in U6 RNA defines a conformational switch required for pre-mRNA splicing. Genes & Dev. 1994;2:221–233. doi: 10.1101/gad.8.2.221. [DOI] [PubMed] [Google Scholar]
- Golas M.M., Sander B., Will C.L., Lührmann R., Stark H., Sander B., Will C.L., Lührmann R., Stark H., Will C.L., Lührmann R., Stark H., Lührmann R., Stark H., Stark H. Molecular architecture of the multiprotein splicing factor SF3b. Science. 2003;5621:980–984. doi: 10.1126/science.1084155. [DOI] [PubMed] [Google Scholar]
- Golas M.M., Sander B., Will C.L., Lührmann R., Stark H., Sander B., Will C.L., Lührmann R., Stark H., Will C.L., Lührmann R., Stark H., Lührmann R., Stark H., Stark H. Major conformational change in the complex SF3b upon integration into the spliceosomal U11/U12 di-snRNP as revealed by electron cryomicroscopy. Mol. Cell. 2005;6:869–883. doi: 10.1016/j.molcel.2005.02.016. [DOI] [PubMed] [Google Scholar]
- Gozani O., Feld R., Reed R., Feld R., Reed R., Reed R. Evidence that sequence-independent binding of highly conserved U2 snRNP proteins upstream of the branch site is required for assembly of spliceosomal complex A. Genes & Dev. 1996;2:233–243. doi: 10.1101/gad.10.2.233. [DOI] [PubMed] [Google Scholar]
- Gozani O., Potashkin J., Reed R., Potashkin J., Reed R., Reed R. A potential role for U2AF–SAP 155 interactions in recruiting U2 snRNP to the branch site. Mol. Cell. Biol. 1998;8:4752–4760. doi: 10.1128/mcb.18.8.4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S., Moxon S., Marshall M., Khanna A., Eddy S.R., Bateman A., Moxon S., Marshall M., Khanna A., Eddy S.R., Bateman A., Marshall M., Khanna A., Eddy S.R., Bateman A., Khanna A., Eddy S.R., Bateman A., Eddy S.R., Bateman A., Bateman A. Rfam: Annotating non-coding RNAs in complete genomes. Nucleic Acids Res. 2005;34 (Database issue):D121–124. doi: 10.1093/nar/gki081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamill S., Pyle A.M., Pyle A.M. The receptor for branch-site docking within a group II intron active site. Mol. Cell. 2006;6:831–840. doi: 10.1016/j.molcel.2006.07.017. [DOI] [PubMed] [Google Scholar]
- Hilliker A.K., Staley J.P., Staley J.P. Multiple functions for the invariant AGC triad of U6 snRNA. RNA. 2004;6:921–928. doi: 10.1261/rna.7310704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A., Fukuda Y., Murata K., Kimura A., Murata K., Kimura A., Kimura A. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 1983;1:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M.H., Frank D.N., Guthrie C., Frank D.N., Guthrie C., Guthrie C. Characterization and functional ordering of Slu7p and Prp17p during the second step of pre-mRNA splicing in yeast. Proc. Natl. Acad. Sci. 1995;21:9687–9691. doi: 10.1073/pnas.92.21.9687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurica M.S., Moore M.J., Moore M.J. Pre-mRNA splicing: Awash in a sea of proteins. Mol. Cell. 2003;1:5–14. doi: 10.1016/s1097-2765(03)00270-3. [DOI] [PubMed] [Google Scholar]
- Konarska M.M., Query C.C., Query C.C. Insights into the mechanisms of splicing: More lessons from the ribosome. Genes & Dev. 2005;19:2255–2260. doi: 10.1101/gad.1363105. [DOI] [PubMed] [Google Scholar]
- Konarska M.M., Vilardell J., Query C.C., Vilardell J., Query C.C., Query C.C. Repositioning of the reaction intermediate within the catalytic center of the spliceosome. Mol. Cell. 2006;4:543–553. doi: 10.1016/j.molcel.2006.01.017. [DOI] [PubMed] [Google Scholar]
- Kramer A., Gruter P., Groning K., Kastner B., Gruter P., Groning K., Kastner B., Groning K., Kastner B., Kastner B. Combined biochemical and electron microscopic analyses reveal the architecture of the mammalian U2 snRNP. J. Cell Biol. 1999;7:1355–1368. doi: 10.1083/jcb.145.7.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesser C.F., Guthrie C., Guthrie C. Mutational analysis of pre-mRNA splicing in Saccharomyces cerevisiae using a sensitive new reporter gene, CUP1. Genetics. 1993;4:851–863. doi: 10.1093/genetics/133.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacMillan A.M., Query C.C., Allerson C.R., Chen S., Verdine G.L., Sharp P.A., Query C.C., Allerson C.R., Chen S., Verdine G.L., Sharp P.A., Allerson C.R., Chen S., Verdine G.L., Sharp P.A., Chen S., Verdine G.L., Sharp P.A., Verdine G.L., Sharp P.A., Sharp P.A. Dynamic association of proteins with the pre-mRNA branch region. Genes & Dev. 1994;24:3008–3020. doi: 10.1101/gad.8.24.3008. [DOI] [PubMed] [Google Scholar]
- Madhani H.D., Guthrie C., Guthrie C. A novel base-pairing interaction between U2 and U6 snRNAs suggests a mechanism for the catalytic activation of the spliceosome. Cell. 1992;5:803–817. doi: 10.1016/0092-8674(92)90556-r. [DOI] [PubMed] [Google Scholar]
- Madhani H.D., Guthrie C., Guthrie C. Genetic interactions between the yeast RNA helicase homolog Prp16 and spliceosomal snRNAs identify candidate ligands for the Prp16 RNA-dependent ATPase. Genetics. 1994a;3:677–687. doi: 10.1093/genetics/137.3.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhani H.D., Guthrie C., Guthrie C. Randomization-selection analysis of snRNAs in vivo: Evidence for a tertiary interaction in the spliceosome. Genes & Dev. 1994b;9:1071–1086. doi: 10.1101/gad.8.9.1071. [DOI] [PubMed] [Google Scholar]
- Mayas R.M., Maita H., Staley J.P., Maita H., Staley J.P., Staley J.P. Exon ligation is proofread by the DExD/H-box ATPase Prp22p. Nat. Struct. Mol. Biol. 2006;6:482–490. doi: 10.1038/nsmb1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPheeters D.S. Interactions of the yeast U6 RNA with the pre-mRNA branch site. RNA. 1996;11:1110–1123. [PMC free article] [PubMed] [Google Scholar]
- Ogle J.M., Murphy F.V., Tarry M.J., Ramakrishnan V., Murphy F.V., Tarry M.J., Ramakrishnan V., Tarry M.J., Ramakrishnan V., Ramakrishnan V. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell. 2002;5:721–732. doi: 10.1016/s0092-8674(02)01086-3. [DOI] [PubMed] [Google Scholar]
- Ogle J.M., Carter A.P., Ramakrishnan V., Carter A.P., Ramakrishnan V., Ramakrishnan V. Insights into the decoding mechanism from recent ribosome structures. Trends Biochem. Sci. 2003;5:259–266. doi: 10.1016/S0968-0004(03)00066-5. [DOI] [PubMed] [Google Scholar]
- Patel A.A., Steitz J.A., Steitz J.A. Splicing double: Insights from the second spliceosome. Nat. Rev. Mol. Cell Biol. 2003;12:960–970. doi: 10.1038/nrm1259. [DOI] [PubMed] [Google Scholar]
- Perriman R.J., Ares M., Jr., Ares M., Jr. Rearrangement of competing U2 RNA helices within the spliceosome promotes multiple steps in splicing. Genes & Dev. 2007 doi: 10.1101/gad.1524307. (this issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perriman R., Barta I., Voeltz G.K., Abelson J., Ares M., Jr., Barta I., Voeltz G.K., Abelson J., Ares M., Jr., Voeltz G.K., Abelson J., Ares M., Jr., Abelson J., Ares M., Jr., Ares M., Jr. ATP requirement for Prp5p function is determined by Cus2p and the structure of U2 small nuclear RNA. Proc. Natl. Acad. Sci. 2003;24:13857–13862. doi: 10.1073/pnas.2036312100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Query C.C., Konarska M.M., Konarska M.M. Suppression of multiple substrate mutations by spliceosomal prp8 alleles suggests functional correlations with ribosomal ambiguity mutants. Mol. Cell. 2004;3:343–354. doi: 10.1016/s1097-2765(04)00217-5. [DOI] [PubMed] [Google Scholar]
- Query C.C., Strobel S.A., Sharp P.A., Strobel S.A., Sharp P.A., Sharp P.A. Three recognition events at the branch-site adenine. EMBO J. 1996;6:1392–1402. [PMC free article] [PubMed] [Google Scholar]
- Ruis B.L., Kivens W.J., Siliciano P.G., Kivens W.J., Siliciano P.G., Siliciano P.G. The interaction between the first and last intron nucleotides in the second step of pre-mRNA splicing is independent of other conserved intron nucleotides. Nucleic Acids Res. 1994;24:5190–5195. doi: 10.1093/nar/22.24.5190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sashital D.G., Cornilescu G., McManus C.J., Brow D.A., Butcher S.E., Cornilescu G., McManus C.J., Brow D.A., Butcher S.E., McManus C.J., Brow D.A., Butcher S.E., Brow D.A., Butcher S.E., Butcher S.E. U2–U6 RNA folding reveals a group II intron-like domain and a four-helix junction. Nat. Struct. Mol. Biol. 2004;12:1237–1242. doi: 10.1038/nsmb863. [DOI] [PubMed] [Google Scholar]
- Schwer B., Gross C.H., Gross C.H. Prp22, a DExH-box RNA helicase, plays two distinct roles in yeast pre-mRNA splicing. EMBO J. 1998;7:2086–2094. doi: 10.1093/emboj/17.7.2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer B., Guthrie C., Guthrie C. PRP16 is an RNA-dependent ATPase that interacts transiently with the spliceosome. Nature. 1991;6309:494–499. doi: 10.1038/349494a0. [DOI] [PubMed] [Google Scholar]
- Schwer B., Guthrie C., Guthrie C. A conformational rearrangement in the spliceosome is dependent on PRP16 and ATP hydrolysis. EMBO J. 1992;13:5033–5039. doi: 10.1002/j.1460-2075.1992.tb05610.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuster E.O., Guthrie C., Guthrie C. Two conserved domains of yeast U2 snRNA are separated by 945 nonessential nucleotides. Cell. 1988;1:41–48. doi: 10.1016/0092-8674(88)90007-4. [DOI] [PubMed] [Google Scholar]
- Sikorski R.S., Boeke J.D., Boeke J.D. In vitro mutagenesis and plasmid shuffling: From cloned gene to mutant yeast. In: Guthrie C., Fink G.R., Fink G.R., editors. Guide to yeast genetics and molecular biology. Academic Press, Inc.; San Diego: 1991. pp. 302–318. [DOI] [PubMed] [Google Scholar]
- Staley J.P., Guthrie C., Guthrie C. Mechanical devices of the spliceosome: Motors, clocks, springs, and things. Cell. 1998;3:315–326. doi: 10.1016/s0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- Steitz T.A., Steitz J.A., Steitz J.A. A general two-metal-ion mechanism for catalytic RNA. Proc. Natl. Acad. Sci. 1993;14:6498–6502. doi: 10.1073/pnas.90.14.6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens S.W., Abelson J., Abelson J. Yeast pre-mRNA splicing: Methods, mechanisms, and machinery. Methods Enzymol. 2002;351:200–220. doi: 10.1016/s0076-6879(02)51849-8. [DOI] [PubMed] [Google Scholar]
- Sun J.S., Manley J.L., Manley J.L. A novel U2–U6 snRNA structure is necessary for mammalian mRNA splicing. Genes & Dev. 1995;7:843–854. doi: 10.1101/gad.9.7.843. [DOI] [PubMed] [Google Scholar]
- Tarn W.-Y., Yario T.A., Steitz J.A., Yario T.A., Steitz J.A., Steitz J.A. U12 snRNA in vertebrates: Evolutionary conservation of 5′ sequences implicated in splicing of pre-mRNAs containing a minor class of introns. RNA. 1995;6:644–656. [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Higgins D.G., Gibson T.J., Higgins D.G., Gibson T.J., Gibson T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umen J.G., Guthrie C., Guthrie C. The second catalytic step of pre-mRNA splicing. RNA. 1995;9:869–885. [PMC free article] [PubMed] [Google Scholar]
- Umen J.G., Guthrie C., Guthrie C. Mutagenesis of the yeast gene PRP8 reveals domains governing the specificity and fidelity of 3′ splice site selection. Genetics. 1996;2:723–739. doi: 10.1093/genetics/143.2.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valadkhan S. snRNAs as the catalysts of pre-mRNA splicing. Curr. Opin. Chem. Biol. 2005;6:603–608. doi: 10.1016/j.cbpa.2005.10.008. [DOI] [PubMed] [Google Scholar]
- Valadkhan S., Manley J.L., Manley J.L. Splicing-related catalysis by protein-free snRNAs. Nature. 2001;6857:701–707. doi: 10.1038/35099500. [DOI] [PubMed] [Google Scholar]
- Valadkhan S., Manley J.L., Manley J.L. Characterization of the catalytic activity of U2 and U6 snRNAs. RNA. 2003;7:892–904. doi: 10.1261/rna.5440303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa T., Guthrie C., Guthrie C. The Isy1p component of the NineTeen complex interacts with the ATPase Prp16p to regulate the fidelity of pre-mRNA splicing. Genes & Dev. 2005;16:1894–1904. doi: 10.1101/gad.1336305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach A., Brachat A., Pohlmann R., Philippsen P., Brachat A., Pohlmann R., Philippsen P., Pohlmann R., Philippsen P., Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;13:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- Wagner J.D., Jankowsky E., Company M., Pyle A.M., Abelson J.N., Jankowsky E., Company M., Pyle A.M., Abelson J.N., Company M., Pyle A.M., Abelson J.N., Pyle A.M., Abelson J.N., Abelson J.N. The DEAH-box protein PRP22 is an ATPase that mediates ATP-dependent mRNA release from the spliceosome and unwinds RNA duplexes. EMBO J. 1998;10:2926–2937. doi: 10.1093/emboj/17.10.2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Wagner J.D., Guthrie C., Wagner J.D., Guthrie C., Guthrie C. The DEAH-box splicing factor Prp16 unwinds RNA duplexes in vitro. Curr. Biol. 1998;8:441–451. doi: 10.1016/s0960-9822(98)70178-2. [DOI] [PubMed] [Google Scholar]
- Wells S.E., Ares M., Jr., Ares M., Jr. Interactions between highly conserved U2 small nuclear RNA structures and Prp5p, Prp9p, Prp11p, and Prp21p proteins are required to ensure integrity of the U2 small nuclear ribonucleoprotein in Saccharomyces cerevisiae. Mol. Cell. Biol. 1994;9:6337–6349. doi: 10.1128/mcb.14.9.6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will C.L., Lührmann R., Lührmann R. Spliceosome structure and function. In: Gesteland R.F., et al., editors. The RNA world. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2006. pp. 369–400. [Google Scholar]
- Will C.L., Schneider C., Reed R., Lührmann R., Schneider C., Reed R., Lührmann R., Reed R., Lührmann R., Lührmann R. Identification of both shared and distinct proteins in the major and minor spliceosomes. Science. 1999;5422:2003–2005. doi: 10.1126/science.284.5422.2003. [DOI] [PubMed] [Google Scholar]
- Will C.L., Schneider C., MacMillan A.M., Katopodis N.F., Neubauer G., Wilm M., Lührmann R., Query C.C., Schneider C., MacMillan A.M., Katopodis N.F., Neubauer G., Wilm M., Lührmann R., Query C.C., MacMillan A.M., Katopodis N.F., Neubauer G., Wilm M., Lührmann R., Query C.C., Katopodis N.F., Neubauer G., Wilm M., Lührmann R., Query C.C., Neubauer G., Wilm M., Lührmann R., Query C.C., Wilm M., Lührmann R., Query C.C., Lührmann R., Query C.C., Query C.C. A novel U2 and U11/U12 snRNP protein that associates with the pre-mRNA branch site. EMBO J. 2001;16:4536–4546. doi: 10.1093/emboj/20.16.4536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D., Field D.J., Tang S.J., Moris A., Bobechko B.P., Friesen J.D., Field D.J., Tang S.J., Moris A., Bobechko B.P., Friesen J.D., Tang S.J., Moris A., Bobechko B.P., Friesen J.D., Moris A., Bobechko B.P., Friesen J.D., Bobechko B.P., Friesen J.D., Friesen J.D. Synthetic lethality of yeast slt mutations with U2 small nuclear RNA mutations suggests functional interactions between U2 and U5 snRNPs that are important for both steps of pre-mRNA splicing. Mol. Cell. Biol. 1998;4:2055–2066. doi: 10.1128/mcb.18.4.2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D., Ares M., Jr., Ares M., Jr. Invariant U2 RNA sequences bordering the branchpoint recognition region are essential for interaction with yeast SF3a and SF3b subunits. Mol. Cell. Biol. 1996;3:818–828. doi: 10.1128/mcb.16.3.818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavanelli M.I., Ares M., Jr., Ares M., Jr. Efficient association of U2 snRNPs with pre-mRNA requires an essential U2 RNA structural element. Genes & Dev. 1991;12B:2521–2533. doi: 10.1101/gad.5.12b.2521. [DOI] [PubMed] [Google Scholar]
- Zavanelli M.I., Britton J.S., Igel A.H., Ares M., Jr., Britton J.S., Igel A.H., Ares M., Jr., Igel A.H., Ares M., Jr., Ares M., Jr. Mutations in an essential U2 small nuclear RNA structure cause cold-sensitive U2 small nuclear ribonucleoprotein function by favoring competing alternative U2 RNA structures. Mol. Cell. Biol. 1994;3:1689–1697. doi: 10.1128/mcb.14.3.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]