Abstract
Transcription by RNA polymerase II (polII) is accompanied by dramatic changes in chromatin structure. Numerous enzymatic activities contribute to these changes, including ATP-dependent nucleosome remodeling enzymes and histone modifying enzymes. Recent studies in budding yeast document a histone modification pathway associated with polII transcription, whereby ubiquitylation of histone H2B leads to methylation of histone H3 on specific lysine residues. Although this series of events appears to be highly conserved among eukaryotes, its mechanistic function in transcription is unknown. Here we document a significant functional divergence between ubiquitylation of H2B and methylation of Lys 4 on histone H3 in the fission yeast Schizosaccharomyces pombe. Loss of H2B ubiquitylation results in defects in cell growth, septation, and nuclear structure, phenotypes not observed in cells lacking H3 Lys 4 methylation. Consistent with these results, gene expression microarray analysis reveals a greater role for H2B ubiquitylation in gene regulation than for H3 Lys 4 methylation. Chromatin immunoprecipitation (ChIP) experiments demonstrate that loss of H2B ubiquitylation alters the distribution of polII and histones in gene coding regions. We propose that ubiquitylation of H2B impacts transcription elongation and nuclear architecture through its effects on chromatin dynamics.
Keywords: Chromatin, histone, methylation, transcription, ubiquitin
Nucleosomes pose a structural barrier to RNA polymerase II (polII) transcription (Izban and Luse 1991; Sims et al. 2004). The challenge posed by nucleosomes to transcribing polII is highlighted by recent genome-wide mapping studies that demonstrate extensive nucleosome occupancy throughout the coding regions of polII genes (Ercan and Simpson 2004; Lee et al. 2004; Bernstein et al. 2005; Yuan et al. 2005). In keeping with these findings, significant evidence points to partial or complete nucleosome disassembly as an important component of the transcription elongation process (Kireeva et al. 2002; Belotserkovskaya et al. 2003; Schwabish and Struhl 2004). Transcription-coupled nucleosome assembly has also been inferred from the observation that certain histone variants are selectively deposited within gene coding regions in a replication-independent manner, although their precise connection to transcription remains unclear (Ahmad and Henikoff 2002; Li et al. 2005; Raisner et al. 2005; H. Zhang et al. 2005). Various conserved histone chaperone complexes are thought to facilitate either disassembly or reassembly of nucleosomes during transcription, including FACT, Spt6, Asf1, HIRA (for the variant H3.3), and SWR-C (for the variant H2A.Z) (Belotserkovskaya et al. 2003; Kaplan et al. 2003; Kobor et al. 2004; Mizuguchi et al. 2004; Tagami et al. 2004; Schwabish and Struhl 2006). The molecular details of the assembly and disassembly process, as well as how it is coordinated with the polII elongation complex, are not understood.
The extent to which other chromatin-modifying activities function with histone chaperones to facilitate polII elongation is also poorly understood (Workman 2006). Of particular interest in this regard is a highly conserved set of histone post-translational modifications that accompany the onset of polII transcription in vivo. These include monoubiquitylation of histone H2B (H2Bub), methylation of specific lysines on histone H3, and acetylation of numerous other lysines in all histone tails (Shilatifard 2006). Chromatin immunoprecipitation (ChIP) experiments have shown that at most polII genes, methylation of histone H3 Lys 4 (H3K4me) and acetylation are concentrated in a relatively small region overlapping the transcription start site (Santos-Rosa et al. 2002; Ng et al. 2003; Bernstein et al. 2005; Guenther et al. 2005; Liu et al. 2005; Pokholok et al. 2005). H2Bub appears to be broadly concentrated in gene coding regions, although its localization pattern has only been determined at a handful of loci (Xiao et al. 2005). Interestingly, there is a strict dependency relationship between H2Bub and H3K4me, such that H2Bub must precede H3K4me (Dover et al. 2002; Sun and Allis 2002). Mutants that are defective in either H2Bub or H3K4me in Saccharomyces cerevisiae share some phenotypes, including loss of telomeric silencing, consistent with a causal relationship between the two modifications (Nislow et al. 1997; Sun and Allis 2002). Furthermore, components of certain histone acetyltransferase complexes can recognize methylated H3K4, consistent with the co-occupancy of these modifications seen by ChIP (Martin et al. 2006; Taverna et al. 2006). These results support a model of a histone modification pathway leading to a certain transcriptional output, the nature of which has yet to be determined (Taverna et al. 2006).
Although this proposed “trans-tail” pathway explains the colocalization of these modifications on chromatin, their mechanistic functions in transcription remain unclear. To study this pathway in greater detail, we have focused on H2Bub, its most upstream component. Conjugation of monoubiquitin to H2B at a conserved C-terminal lysine (K119 in fission yeast, K123 in budding yeast, or K120 in mammals) has long been known to be associated with active transcription by polII (Jason et al. 2002). Several lines of evidence have suggested that H2Bub is connected to polII elongation. In budding yeast, Rad6, the E2 ubiquitin-conjugating enzyme for H2B, is specifically associated with the Ser 5 phosphorylated form of polII that is committed to elongation (Xiao et al. 2005). Also, mutants lacking H2Bub show genetic interactions with known transcription elongation mutants (Xiao et al. 2005). Recent work using a highly purified mammalian in vitro transcription system links H2Bub to the function of the FACT histone chaperone complex (Pavri et al. 2006).
Despite these connections, many questions surrounding the function of H2Bub remain unanswered. This is highlighted by the fact that S. cerevisiae mutants lacking H2Bub have few discernable phenotypes and display no general transcription defects (Robzyk et al. 2000; X. Zhang et al. 2005). The lack of endogenous target genes has precluded a mechanistic analysis of the role of this modification in transcription in vivo. Furthermore, the extent to which its potential functions in transcription overlap with, or diverge from, those of H3K4me in vivo has not been determined. Thus, it is possible that the trans-tail pathway, while tightly correlated with active polII transcription, is dispensable for transcription per se.
We have analyzed the function of H2Bub in the fission yeast, Schizosacchromyces pombe. By identifying and analyzing genes whose expression depends on H2Bub, we show that the primary function of H2Bub in transcription is to stimulate transcription elongation. Further, we argue that this function involves control of nucleosome dynamics. Importantly, we show that H2Bub carries out its role in transcription independently of H3K4me.
Results
Ubiquitylation of H2B is required for normal growth in S. pombe
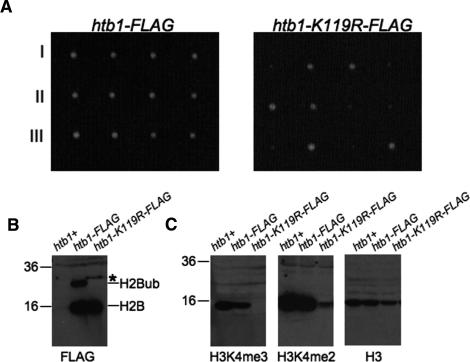
To study the function of H2Bub in S. pombe, we first tagged the htb1+ gene, the lone S. pombe gene encoding H2B, with a single Flag epitope at its C terminus (htb1-Flag). In parallel, we constructed a similar strain in which the conserved ubiquitin acceptor site was mutated to arginine (htb1-K119R-Flag). Both strains were initially generated as heterozygous diploids, which were then sporulated to derive haploid tagged strains. We noticed that whereas the htb1-Flag strain grew at the same rate as the wild type, the htb1-K119R-Flag strain grew slowly at 30°C. Backcrossing of both strains to a wild-type parent confirmed the presence of a growth defect in the htb1-K119R-Flag strain that was linked to the kanamycin resistance marker used to integrate the Flag tag (Fig. 1A).
Figure 1.
Ubiquitylation of H2B is present in S. pombe and is required for H3K4me and for normal growth. (A) Either the htb1-Flag or htb1-K119R-Flag strain was crossed to an untagged wild-type strain; tetrads were dissected on YES media. Shown are three tetrads from each cross. The slow-growing colonies in the htb1-K119R-Flag cross were all G418-resistant. (B) Anti-Flag Western blots on whole-cell extracts prepared from an untagged strain (htb1+) and the indicated tagged strains. Bands corresponding to H2B-Flag and its ubiquitylated form are indicated on the right. On the left are molecular weight markers (in kilodaltons). The asterisk denotes a probable H2B–SUMO conjugate. (C) Extracts from B were probed with the indicated antibodies.
Anti-Flag Western blots on whole-cell extracts prepared from wild-type, htb1-Flag, and htb1-K119R-Flag strains confirmed the presence of Flag-tagged H2B at the expected size in both htb1-Flag and htb1-K119R-Flag (see Fig. 1B, lanes 2,3). Extracts from the htb1-Flag strain also showed a slower-migrating band at the size predicted for H2Bub. This band was absent from htb1-K119R-Flag extracts (Fig. 1B, cf. lanes 2 and 3). We note the presence of a band at a slightly higher molecular weight than the putative H2Bub band in the htb1-K119R-Flag strain (Fig. 1A, asterisk). This likely corresponds to a SUMO-conjugated form of H2B, as has been observed previously in S. cerevisiae (Nathan et al. 2006). Blotting against total histone H3 showed that the extracts were equally loaded (Fig. 1C).
Excision of the putative H2Bub band from a Coommassie-stained gel and analysis by tandem mass spectrometry confirmed its identification as a monoubiquitylated form of H2B, and confirmed the ubiquitin attachment site as H2B Lys 119 (Supplementary Fig. 1). Therefore, H2Bub is present in S. pombe and is required for normal growth, in contrast to what has been found in S. cerevisiae. However, both share a requirement for H2Bub in methylation of H3K4 (Fig. 1C), in agreement with previous results (Roguev et al. 2003; Maruyama et al. 2006).
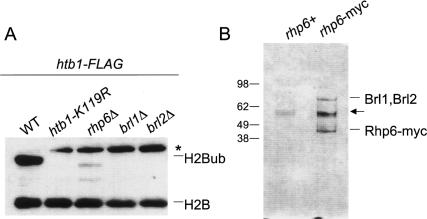
Knockouts of conserved components of the H2Bub pathway were crossed to the htb1-Flag strain to monitor their effects on H2Bub. As expected from previous work, a deletion of rhp6+, encoding the S. pombe homolog of the RAD6 ubiquitin conjugating E2 enzyme, resulted in a loss of H2Bub and H3K4me (Fig. 2A; Supplementary Fig. 2; Roguev et al. 2003; Maruyama et al. 2006).
Figure 2.
H2Bub in S. pombe is mediated by conserved enzymes. (A) Strains carrying both the htb1-Flag allele and one of the indicated mutations were analyzed by anti-Flag Western blot. Bands corresponding to H2B-Flag and its ubiquitylated form are indicated on the right. The asterisk denotes a probable H2B–SUMO conjugate. (B) Whole-cell extracts from the indicated strains were subjected to anti-myc immunoprecipitations, and the bound fractions were analyzed by SDS-PAGE and Coommassie staining. Bands identified by tandem mass spectrometry or by anti-myc Western blotting are denoted on the right (Brl1 and Brl2 have nearly identical molecular masses and thus were not resolved). The arrow denotes a contaminating protein. On the left are molecular weight markers (in kilodaltons).
Ubiquitylation of H2B in S. cerevisiae and metazoan systems also requires the RING finger E3 ubiquitin ligase BRE1 (called RNF20 or hBRE1 in human cells) (Hwang et al. 2003; Wood et al. 2003; Kim et al. 2005; Zhu et al. 2005). Deletion of either of the two S. pombe genes similar to BRE1 also resulted in loss of H2Bub and H3K4me (Fig. 2A). We have named these genes brl1+ (SPCC1919.15) and brl2+ (SPCC970.10c) (for BRE1-like). Strains that were deleted for any of rhp6+, brl1+, or brl2+ showed slow-growth phenotypes, further arguing that H2Bub is required for normal growth (data not shown). The rhp6Δ strain was particularly slow-growing, consistent with the loss of ubiquitylation of other known Rhp6 targets (Takeda and Yanagida 2005).
We were intrigued by the fact that both Brl1 and Brl2 proteins were required for H2Bub. To further characterize the functional connection between these proteins and Rhp6, rhp6+ was tagged at its endogenous locus with a 13-myc epitope and purified, and copurifying proteins were identified by Coommassie staining and tandem mass spectrometry (Fig. 2B; data not shown). We found that both Brl1 and Brl2 copurified with Rhp6, suggesting that Rhp6 acts in concert with both Brl1 and Brl2 to promote H2Bub (Brl1 and Brl2 have near-identical molecular masses and therefore were not resolved on the gel) (see Fig. 2B).
Functional divergence of H2B ubiquitylation and H3K4 methylation
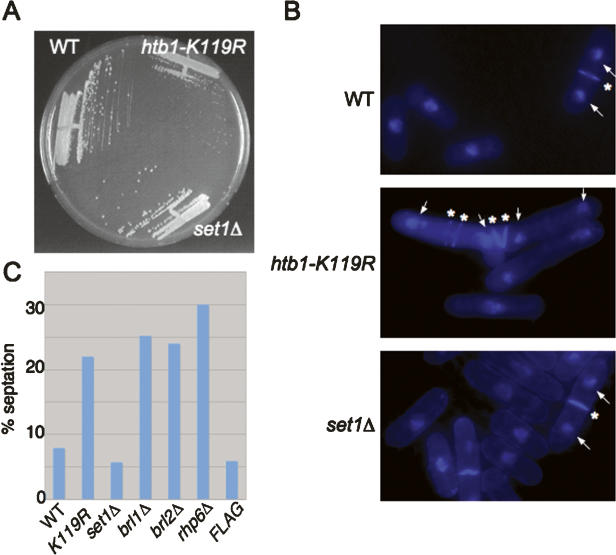
Given that H2Bub is needed for methylation of H3K4, we wished to know whether the growth defect observed in the htb1-K119R-Flag strain was due to loss of downstream methylation. We constructed untagged, isogenic wild-type and htb1-K119R strains and compared them to a strain deleted for the set1+ gene, encoding the S. pombe homolog of the H3K4 methyltransferase (Noma and Grewal 2002). In keeping with previous results in S. cerevisiae and S. pombe, Western blots showed that the set1Δ strain lacked all H3K4me but was proficient in H2Bub (see Supplementary Fig. 2; data not shown). This strain also grew at the wild-type rate, in contrast to the htb1-K119R strain (Fig. 3A). Therefore, at least some functions of H2Bub are independent of H3K4me in S. pombe.
Figure 3.
Functional divergence of H2Bub and H3K4me. (A) The indicated strains were streaked onto YES media and grown for 3 d at 30°C. (B) The indicated strains were grown to log phase in YES media, fixed, stained with DAPI and calcofluor, and photographed. For representative cells in each panel, white arrows denote DAPI-stained nuclei and white asterisks denote calcofluor staining septa. (C) The indicated strains were grown to log phase in YES media, fixed briefly, and stained with calcofluor. Percentages are based on counts of 100–200 cells.
Microscopic examination of htb1-K119R cells grown at 30°C showed an abnormal morphology: Cells were large and tended to associate in clumps. Staining with DAPI (diamino-phenylindole) and calcofluor revealed occasional cells with multiple compartments enclosed by septa, some containing a single nucleus (Fig. 3B, middle panel). Cells in which the nuclei were separated by multiple septa were also observed (Fig. 3B, middle panel). These phenotypes are indicative of cell separation defects (Simanis 2003). Overall, mutants lacking H2Bub showed a two- to threefold increase in percentage of cells containing septa in an asynchronous culture (Fig. 3C). Importantly, set1Δ cells were morphologically normal, indicating that the htb1-K119R defects were not due to lack of H3K4me.
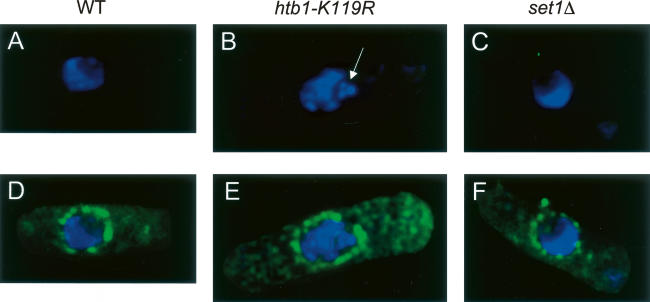
We also observed aberrant nuclear morphology in htb1-K119R cells. Wild-type, htb1-K119R, and set1Δ cells were stained with DAPI and an antibody recognizing the nuclear pore complex and examined using deconvolution microscopy. Nuclei in wild-type and set1Δ cells showed a crescent-shaped DAPI-staining region occupying roughly half of the area bounded by the nuclear pore staining (Fig. 4A,C,D,F). However, among htb1-K119R cells, we found examples of nuclei containing fragmented DAPI-stained material inside the nucleus (Fig. 4B,E), indicating a defect in nuclear architecture.
Figure 4.
Abnormal nuclear morphology in the htb1-K119R mutant. Wild-type, htb1-K119R, and set1Δ cells were stained with a nuclear pore antibody and DAPI. Shown are deconvolved images taken using a DeltaVision microscope equipped with a CCD camera. (A–C) DAPI stain alone. (D–F) DAPI stain merged with nuclear pore stain. The white arrow in B denotes a fragment of nuclear, DAPI-stained material in the htb1-K119R mutant cell.
We then turned to microarray analysis to determine whether the phenotypes observed in the mutants were correlated with specific defects in gene expression. In assigning the total number of genes that are affected by each of the mutations, we adopted a cutoff such that genes whose expression changed by at least twofold in two independent hybridizations were included. We consider this to be a stringent cutoff, since in a typical experiment ∼90% of genes that satisfied this requirement experienced expression level changes of threefold or less (see Supplementary Tables 2–4). All analyses were conducted on asynchronous cultures grown in rich medium at 30°C.
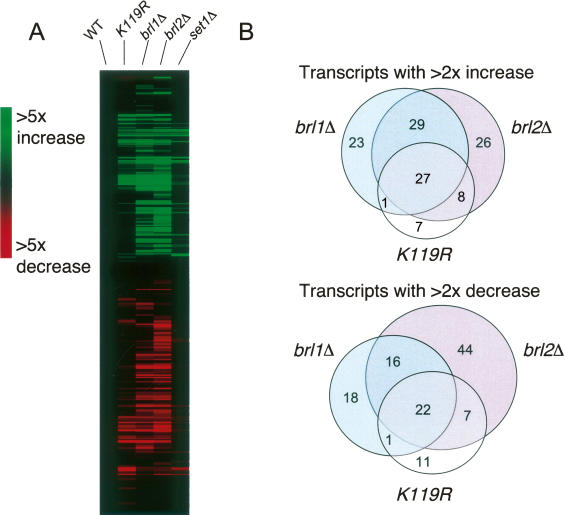
Affected genes in all the mutants were clustered using hierarchical clustering analysis (a representative example of which is shown in Fig. 5A). The htb1-K119R mutant had altered expression of 84 genes (43 increased and 41 decreased). Surprisingly, the expression of only 12 genes was found to reproducibly change in the set1Δ mutant (nine increased and three decreased). Of these, six were also found in the htb1-K119R data sets (five of nine for increased, one of three for decreased), as expected from the loss of H3K4me in the htb1-K119R mutant. These results corroborate the phenotypic data described above and demonstrate that H2Bub has gene regulatory functions that are independent of H3K4me.
Figure 5.
Gene expression microarray analysis of mutants defective in H2Bub and H3K4me. (A) Transcripts whose levels were altered by at least twofold in one or more of the mutants were clustered using a hierarchical clustering algorithm. Shown is a representative cluster diagram from one of three independent experiments. Values from a wild-type control strain were subtracted from all data sets (WT column). (B) Venn diagrams showing overlap between htb1-K119R, brl1Δ, and brl2Δ data sets.
We also determined the gene expression profiles of brl1Δ and brl2Δ mutants. Both of these mutants affected a broader spectrum of genes than the htb1-K119R mutant (see Fig. 5A,B). However, as illustrated in Figure 5B, highly significant overlaps were found between data sets for htb1-K119R and brl1Δ (28 of 95 and 23 of 75; p = 1.36 × 10−41 and 1.6 × 10−35 for increased and decreased genes, respectively), htb1-K119R and brl2Δ (35 of 98 and 29 of 101; p = 1.06 × 10−56 and 2.87 × 10−43), and brl1Δ and brl2Δ (56 of 114 and 38 of 108; p = 9.97 × 10−89 and 2.22 × 10−56). In all, 49 of the 84 genes altered in the htb1-K119R mutant were also altered in both brl1Δ and brl2Δ. These data argue that Brl1 and Brl2 proteins function together through the ubiquitylation of H2B. Together with the physical interaction data presented above (Fig. 2B), these data suggest that a Rhp6–Brl1–Brl2 complex mediates most H2B ubiquitylation in vivo. Also, since the functions of Brl1 and Brl2 cannot be accounted for completely by H2Bub, they likely have other ubiquitylation targets that are important for gene regulation.
The affected genes in our data sets overlapped significantly with two functional categories annotated in the S. pombe genome database. These are the amino acid permeases, which are represented in genes repressed by brl2Δ (p = 0.00359), and membrane transporters, which are represented in genes repressed by htb1-K119R (p = 0.00236) and brl2Δ (p = 0.0482). Thus, H2Bub may have an especially important role in activating genes involved in membrane functions, which would be consistent with its role in septation. We also observed a broad and significant overlap between the expression profiles of mutants affecting H2Bub and genes whose expression is altered in response to various forms of stress (summarized in Supplementary Table 5; Chen et al. 2003). This could reflect a specific role of H2Bub in the stress response, or an indirect effect of other phenotypes in the mutants.
The pronounced cell separation phenotypes of mutants affecting H2Bub prompted us to compare our data sets to those generated for known S. pombe septation mutants (Lee et al. 2005). Interestingly, we observed modest but significant overlaps between genes induced in brl1Δ and brl2Δ mutants and those induced in the sep15Δ mutant grown at the semipermissive temperature (p = 0.044 for brl1Δ and p = 0.000239 for brl2Δ). For brl2Δ, overlap was also seen with sep10Δ (p = 0.038). The sep10+ and sep15+ genes encode subunits of Mediator, a complex that facilitates communication between transcriptional activators and RNA polII (Zilahi et al. 2000; Szilagyi et al. 2002). These data implicate ubiquitylation by Brl1 and Brl2 in a repressive function of the Mediator complex.
Surprisingly, induced genes in our data sets also showed significant overlap with genes induced in clr6-1 and ago1Δ mutants, which are defective in heterochromatic gene silencing (Hansen et al. 2005). The overlap was observed for all of the mutants that we tested, including set1Δ. This implies that both H2Bub and H3K4me play some role, either direct or indirect, in repression of gene expression by some gene silencing factors.
Ubiquitylation of H2B is required for normal transcription elongation in vivo
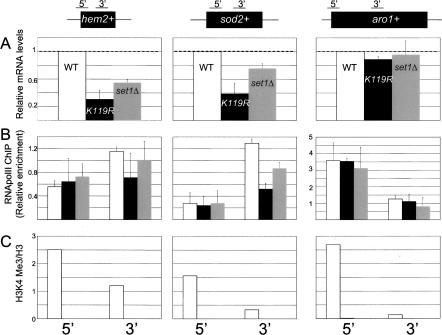
To arrive at a mechanistic understanding of how H2Bub and H3K4me regulate gene expression, we analyzed individual target loci using ChIP. We focused our analysis on two genes whose expression is repressed in the htb1-K119R and set1Δ mutants: hem2+, involved in heme biosynthesis, and sod2+, involved in sodium transport. These genes were chosen since they were reproducibly repressed in the microarray experiments (see Supplementary Tables 2–4). Quantitative PCR (Q-PCR) analysis and Northern blotting confirmed that these genes are bona fide targets (see Fig. 6A; Supplementary Fig. 3). As a control, we also analyzed the aro1+ gene, whose expression is unaffected by either mutation (see Fig. 6A; Supplementary Fig. 3).
Figure 6.
ChIP analysis of individual genes regulated by htb1-K119R and set1Δ. (A) Total RNA isolated from each strain was analyzed by Q-RT–PCR using the 3′ primer pair for each gene indicated at the top. The data are represented as mRNA levels relative to the wild type, with wild-type levels arbitrarily set to 1 (denoted by dotted lines). Error bars denote standard deviations derived from at least two independent RNA isolations. (B) ChIP for RNA polII was performed on each strain as described in Materials and Methods. Data are expressed as enrichment relative to a region of the control gene cdc25+. Error bars denote standard deviations from at least two experiments. The primer pairs at the bottom of the figure correspond to the regions of each gene diagrammed at the top. Bar colors are as in A. (C) ChIP for H3K4me3. Primer pairs for PCR are denoted at the bottom. Data are representative of two to three independent experiments and are expressed as in B, except they are further normalized to enrichment in total histone H3 immunoprecipitations done in parallel. Bar colors are as in A.
We monitored occupancy of polII and histone H3 trimethylated on K4 (H3K4me3) by ChIP at aro1+, hem2+, and sod2+, both at their 5′ ends and at a region downstream (Fig. 6B,C). As has been observed in budding yeast and metazoan systems, H3K4me3 was greatly enriched at the 5′ ends of each gene and was completely dependent on both H2Bub and set1+ (Fig. 6C). We note that similar enrichment was observed at every active gene we tested, regardless of its dependence on H2Bub or H3K4me for expression (Supplementary Fig. 4). In addition, very little enrichment was observed at inactive genes (data not shown), in agreement with the idea that H3K4me generally marks the 5′ end of active genes (Bernstein et al. 2005; Guenther et al. 2005; Liu et al. 2005; Pokholok et al. 2005).
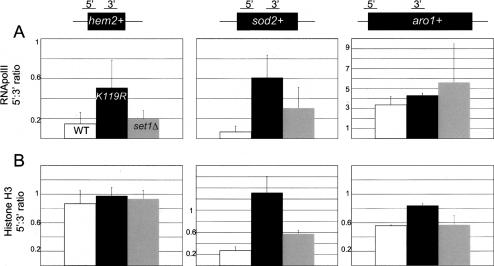
Although only subtle differences in polII occupancy could be discerned between the wild-type and either mutant strain at a given location, we noticed that the htb1-K119R mutation caused a specific decrease in polIII occupancy at the 3′ ends of hem2+ and sod2+ genes (Fig. 6B). This is also evident when comparing the ratios of polII ChIP signals from the 5′ and 3′ probes within each gene between the different strains (Fig. 7A). For hem2+ and sod2+, the 5′:3′ ratios increase approximately threefold and approximately sixfold, respectively, in the htb1-K119R mutant relative to the wild type (Fig. 7A). Importantly, these effects correlate with the observed effects on gene expression, since they are much weaker in the set1Δ mutant and at the aro1+ control gene. They are also absent in two additional genes that are not affected by the htb1-K119R mutation, cdc25+ and psu1+ (Supplementary Fig. 4). Since the occupancy of polIII at H2Bub target genes is specifically affected at their 3′ ends in the htb1-K119R mutant (Fig. 6B), H2Bub likely plays a role in polII transcription elongation. Moreover, this function is independent of H3K4me.
Figure 7.
Repression of gene expression in the htb1-K119R mutant is correlated with a transcription elongation defect. (A) Ratios of RNA polII ChIP signals for the 5′ primer pair to the 3′ primer pair are shown for each gene, as diagrammed at the top. Bar colors are denoted in the top left panel. Error bars denote standard deviations from two to three independent immunoprecipitations. (B) As in A for histone H3 ChIP.
Evidence linking H2B ubiquitylation to chromatin assembly
To explore the mechanism by which H2Bub enhances transcription elongation, we also derived 5′:3′ distribution ratios at the same genes for histone H3. At the sod2+ gene, we found that H3 distribution is skewed toward the 5′ end in htb1-K119R relative to wild type, similar to what we found for polII (Fig. 7B, right). The set1Δ mutation caused a much smaller change in distribution. This shows that the transcription elongation defect at sod2+ in the htb1-K119R mutant is accompanied by dramatic chromatin changes at the locus, and implies that H2Bub helps to alter nucleosome occupancy in some gene coding regions to accommodate elongating polII. Interestingly, histone H3 distribution at hem2+ is unchanged in either mutant, suggesting that the elongation function of H2Bub is not strictly dependent on changes in nucleosome occupancy.
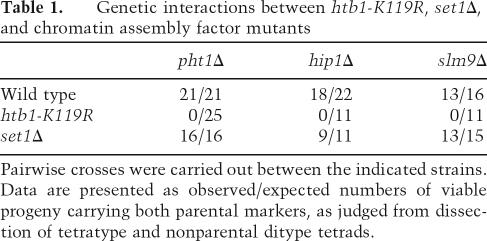
We sought genetic evidence that H2Bub acts at least in part through a chromatin assembly pathway coupled to polII transcription. The highly conserved histone variant H2A.Z, encoded by the pht1+ gene in S. pombe, is assembled into nucleosomes in an S-phase-independent manner and plays a role in transcription that remains poorly understood (Carr et al. 1994; Henikoff et al. 2004). Interestingly, its deposition pattern within polII coding regions bears a striking resemblance to that of H2Bub and H3K4me, in that it is concentrated near the 5′ end (Raisner et al. 2005). We examined the relationship between H2Bub and H2A.Z deposition by crossing the htb1-K119R mutant to a pht1Δ mutant. We found that double mutants could not be recovered from this cross, indicating that the two mutations are synthetically lethal (see Table 1). Double mutants between pht1Δ and set1Δ were readily recovered, indicating that the synthetic lethality is unrelated to H3K4me. This is consistent with previous work in budding yeast that identified rad6Δ and bre1Δ as synthetically lethal with htz1Δ (Hwang et al. 2003). The requirement for H2A.Z in the htb1-K119R mutant indicates that H2Bub function is related to S-phase-independent chromatin assembly.
Table 1.
Genetic interactions between htb1-K119R, set1Δ, and chromatin assembly factor mutants
Pairwise crosses were carried out between the indicated strains. Data are presented as observed/expected numbers of viable progeny carrying both parental markers, as judged from dissection of tetratype and nonparental ditype tetrads.
We suspected that a specific histone chaperone complex may function through H2Bub to enhance polII elongation. In mammalian cells, the histone chaperone HIRA has been implicated in replication-independent incorporation of histone H3 into chromatin (Tagami et al. 2004). Homologs of HIRA in budding yeast and fission yeast function in histone gene regulation, DNA repair, gene silencing, and chromosome segregation, but have not been assigned a general role in transcription (Spector et al. 1997; Kaufman et al. 1998; Sharp et al. 2002; Blackwell et al. 2004). We attempted to combine the htb1-K119R mutant with a deletion of hip1+, the gene encoding the HIRA homolog in S. pombe. We found that we were unable to recover double mutants from this cross, indicating that the two mutations are synthetically lethal. This was also true for a deletion of the slm9+ gene, which encodes a protein that interacts with Hip1 and is similar to the budding yeast HIR2 gene (Blackwell et al. 2004). Thus, it is likely that H2Bub plays a role in chromatin assembly. Neither hip1Δ nor slm9Δ was lethal in combination with set1Δ, indicating that the chromatin assembly function of H2Bub is independent of H3K4me (see Table 1). We note that all crosses involving hip1Δ and slm9Δ strains gave rise to ∼10% inviable spores (see Table 1). These results suggest a role for H2Bub in chromatin assembly during transcription.
Recent studies have suggested a connection between H2Bub function in transcription elongation and the FACT chromatin assembly complex (Pavri et al. 2006). We tested whether FACT occupancy at H2Bub target genes was affected by the htb1-K119R and set1Δ mutations using strains carrying an epitope-tagged FACT subunit (Supplementary Fig. 5). Western blots confirmed that Spt16-myc protein levels were unchanged in the mutant strains (Supplementary Fig. 5B). Using ChIP, we found a modest (approximately twofold) decrease in Spt16-myc binding to the 3′ end of the H2Bub-regulated sod2+ gene in the htb1-K119R mutant (Supplementary Fig. 5A, middle panel), consistent with the transcription elongation defect noted previously (Figs. 6, 7). However, FACT occupancy was unaffected at other loci tested (Supplementary Fig. 5A; data not shown), suggesting that H2Bub is not generally required for FACT binding to chromatin.
Discussion
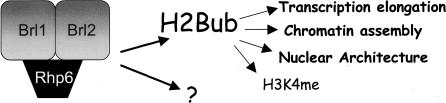
Our data link the ubiquitylation of histone H2B to nucleosome dynamics and transcription elongation in vivo. Collectively, our results provide a mechanistic explanation for the correlation between H2Bub and polII transcription documented previously. Importantly, we demonstrate that at least part of the mechanistic role of H2Bub in polII transcription does not involve H3K4me, despite the causal link between the two modifications. Our data are consistent with the idea that H2Bub participates in at least two independent transcriptional regulatory pathways: one that has mechanistic consequences for transcription elongation and is required for normal cell growth and morphology, and another that leads to H3K4me and downstream events that have yet to be uncovered (Fig. 8).
Figure 8.
Schematic diagram depicting the functional outcomes of H2Bub. Functions described in this report that are independent of H3K4me are in bold. The question mark (?) refers to additional, unidentified targets of the Rhp6–Brl1–Brl2 complex.
Much of the enzymatic machinery responsible for H2Bub appears to be conserved from yeast to human cells. Our study provides the first characterization of the S. pombe homologs of budding yeast BRE1, encoding the ubiquitin ligase for H2B. We found that knockouts of either brl1+ or brl2+ abolish H2Bub and result in similar phenotypes to those observed in the htb1-K119R mutant (Figs. 2A, 3C). This means that both of these RING finger proteins are individually required for H2Bub. Further, our physical interaction results, combined with the highly overlapping microarray profiles for both knockouts, strongly suggest that these proteins function together in the same complex (Figs. 2B, 5B; summarized in Fig. 8). Interestingly, the two human proteins most similar to Bre1, RNF20/hBRE1 and RNF40, are also found in a complex, although whether both are required for H2Bub in vivo has yet to be determined (Zhu et al. 2005). Direct association of functionally related RING finger proteins may be a general feature of ubiquitin ligase complexes that act on chromatin substrates, since this is also true of the Ring1b–Bmi1 and BRCA1–BARD1 complexes involved in H2A ubiquitylation in human cells (Brzovic et al. 2001; Buchwald et al. 2006). Detailed biochemical analyses of the activity of the Ring1b–Bmi1 complex have shown that Ring1b is primarily responsible for ubiquitin transfer to H2A, whereas Bmi1 functions to stimulate Ring1b activity (Buchwald et al. 2006). It remains to be seen whether the dual RING finger complexes involved in H2Bub have a similar functional organization.
In S. cerevisiae, the conserved ubiquitin-conjugating enzyme Rad6 mediates H2Bub (Robzyk et al. 2000). We have found that the S. pombe Rad6 homolog Rhp6 plays a similar role, in agreement with the findings of others (Roguev et al. 2003; Maruyama et al. 2006). In addition, we have found that Rhp6 physically interacts with both Brl1 and Brl2 (Fig. 2B). The human genome encodes two Rad6 homologs, hRAD6A and hRAD6B, but a recent report identified a different ubiquitin-conjugating enzyme, UbcH6, as the functional complement of the RNF20–RNF40 complex (Zhu et al. 2005). This may suggest a divergence between yeast and mammalian cells with respect to the H2Bub pathway, although the role of hRAD6 proteins in this pathway has yet to be extensively tested in vivo.
Our microarray analysis shows that only a small subset of genes in the S. pombe genome is significantly dependent on H2Bub or H3K4me for normal expression. This raises the question of what characteristics render a gene sensitive to regulation by H2Bub. One possibility is that certain gene-specific transcriptional regulators have a particular requirement for H2Bub for their function. This model is consistent with the finding that functionally related genes were enriched among the htb1-K119R-repressed genes, and is further supported by a recent study demonstrating that a mutation in the Drosophila BRE1 homolog dBRE causes specific defects related to Notch signaling (Bray et al. 2005). Another possibility is that the chromatin structure at certain loci, perhaps dictated by their underlying DNA sequence, is inherently unfavorable to polII transcription elongation, and thus would require the added fluidity conferred by H2Bub.
Although our analysis has focused on genes that are positively regulated by H2Bub, we found that equivalent numbers of genes were negatively regulated in our mutants. ChIP experiments at some of these genes showed that H3K4me is not enriched at these loci, even in the wild-type strain (data not shown). This negative regulation could either result from instances of H2Bub that are not associated with H3K4me, or from indirect effects. A handful of the genes that were induced in the H2Bub mutants were also induced in clr6-1 and ago1Δ mutants that are defective in heterochromatic gene silencing (Grewal and Moazed 2003). Importantly, the overlap also held for the set1Δ mutant, arguing that some of these effects are related to loss of H3K4me. The set1Δ and htb1-K119R mutants have been shown previously to be proficient in heterochromatic gene silencing (Noma and Grewal 2002; Jia et al. 2005; E. Hong and D. Moazed, pers. comm.), suggesting that the overlap is unlikely to result from a direct role for H3K4me in gene silencing. Instead, it likely reflects a heterochromatin “spreading” effect, as has been proposed to explain the loss of gene silencing in H3K4me-defective mutants in S. cerevisiae (Katan-Khaykovich and Struhl 2005).
Our experiments suggest that H3K4me has very little importance for gene regulation on its own in S. pombe. We emphasize, however, that the experiments presented here were conducted with cells grown in rich medium at 30°C, and that H3K4me may have specialized functions in gene regulation that are not apparent under these conditions.
It is possible that methylation of another histone lysine is affected in the absence of H2Bub in fission yeast. We note, however, that H3 Lys 79, which is methylated in an H2Bub-dependent manner in budding yeast and human cells (Briggs et al. 2002; Ng et al. 2002; Kim et al. 2005), is not detectably methylated in S. pombe, and a homolog of the H3K79 methyltransferase Dot1 is not encoded in its genome (J.C. Tanny and C.D. Allis, unpubl.). Also, no change in methylation of H3K36 or H4K20 is detected in htb1-K119R or set1Δ strains (data not shown).
Mutants defective in H2Bub showed defects in cell separation (Fig. 3B). These phenotypes are reminiscent of the septation defects observed in some known sep mutants, including sep10Δ and sep15Δ, and further, we observed some overlap between the microarray data sets for brl1Δ and brl2Δ and those for sep10Δ and sep15Δ. It is interesting that a number of gene products thought to be generally involved in polII transcription, such as Mediator components and histone ubiquitylation enzymes, appear to preferentially regulate specific sets of genes involved in cell separation. This is also true for the kinases Mcs6 and Cdk9 and the RING finger protein Pmh1, all involved in phosphorylation of the polII C-terminal domain (CTD) (Pei and Shuman 2003; Lee et al. 2005). In budding yeast, CTD phosphorylation has been shown to be required for H2Bub and subsequent H3K4me (Laribee et al. 2005; Wood et al. 2005), and phospho-CTD forms of polII colocalize with H3K4me by ChIP in human cells (Bernstein et al. 2005). These data could indicate a conserved pathway that links some functions of Mediator and polII CTD phosphorylation with H2Bub and chromatin assembly.
In addition to the septation defects observed in htb1-K119R mutants, some nuclei in these cells showed a fragmented pattern of DAPI staining (Fig. 4B,E). It is unlikely that this pattern is related to mitotic chromosome segregation defects, since previous studies failed to detect effects on integrity of centromeric chromatin in a rhp6Δ mutant (Maruyama et al. 2006). We also did not observe fragmented DAPI staining patterns in nuclei of a hip1Δ mutant, arguing against their being a result of defective chromatin assembly per se (data not shown). The crystal structure of the nucleosome predicts that the C terminus of H2B is exposed on the nucleosome surface (White et al. 2001), raising the intriguing possibility that the htb1-K119R mutant may cause global defects in internucleosome or other interactions that contribute to chromatin architecture.
What is the molecular mechanism by which H2Bub exerts its biological effects? Our ChIP experiments implicate H2Bub in control of transcription elongation and chromatin structure at a subset of genes (Fig. 6). The htb1-K119R mutant also showed lethal genetic interactions with other mutants known to affect chromatin assembly (Table 1). Thus, we propose that H2Bub directly promotes chromatin assembly and/or disassembly processes that accompany polII transcription elongation. Recent in vitro studies also support a direct role for ubiquitylation in transcription on chromatin templates. First, transcriptional activation by LEF-1–β-catenin on chromatin templates, but not on naked DNA, could be blocked by addition of a strong ubiquitin-binding protein. This affect was reversed upon addition of free ubiquitin, arguing that ubiquitin is specifically needed for activated transcription on chromatin templates (Sierra et al. 2006). More recently, a highly purified polII in vitro transcription system was used to reconstitute activation by the retinoic acid receptor on a chromatin template (Pavri et al. 2006). Importantly, addition of FACT, RNF20–RNF40 complex, and ubiquitin had a cooperative affect on transcription activation. Since our ChIP studies did not detect a general defect in chromatin association of the FACT complex in the absence of H2Bub (Supplementary Fig. 5A), this cooperativity could be due either to locus-specific effects on binding or to a different effect on FACT activity.
Protein monoubiquitylation plays well-documented roles in molecular recognition in a number of biological processes, notably endosomal protein sorting and DNA damage repair (Hoege et al. 2002; Alam et al. 2006). Our results suggest an additional role in transcription-coupled chromatin assembly, and lay the groundwork for study of potentially new modes of ubiquitin recognition.
Materials and methods
Yeast strains and media
Yeast strains used in this study are listed in Supplementary Table 1. All knockout strains and tagged strains were constructed using standard techniques (Bahler et al. 1998; Sato et al. 2005). Strains bearing the Flag epitope used plasmid pDM713 as template to generate the transforming PCR products (Tanny et al. 2004). To generate the htb1-K119R-Flag strain, an A-to-G substitution was introduced at codon 119 in the forward primer used in the transforming PCR reactions. To construct strains with combinations of markers, haploid strains with the individual markers were crossed and tetrads were dissected to isolate haploid progeny with both parental markers. Rich media was YE-supplemented with adenine, uracil, leucine, and histidine (YES) (Moreno et al. 1991).
Immunoprecipitations and Western blots
Large-scale immunoprecipitation of Rhp6-myc was performed as described with minor modifications (Tackett et al. 2005). Briefly, 3 L of culture grown in YES to 5 × 106 cells/mL was harvested and flash-frozen in small chunks. Cells were lysed under cryogenic conditions as described (Tackett et al. 2005), and frozen cell powder was resuspended in 7 mL of buffer L (20 mM HEPES at pH 7.6, 250 mM KCl, 5% glycerol, 2 mM MgCl2, 0.1 mM EDTA, 10 mM β-glycerophosphate, 0.1% Nonidet P-40, 1 mM phenylmethylsulfonyl fluoride [PMSF], protease inhibitor cocktail [Roche]). All subsequent steps were carried out at 4°C. Extracts were centrifuged for 15 min at 15,000g, and supernatants were incubated with 0.2 mL of anti-myc resin (Sigma) that was prewashed in buffer L. After 2 h, the resin was collected by centrifugation and transferred to a small column (Bio-Rad). The resin was washed with 10 mL of buffer L and then eluted with 1 mL of 0.5 M NH4OH/0.5 mM EDTA for 10 min at room temperature. Eluates were lyophilized, resuspended in 1× sample buffer, and analyzed by electrophoresis on 4%–12% gradient acrylamide gels (Novex) followed by Coommassie staining.
Extracts for Western blots were prepared using the TCA lysis method as described (Kao and Osley 2003). SDS-PAGE, transfer to PVDF, and blotting were carried out as described (Cheung et al. 2005). Anti-Flag antibody was purchased from Sigma, anti-H3K4me antibodies and anti-myc (clone 9E10) were from Millipore, anti-HA (clone 12CA5) was from the Sloan Kettering Monoclonal Antibody Core Facility, and anti-H3 was from Abcam. The tubulin antibody TAT1 was kindly provided by P. Nurse.
Microscopy and immunofluorescence
For DAPI/calcofluor staining, cells were fixed by addition of 1/10th culture volume of 37% formaldehyde and incubated with shaking for 15 min. Cells were pelleted, washed twice with dH2O, and mixed with an equal volume of 10 μg/mL DAPI + calcofluor on a glass slide. Photographs were taken using a Zeiss Axioscop 2 microscope.
Immunofluorescence with the nuclear pore antibody (mAb414, kindly provided by G. Blobel) was performed as described (Hagan and Ayscough 1999). A 1:1000 dilution of primary antibody and 1:250 dilution of FITC-conjugated secondary antibody (Jackson Laboratories) were employed. Images were captured on a DeltaVision microscope (Applied Precision/Olympus) using a cooled-charge device (CCD) camera and deconvolved using SoftWorx software.
Microarrays and Q-RT–PCR analysis
Total RNA was extracted from 50 mL of YES cultures using hot phenol and purified further using Qiagen RNeasy columns. For Q-RT–PCR, 0.1 μg of total RNA was reverse-transcribed with an oligo-dT primer using the Invitrogen SuperScript III first-strand synthesis kit. Resulting cDNA was quantified by real-time PCR using a SYBR Green mix (Applied Biosystems) in a Stratagene Mx3000P instrument.
Gene expression microarray analysis was performed in the laboratory of Bruce Futcher and Janet Leatherwood at State University of New York at Stony Brook (Stony Brook, NY) as previously described (Oliva et al. 2005). In each experiment, samples from wild-type (htb1-kanMX6) (see Supplementary Table 1) or mutant strains were cohybridized with a sample prepared from a reference wild-type strain (no antibiotic resistance marker). Clustering analysis used a hierarchical clustering algorithm. Genespring software (Agilent) was used to derive p-values (hypergeometric probabilities).
ChIP
ChIP was performed as described (Huang and Moazed 2003) with modifications. Briefly, 50 mL of YES cultures were fixed with 1% formaldehyde, harvested, and flash frozen. Cells were lysed by bead-beating in 0.4 mL of immunoprecipitation buffer (50 mM HEPES at pH 7.6, 500 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% SDS, 0.1% sodium deoxycholate, 1 mM PMSF, protease inhibitor cocktail [Roche]). Extracts were diluted to 2 mL with immunoprecipitation buffer and sonicated for seven cycles (30 sec on, 60 sec off) in a water bath sonicator (Diagenode) at 4°C. After clearing by centrifugation, 100 μL of extract were incubated with 1 μL of antibody overnight at 4°C. Anti-myc, anti-HA, anti-H3K4me3, and anti-H3 were described above. Anti-RNApolII (clone 8WG16) was purchased from Millipore. Immune complexes were recovered with 15 μL of protein A or protein G agarose beads (Pierce), then washed, eluted, and purified as described (Huang and Moazed 2003). ChIP DNA (1.5 μL) and input DNA (1.5 μL of a 1/100 dilution) samples were analyzed by real-time PCR with specific primer pairs. Enrichment of a particular sequence in the immunoprecipitations was calculated relative to input after subtraction of enrichment obtained from a negative control immunoprecipitation (no antibody). Details regarding primers used for PCR are provided in the Supplemental Material.
Acknowledgments
We thank Paul Nurse, A.F.S. Stewart, and Simon Whitehall for strains; Paul Nurse for the TAT1 antibody; Gunther Blobel for the mAb414 antibody; Julie Huang, Jaehoon Kim, Tom Milne, Mary Ann Osley, Alex Ruthenberg, David Shechter, Sean Taverna, and Jenny Wu for critical reading of the manuscript; Jenny Wu, Yasha Zhurinsky, and Atanas Khaykov for helpful advice about S. pombe; members of the Chait laboratory for assistance with cryolysis; Bruce Futcher and Janet Leatherwood for performing the microarray experiments; Mary Ann Osley for sharing results prior to publication; and members of the Allis and Nurse labs for many stimulating discussions. J.C.T. is funded by a grant from the Leukemia and Lymphoma Society (reference: 5335-06). This work was supported by grants to C.D.A. from the National Institutes of Health (GM40922, GM53512, and GM63959).
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1516207
References
- Ahmad K., Henikoff S., Henikoff S. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell. 2002;9:1191–1200. doi: 10.1016/s1097-2765(02)00542-7. [DOI] [PubMed] [Google Scholar]
- Alam S.L., Langelier C., Whitby F.G., Koirala S., Robinson H., Hill C.P., Sundquist W.I., Langelier C., Whitby F.G., Koirala S., Robinson H., Hill C.P., Sundquist W.I., Whitby F.G., Koirala S., Robinson H., Hill C.P., Sundquist W.I., Koirala S., Robinson H., Hill C.P., Sundquist W.I., Robinson H., Hill C.P., Sundquist W.I., Hill C.P., Sundquist W.I., Sundquist W.I. Structural basis for ubiquitin recognition by the human ESCRT-II EAP45 GLUE domain. Nat. Struct. Mol. Biol. 2006;13:1029–1030. doi: 10.1038/nsmb1160. [DOI] [PubMed] [Google Scholar]
- Bahler J., Wu J.Q., Longtine M.S., Shah N.G., McKenzie A., III, Steever A.B., Wach A., Philippsen P., Pringle J.R., Wu J.Q., Longtine M.S., Shah N.G., McKenzie A., III, Steever A.B., Wach A., Philippsen P., Pringle J.R., Longtine M.S., Shah N.G., McKenzie A., III, Steever A.B., Wach A., Philippsen P., Pringle J.R., Shah N.G., McKenzie A., III, Steever A.B., Wach A., Philippsen P., Pringle J.R., McKenzie A., III, Steever A.B., Wach A., Philippsen P., Pringle J.R., Steever A.B., Wach A., Philippsen P., Pringle J.R., Wach A., Philippsen P., Pringle J.R., Philippsen P., Pringle J.R., Pringle J.R. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Belotserkovskaya R., Oh S., Bondarenko V.A., Orphanides G., Studitsky V.M., Reinberg D., Oh S., Bondarenko V.A., Orphanides G., Studitsky V.M., Reinberg D., Bondarenko V.A., Orphanides G., Studitsky V.M., Reinberg D., Orphanides G., Studitsky V.M., Reinberg D., Studitsky V.M., Reinberg D., Reinberg D. FACT facilitates transcription-dependent nucleosome alteration. Science. 2003;301:1090–1093. doi: 10.1126/science.1085703. [DOI] [PubMed] [Google Scholar]
- Bernstein B.E., Kamal M., Lindblad-Toh K., Bekiranov S., Bailey D.K., Huebert D.J., McMahon S., Karlsson E.K., Kulbokas E.J., III, Gingeras T.R., Kamal M., Lindblad-Toh K., Bekiranov S., Bailey D.K., Huebert D.J., McMahon S., Karlsson E.K., Kulbokas E.J., III, Gingeras T.R., Lindblad-Toh K., Bekiranov S., Bailey D.K., Huebert D.J., McMahon S., Karlsson E.K., Kulbokas E.J., III, Gingeras T.R., Bekiranov S., Bailey D.K., Huebert D.J., McMahon S., Karlsson E.K., Kulbokas E.J., III, Gingeras T.R., Bailey D.K., Huebert D.J., McMahon S., Karlsson E.K., Kulbokas E.J., III, Gingeras T.R., Huebert D.J., McMahon S., Karlsson E.K., Kulbokas E.J., III, Gingeras T.R., McMahon S., Karlsson E.K., Kulbokas E.J., III, Gingeras T.R., Karlsson E.K., Kulbokas E.J., III, Gingeras T.R., Kulbokas E.J., III, Gingeras T.R., Gingeras T.R., et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Blackwell C., Martin K.A., Greenall A., Pidoux A., Allshire R.C., Whitehall S.K., Martin K.A., Greenall A., Pidoux A., Allshire R.C., Whitehall S.K., Greenall A., Pidoux A., Allshire R.C., Whitehall S.K., Pidoux A., Allshire R.C., Whitehall S.K., Allshire R.C., Whitehall S.K., Whitehall S.K. The Schizosaccharomyces pombe HIRA-like protein Hip1 is required for the periodic expression of histone genes and contributes to the function of complex centromeres. Mol. Cell. Biol. 2004;24:4309–4320. doi: 10.1128/MCB.24.10.4309-4320.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray S., Musisi H., Bienz M., Musisi H., Bienz M., Bienz M. Bre1 is required for Notch signaling and histone modification. Dev. Cell. 2005;8:279–286. doi: 10.1016/j.devcel.2004.11.020. [DOI] [PubMed] [Google Scholar]
- Briggs S.D., Xiao T., Sun Z.W., Caldwell J.A., Shabanowitz J., Hunt D.F., Allis C.D., Strahl B.D., Xiao T., Sun Z.W., Caldwell J.A., Shabanowitz J., Hunt D.F., Allis C.D., Strahl B.D., Sun Z.W., Caldwell J.A., Shabanowitz J., Hunt D.F., Allis C.D., Strahl B.D., Caldwell J.A., Shabanowitz J., Hunt D.F., Allis C.D., Strahl B.D., Shabanowitz J., Hunt D.F., Allis C.D., Strahl B.D., Hunt D.F., Allis C.D., Strahl B.D., Allis C.D., Strahl B.D., Strahl B.D. Gene silencing: Trans-histone regulatory pathway in chromatin. Nature. 2002;418:498. doi: 10.1038/nature00970. [DOI] [PubMed] [Google Scholar]
- Brzovic P.S., Rajagopal P., Hoyt D.W., King M.C., Klevit R.E., Rajagopal P., Hoyt D.W., King M.C., Klevit R.E., Hoyt D.W., King M.C., Klevit R.E., King M.C., Klevit R.E., Klevit R.E. Structure of a BRCA1–BARD1 heterodimeric RING–RING complex. Nat. Struct. Biol. 2001;8:833–837. doi: 10.1038/nsb1001-833. [DOI] [PubMed] [Google Scholar]
- Buchwald G., van der Stoop P., Weichenrieder O., Perrakis A., van Lohuizen M., Sixma T.K., van der Stoop P., Weichenrieder O., Perrakis A., van Lohuizen M., Sixma T.K., Weichenrieder O., Perrakis A., van Lohuizen M., Sixma T.K., Perrakis A., van Lohuizen M., Sixma T.K., van Lohuizen M., Sixma T.K., Sixma T.K. Structure and E3-ligase activity of the Ring–Ring complex of polycomb proteins Bmi1 and Ring1b. EMBO J. 2006;25:2465–2474. doi: 10.1038/sj.emboj.7601144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr A.M., Dorrington S.M., Hindley J., Phear G.A., Aves S.J., Nurse P., Dorrington S.M., Hindley J., Phear G.A., Aves S.J., Nurse P., Hindley J., Phear G.A., Aves S.J., Nurse P., Phear G.A., Aves S.J., Nurse P., Aves S.J., Nurse P., Nurse P. Analysis of a histone H2A variant from fission yeast: Evidence for a role in chromosome stability. Mol. Gen. Genet. 1994;245:628–635. doi: 10.1007/BF00282226. [DOI] [PubMed] [Google Scholar]
- Chen D., Toone W.M., Mata J., Lyne R., Burns G., Kivinen K., Brazma A., Jones N., Bahler J., Toone W.M., Mata J., Lyne R., Burns G., Kivinen K., Brazma A., Jones N., Bahler J., Mata J., Lyne R., Burns G., Kivinen K., Brazma A., Jones N., Bahler J., Lyne R., Burns G., Kivinen K., Brazma A., Jones N., Bahler J., Burns G., Kivinen K., Brazma A., Jones N., Bahler J., Kivinen K., Brazma A., Jones N., Bahler J., Brazma A., Jones N., Bahler J., Jones N., Bahler J., Bahler J. Global transcriptional responses of fission yeast to environmental stress. Mol. Biol. Cell. 2003;14:214–229. doi: 10.1091/mbc.E02-08-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung W.L., Turner F.B., Krishnamoorthy T., Wolner B., Ahn S.H., Foley M., Dorsey J.A., Peterson C.L., Berger S.L., Allis C.D., Turner F.B., Krishnamoorthy T., Wolner B., Ahn S.H., Foley M., Dorsey J.A., Peterson C.L., Berger S.L., Allis C.D., Krishnamoorthy T., Wolner B., Ahn S.H., Foley M., Dorsey J.A., Peterson C.L., Berger S.L., Allis C.D., Wolner B., Ahn S.H., Foley M., Dorsey J.A., Peterson C.L., Berger S.L., Allis C.D., Ahn S.H., Foley M., Dorsey J.A., Peterson C.L., Berger S.L., Allis C.D., Foley M., Dorsey J.A., Peterson C.L., Berger S.L., Allis C.D., Dorsey J.A., Peterson C.L., Berger S.L., Allis C.D., Peterson C.L., Berger S.L., Allis C.D., Berger S.L., Allis C.D., Allis C.D. Phosphorylation of histone H4 serine 1 during DNA damage requires casein kinase II in S. cerevisiae. Curr. Biol. 2005;15:656–660. doi: 10.1016/j.cub.2005.02.049. [DOI] [PubMed] [Google Scholar]
- Dover J., Schneider J., Tawiah-Boateng M.A., Wood A., Dean K., Johnston M., Shilatifard A., Schneider J., Tawiah-Boateng M.A., Wood A., Dean K., Johnston M., Shilatifard A., Tawiah-Boateng M.A., Wood A., Dean K., Johnston M., Shilatifard A., Wood A., Dean K., Johnston M., Shilatifard A., Dean K., Johnston M., Shilatifard A., Johnston M., Shilatifard A., Shilatifard A. Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J. Biol. Chem. 2002;277:28368–28371. doi: 10.1074/jbc.C200348200. [DOI] [PubMed] [Google Scholar]
- Ercan S., Simpson R.T., Simpson R.T. Global chromatin structure of 45,000 base pairs of chromosome III in a- and α-cell yeast and during mating-type switching. Mol. Cell. Biol. 2004;24:10026–10035. doi: 10.1128/MCB.24.22.10026-10035.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal S.I., Moazed D., Moazed D. Heterochromatin and epigenetic control of gene expression. Science. 2003;301:798–802. doi: 10.1126/science.1086887. [DOI] [PubMed] [Google Scholar]
- Guenther M.G., Jenner R.G., Chevalier B., Nakamura T., Croce C.M., Canaani E., Young R.A., Jenner R.G., Chevalier B., Nakamura T., Croce C.M., Canaani E., Young R.A., Chevalier B., Nakamura T., Croce C.M., Canaani E., Young R.A., Nakamura T., Croce C.M., Canaani E., Young R.A., Croce C.M., Canaani E., Young R.A., Canaani E., Young R.A., Young R.A. Global and Hox-specific roles for the MLL1 methyltransferase. Proc. Natl. Acad. Sci. 2005;102:8603–8608. doi: 10.1073/pnas.0503072102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan I.M., Ayscough K.R., Ayscough K.R. Fluorescence microscopy in yeast. In: Allan V., editor. Protein localization by fluorescence microscopy: A practical approach. Oxford University Press; Oxford, UK: 1999. [Google Scholar]
- Hansen K.R., Burns G., Mata J., Volpe T.A., Martienssen R.A., Bahler J., Thon G., Burns G., Mata J., Volpe T.A., Martienssen R.A., Bahler J., Thon G., Mata J., Volpe T.A., Martienssen R.A., Bahler J., Thon G., Volpe T.A., Martienssen R.A., Bahler J., Thon G., Martienssen R.A., Bahler J., Thon G., Bahler J., Thon G., Thon G. Global effects on gene expression in fission yeast by silencing and RNA interference machineries. Mol. Cell. Biol. 2005;25:590–601. doi: 10.1128/MCB.25.2.590-601.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S., Furuyama T., Ahmad K., Furuyama T., Ahmad K., Ahmad K. Histone variants, nucleosome assembly and epigenetic inheritance. Trends Genet. 2004;20:320–326. doi: 10.1016/j.tig.2004.05.004. [DOI] [PubMed] [Google Scholar]
- Hoege C., Pfander B., Moldovan G.L., Pyrowolakis G., Jentsch S., Pfander B., Moldovan G.L., Pyrowolakis G., Jentsch S., Moldovan G.L., Pyrowolakis G., Jentsch S., Pyrowolakis G., Jentsch S., Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- Huang J., Moazed D., Moazed D. Association of the RENT complex with nontranscribed and coding regions of rDNA and a regional requirement for the replication fork block protein Fob1 in rDNA silencing. Genes & Dev. 2003;17:2162–2176. doi: 10.1101/gad.1108403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang W.W., Venkatasubrahmanyam S., Ianculescu A.G., Tong A., Boone C., Madhani H.D., Venkatasubrahmanyam S., Ianculescu A.G., Tong A., Boone C., Madhani H.D., Ianculescu A.G., Tong A., Boone C., Madhani H.D., Tong A., Boone C., Madhani H.D., Boone C., Madhani H.D., Madhani H.D. A conserved RING finger protein required for histone H2B monoubiquitination and cell size control. Mol. Cell. 2003;11:261–266. doi: 10.1016/s1097-2765(02)00826-2. [DOI] [PubMed] [Google Scholar]
- Izban M.G., Luse D.S., Luse D.S. Transcription on nucleosomal templates by RNA polymerase II in vitro: Inhibition of elongation with enhancement of sequence-specific pausing. Genes & Dev. 1991;5:683–696. doi: 10.1101/gad.5.4.683. [DOI] [PubMed] [Google Scholar]
- Jason L.J., Moore S.C., Lewis J.D., Lindsey G., Ausio J., Moore S.C., Lewis J.D., Lindsey G., Ausio J., Lewis J.D., Lindsey G., Ausio J., Lindsey G., Ausio J., Ausio J. Histone ubiquitination: A tagging tail unfolds? Bioessays. 2002;24:166–174. doi: 10.1002/bies.10038. [DOI] [PubMed] [Google Scholar]
- Jia S., Kobayashi R., Grewal S.I., Kobayashi R., Grewal S.I., Grewal S.I. Ubiquitin ligase component Cul4 associates with Clr4 histone methyltransferase to assemble heterochromatin. Nat. Cell Biol. 2005;7:1007–1013. doi: 10.1038/ncb1300. [DOI] [PubMed] [Google Scholar]
- Kao C.F., Osley M.A., Osley M.A. In vivo assays to study histone ubiquitylation. Methods. 2003;31:59–66. doi: 10.1016/s1046-2023(03)00088-4. [DOI] [PubMed] [Google Scholar]
- Kaplan C.D., Laprade L., Winston F., Laprade L., Winston F., Winston F. Transcription elongation factors repress transcription initiation from cryptic sites. Science. 2003;301:1096–1099. doi: 10.1126/science.1087374. [DOI] [PubMed] [Google Scholar]
- Katan-Khaykovich Y., Struhl K., Struhl K. Heterochromatin formation involves changes in histone modifications over multiple cell generations. EMBO J. 2005;24:2138–2149. doi: 10.1038/sj.emboj.7600692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman P.D., Cohen J.L., Osley M.A., Cohen J.L., Osley M.A., Osley M.A. Hir proteins are required for position-dependent gene silencing in Saccharomyces cerevisiae in the absence of chromatin assembly factor I. Mol. Cell. Biol. 1998;18:4793–4806. doi: 10.1128/mcb.18.8.4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Hake S.B., Roeder R.G., Hake S.B., Roeder R.G., Roeder R.G. The human homolog of yeast BRE1 functions as a transcriptional coactivator through direct activator interactions. Mol. Cell. 2005;20:759–770. doi: 10.1016/j.molcel.2005.11.012. [DOI] [PubMed] [Google Scholar]
- Kireeva M.L., Walter W., Tchernajenko V., Bondarenko V., Kashlev M., Studitsky V.M., Walter W., Tchernajenko V., Bondarenko V., Kashlev M., Studitsky V.M., Tchernajenko V., Bondarenko V., Kashlev M., Studitsky V.M., Bondarenko V., Kashlev M., Studitsky V.M., Kashlev M., Studitsky V.M., Studitsky V.M. Nucleosome remodeling induced by RNA polymerase II: Loss of the H2A/H2B dimer during transcription. Mol. Cell. 2002;9:541–552. doi: 10.1016/s1097-2765(02)00472-0. [DOI] [PubMed] [Google Scholar]
- Kobor M.S., Venkatasubrahmanyam S., Meneghini M.D., Gin J.W., Jennings J.L., Link A.J., Madhani H.D., Rine J., Venkatasubrahmanyam S., Meneghini M.D., Gin J.W., Jennings J.L., Link A.J., Madhani H.D., Rine J., Meneghini M.D., Gin J.W., Jennings J.L., Link A.J., Madhani H.D., Rine J., Gin J.W., Jennings J.L., Link A.J., Madhani H.D., Rine J., Jennings J.L., Link A.J., Madhani H.D., Rine J., Link A.J., Madhani H.D., Rine J., Madhani H.D., Rine J., Rine J. A protein complex containing the conserved Swi2/Snf2-related ATPase Swr1p deposits histone variant H2A.Z into euchromatin. PLoS Biol. 2004;2:E131. doi: 10.1371/journal.pbio.0020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laribee R.N., Krogan N.J., Xiao T., Shibata Y., Hughes T.R., Greenblatt J.F., Strahl B.D., Krogan N.J., Xiao T., Shibata Y., Hughes T.R., Greenblatt J.F., Strahl B.D., Xiao T., Shibata Y., Hughes T.R., Greenblatt J.F., Strahl B.D., Shibata Y., Hughes T.R., Greenblatt J.F., Strahl B.D., Hughes T.R., Greenblatt J.F., Strahl B.D., Greenblatt J.F., Strahl B.D., Strahl B.D. BUR kinase selectively regulates H3 K4 trimethylation and H2B ubiquitylation through recruitment of the PAF elongation complex. Curr. Biol. 2005;15:1487–1493. doi: 10.1016/j.cub.2005.07.028. [DOI] [PubMed] [Google Scholar]
- Lee C.K., Shibata Y., Rao B., Strahl B.D., Lieb J.D., Shibata Y., Rao B., Strahl B.D., Lieb J.D., Rao B., Strahl B.D., Lieb J.D., Strahl B.D., Lieb J.D., Lieb J.D. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat. Genet. 2004;36:900–905. doi: 10.1038/ng1400. [DOI] [PubMed] [Google Scholar]
- Lee K.M., Miklos I., Du H., Watt S., Szilagyi Z., Saiz J.E., Madabhushi R., Penkett C.J., Sipiczki M., Bahler J., Miklos I., Du H., Watt S., Szilagyi Z., Saiz J.E., Madabhushi R., Penkett C.J., Sipiczki M., Bahler J., Du H., Watt S., Szilagyi Z., Saiz J.E., Madabhushi R., Penkett C.J., Sipiczki M., Bahler J., Watt S., Szilagyi Z., Saiz J.E., Madabhushi R., Penkett C.J., Sipiczki M., Bahler J., Szilagyi Z., Saiz J.E., Madabhushi R., Penkett C.J., Sipiczki M., Bahler J., Saiz J.E., Madabhushi R., Penkett C.J., Sipiczki M., Bahler J., Madabhushi R., Penkett C.J., Sipiczki M., Bahler J., Penkett C.J., Sipiczki M., Bahler J., Sipiczki M., Bahler J., Bahler J., et al. Impairment of the TFIIH-associated CDK-activating kinase selectively affects cell cycle-regulated gene expression in fission yeast. Mol. Biol. Cell. 2005;16:2734–2745. doi: 10.1091/mbc.E04-11-0982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Pattenden S.G., Lee D., Gutierrez J., Chen J., Seidel C., Gerton J., Workman J.L., Pattenden S.G., Lee D., Gutierrez J., Chen J., Seidel C., Gerton J., Workman J.L., Lee D., Gutierrez J., Chen J., Seidel C., Gerton J., Workman J.L., Gutierrez J., Chen J., Seidel C., Gerton J., Workman J.L., Chen J., Seidel C., Gerton J., Workman J.L., Seidel C., Gerton J., Workman J.L., Gerton J., Workman J.L., Workman J.L. Preferential occupancy of histone variant H2AZ at inactive promoters influences local histone modifications and chromatin remodeling. Proc. Natl. Acad. Sci. 2005;102:18385–18390. doi: 10.1073/pnas.0507975102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C.L., Kaplan T., Kim M., Buratowski S., Schreiber S.L., Friedman N., Rando O.J., Kaplan T., Kim M., Buratowski S., Schreiber S.L., Friedman N., Rando O.J., Kim M., Buratowski S., Schreiber S.L., Friedman N., Rando O.J., Buratowski S., Schreiber S.L., Friedman N., Rando O.J., Schreiber S.L., Friedman N., Rando O.J., Friedman N., Rando O.J., Rando O.J. Single-nucleosome mapping of histone modifications in S. cerevisiae. PLoS Biol. 2005;3:e328. doi: 10.1371/journal.pbio.0030328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin D.G., Baetz K., Shi X., Walter K.L., MacDonald V.E., Wlodarski M.J., Gozani O., Hieter P., Howe L., Baetz K., Shi X., Walter K.L., MacDonald V.E., Wlodarski M.J., Gozani O., Hieter P., Howe L., Shi X., Walter K.L., MacDonald V.E., Wlodarski M.J., Gozani O., Hieter P., Howe L., Walter K.L., MacDonald V.E., Wlodarski M.J., Gozani O., Hieter P., Howe L., MacDonald V.E., Wlodarski M.J., Gozani O., Hieter P., Howe L., Wlodarski M.J., Gozani O., Hieter P., Howe L., Gozani O., Hieter P., Howe L., Hieter P., Howe L., Howe L. The Yng1p plant homeodomain finger is a methyl-histone binding module that recognizes lysine 4-methylated histone H3. Mol. Cell. Biol. 2006;26:7871–7879. doi: 10.1128/MCB.00573-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama T., Nakamura T., Hayashi T., Yanagida M., Nakamura T., Hayashi T., Yanagida M., Hayashi T., Yanagida M., Yanagida M. Histone H2B mutations in inner region affect ubiquitination, centromere function, silencing and chromosome segregation. EMBO J. 2006;25:2420–2431. doi: 10.1038/sj.emboj.7601110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi G., Shen X., Landry J., Wu W.H., Sen S., Wu C., Shen X., Landry J., Wu W.H., Sen S., Wu C., Landry J., Wu W.H., Sen S., Wu C., Wu W.H., Sen S., Wu C., Sen S., Wu C., Wu C. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science. 2004;303:343–348. doi: 10.1126/science.1090701. [DOI] [PubMed] [Google Scholar]
- Moreno S., Klar A., Nurse P., Klar A., Nurse P., Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Nathan D., Ingvarsdottir K., Sterner D.E., Bylebyl G.R., Dokmanovic M., Dorsey J.A., Whelan K.A., Krsmanovic M., Lane W.S., Meluh P.B., Ingvarsdottir K., Sterner D.E., Bylebyl G.R., Dokmanovic M., Dorsey J.A., Whelan K.A., Krsmanovic M., Lane W.S., Meluh P.B., Sterner D.E., Bylebyl G.R., Dokmanovic M., Dorsey J.A., Whelan K.A., Krsmanovic M., Lane W.S., Meluh P.B., Bylebyl G.R., Dokmanovic M., Dorsey J.A., Whelan K.A., Krsmanovic M., Lane W.S., Meluh P.B., Dokmanovic M., Dorsey J.A., Whelan K.A., Krsmanovic M., Lane W.S., Meluh P.B., Dorsey J.A., Whelan K.A., Krsmanovic M., Lane W.S., Meluh P.B., Whelan K.A., Krsmanovic M., Lane W.S., Meluh P.B., Krsmanovic M., Lane W.S., Meluh P.B., Lane W.S., Meluh P.B., Meluh P.B., et al. Histone sumoylation is a negative regulator in Saccharomyces cerevisiae and shows dynamic interplay with positive-acting histone modifications. Genes & Dev. 2006;20:966–976. doi: 10.1101/gad.1404206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng H.H., Xu R.M., Zhang Y., Struhl K., Xu R.M., Zhang Y., Struhl K., Zhang Y., Struhl K., Struhl K. Ubiquitination of histone H2B by Rad6 is required for efficient Dot1-mediated methylation of histone H3 lysine 79. J. Biol. Chem. 2002;277:34655–34657. doi: 10.1074/jbc.C200433200. [DOI] [PubMed] [Google Scholar]
- Ng H.H., Robert F., Young R.A., Struhl K., Robert F., Young R.A., Struhl K., Young R.A., Struhl K., Struhl K. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol. Cell. 2003;11:709–719. doi: 10.1016/s1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- Nislow C., Ray E., Pillus L., Ray E., Pillus L., Pillus L. SET1, a yeast member of the trithorax family, functions in transcriptional silencing and diverse cellular processes. Mol. Biol. Cell. 1997;8:2421–2436. doi: 10.1091/mbc.8.12.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noma K., Grewal S.I., Grewal S.I. Histone H3 lysine 4 methylation is mediated by Set1 and promotes maintenance of active chromatin states in fission yeast. Proc. Natl. Acad. Sci. 2002;99 (Suppl. 4):16438–16445. doi: 10.1073/pnas.182436399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliva A., Rosebrock A., Ferrezuelo F., Pyne S., Chen H., Skiena S., Futcher B., Leatherwood J., Rosebrock A., Ferrezuelo F., Pyne S., Chen H., Skiena S., Futcher B., Leatherwood J., Ferrezuelo F., Pyne S., Chen H., Skiena S., Futcher B., Leatherwood J., Pyne S., Chen H., Skiena S., Futcher B., Leatherwood J., Chen H., Skiena S., Futcher B., Leatherwood J., Skiena S., Futcher B., Leatherwood J., Futcher B., Leatherwood J., Leatherwood J. The cell cycle-regulated genes of Schizosaccharomyces pombe. PLoS Biol. 2005;3:e225. doi: 10.1371/journal.pbio.0030225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavri R., Zhu B., Li G., Trojer P., Mandal S., Shilatifard A., Reinberg D., Zhu B., Li G., Trojer P., Mandal S., Shilatifard A., Reinberg D., Li G., Trojer P., Mandal S., Shilatifard A., Reinberg D., Trojer P., Mandal S., Shilatifard A., Reinberg D., Mandal S., Shilatifard A., Reinberg D., Shilatifard A., Reinberg D., Reinberg D. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell. 2006;125:703–717. doi: 10.1016/j.cell.2006.04.029. [DOI] [PubMed] [Google Scholar]
- Pei Y., Shuman S., Shuman S. Characterization of the Schizosaccharomyces pombe Cdk9/Pch1 protein kinase: Spt5 phosphorylation, autophosphorylation, and mutational analysis. J. Biol. Chem. 2003;278:43346–43356. doi: 10.1074/jbc.M307319200. [DOI] [PubMed] [Google Scholar]
- Pokholok D.K., Harbison C.T., Levine S., Cole M., Hannett N.M., Lee T.I., Bell G.W., Walker K., Rolfe P.A., Herbolsheimer E., Harbison C.T., Levine S., Cole M., Hannett N.M., Lee T.I., Bell G.W., Walker K., Rolfe P.A., Herbolsheimer E., Levine S., Cole M., Hannett N.M., Lee T.I., Bell G.W., Walker K., Rolfe P.A., Herbolsheimer E., Cole M., Hannett N.M., Lee T.I., Bell G.W., Walker K., Rolfe P.A., Herbolsheimer E., Hannett N.M., Lee T.I., Bell G.W., Walker K., Rolfe P.A., Herbolsheimer E., Lee T.I., Bell G.W., Walker K., Rolfe P.A., Herbolsheimer E., Bell G.W., Walker K., Rolfe P.A., Herbolsheimer E., Walker K., Rolfe P.A., Herbolsheimer E., Rolfe P.A., Herbolsheimer E., Herbolsheimer E., et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- Raisner R.M., Hartley P.D., Meneghini M.D., Bao M.Z., Liu C.L., Schreiber S.L., Rando O.J., Madhani H.D., Hartley P.D., Meneghini M.D., Bao M.Z., Liu C.L., Schreiber S.L., Rando O.J., Madhani H.D., Meneghini M.D., Bao M.Z., Liu C.L., Schreiber S.L., Rando O.J., Madhani H.D., Bao M.Z., Liu C.L., Schreiber S.L., Rando O.J., Madhani H.D., Liu C.L., Schreiber S.L., Rando O.J., Madhani H.D., Schreiber S.L., Rando O.J., Madhani H.D., Rando O.J., Madhani H.D., Madhani H.D. Histone variant H2A.Z marks the 5′ ends of both active and inactive genes in euchromatin. Cell. 2005;123:233–248. doi: 10.1016/j.cell.2005.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robzyk K., Recht J., Osley M.A., Recht J., Osley M.A., Osley M.A. Rad6-dependent ubiquitination of histone H2B in yeast. Science. 2000;287:501–504. doi: 10.1126/science.287.5452.501. [DOI] [PubMed] [Google Scholar]
- Roguev A., Schaft D., Shevchenko A., Aasland R., Stewart A.F., Schaft D., Shevchenko A., Aasland R., Stewart A.F., Shevchenko A., Aasland R., Stewart A.F., Aasland R., Stewart A.F., Stewart A.F. High conservation of the Set1/Rad6 axis of histone 3 lysine 4 methylation in budding and fission yeasts. J. Biol. Chem. 2003;278:8487–8493. doi: 10.1074/jbc.M209562200. [DOI] [PubMed] [Google Scholar]
- Santos-Rosa H., Schneider R., Bannister A.J., Sherriff J., Bernstein B.E., Emre N.C., Schreiber S.L., Mellor J., Kouzarides T., Schneider R., Bannister A.J., Sherriff J., Bernstein B.E., Emre N.C., Schreiber S.L., Mellor J., Kouzarides T., Bannister A.J., Sherriff J., Bernstein B.E., Emre N.C., Schreiber S.L., Mellor J., Kouzarides T., Sherriff J., Bernstein B.E., Emre N.C., Schreiber S.L., Mellor J., Kouzarides T., Bernstein B.E., Emre N.C., Schreiber S.L., Mellor J., Kouzarides T., Emre N.C., Schreiber S.L., Mellor J., Kouzarides T., Schreiber S.L., Mellor J., Kouzarides T., Mellor J., Kouzarides T., Kouzarides T. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- Sato M., Dhut S., Toda T., Dhut S., Toda T., Toda T. New drug-resistant cassettes for gene disruption and epitope tagging in Schizosaccharomyces pombe. Yeast. 2005;22:583–591. doi: 10.1002/yea.1233. [DOI] [PubMed] [Google Scholar]
- Schwabish M.A., Struhl K., Struhl K. Evidence for eviction and rapid deposition of histones upon transcriptional elongation by RNA polymerase II. Mol. Cell. Biol. 2004;24:10111–10117. doi: 10.1128/MCB.24.23.10111-10117.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwabish M.A., Struhl K., Struhl K. Asf1 mediates histone eviction and deposition during elongation by RNA polymerase II. Mol. Cell. 2006;22:415–422. doi: 10.1016/j.molcel.2006.03.014. [DOI] [PubMed] [Google Scholar]
- Sharp J.A., Franco A.A., Osley M.A., Kaufman P.D., Franco A.A., Osley M.A., Kaufman P.D., Osley M.A., Kaufman P.D., Kaufman P.D. Chromatin assembly factor I and Hir proteins contribute to building functional kinetochores in S. cerevisiae. Genes & Dev. 2002;16:85–100. doi: 10.1101/gad.925302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilatifard A. Chromatin modifications by methylation and ubiquitination: Implications in the regulation of gene expression. Annu. Rev. Biochem. 2006;75:243–269. doi: 10.1146/annurev.biochem.75.103004.142422. [DOI] [PubMed] [Google Scholar]
- Sierra J., Yoshida T., Joazeiro C.A., Jones K.A., Yoshida T., Joazeiro C.A., Jones K.A., Joazeiro C.A., Jones K.A., Jones K.A. The APC tumor suppressor counteracts β-catenin activation and H3K4 methylation at Wnt target genes. Genes & Dev. 2006;20:586–600. doi: 10.1101/gad.1385806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simanis V. The mitotic exit and septation initiation networks. J. Cell Sci. 2003;116:4261–4262. doi: 10.1242/jcs.00777. [DOI] [PubMed] [Google Scholar]
- Sims R.J., III, Belotserkovskaya R., Reinberg D., Belotserkovskaya R., Reinberg D., Reinberg D. Elongation by RNA polymerase II: The short and long of it. Genes & Dev. 2004;18:2437–2468. doi: 10.1101/gad.1235904. [DOI] [PubMed] [Google Scholar]
- Spector M.S., Raff A., DeSilva H., Lee K., Osley M.A., Raff A., DeSilva H., Lee K., Osley M.A., DeSilva H., Lee K., Osley M.A., Lee K., Osley M.A., Osley M.A. Hir1p and Hir2p function as transcriptional corepressors to regulate histone gene transcription in the Saccharomyces cerevisiae cell cycle. Mol. Cell. Biol. 1997;17:545–552. doi: 10.1128/mcb.17.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z.W., Allis C.D., Allis C.D. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature. 2002;418:104–108. doi: 10.1038/nature00883. [DOI] [PubMed] [Google Scholar]
- Szilagyi Z., Grallert A., Zilahi E., Sipiczki M., Grallert A., Zilahi E., Sipiczki M., Zilahi E., Sipiczki M., Sipiczki M. Isolation and characterization of fission yeast genes involved in transcription regulation of cell cycle events (a short communication) Acta Microbiol. Immunol. Hung. 2002;49:285–287. doi: 10.1556/AMicr.49.2002.2-3.16. [DOI] [PubMed] [Google Scholar]
- Tackett A.J., Dilworth D.J., Davey M.J., O’Donnell M., Aitchison J.D., Rout M.P., Chait B.T., Dilworth D.J., Davey M.J., O’Donnell M., Aitchison J.D., Rout M.P., Chait B.T., Davey M.J., O’Donnell M., Aitchison J.D., Rout M.P., Chait B.T., O’Donnell M., Aitchison J.D., Rout M.P., Chait B.T., Aitchison J.D., Rout M.P., Chait B.T., Rout M.P., Chait B.T., Chait B.T. Proteomic and genomic characterization of chromatin complexes at a boundary. J. Cell Biol. 2005;169:35–47. doi: 10.1083/jcb.200502104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagami H., Ray-Gallet D., Almouzni G., Nakatani Y., Ray-Gallet D., Almouzni G., Nakatani Y., Almouzni G., Nakatani Y., Nakatani Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004;116:51–61. doi: 10.1016/s0092-8674(03)01064-x. [DOI] [PubMed] [Google Scholar]
- Takeda K., Yanagida M., Yanagida M. Regulation of nuclear proteasome by Rhp6/Ubc2 through ubiquitination and destruction of the sensor and anchor Cut8. Cell. 2005;122:393–405. doi: 10.1016/j.cell.2005.05.023. [DOI] [PubMed] [Google Scholar]
- Tanny J.C., Kirkpatrick D.S., Gerber S.A., Gygi S.P., Moazed D., Kirkpatrick D.S., Gerber S.A., Gygi S.P., Moazed D., Gerber S.A., Gygi S.P., Moazed D., Gygi S.P., Moazed D., Moazed D. Budding yeast silencing complexes and regulation of Sir2 activity by protein-protein interactions. Mol. Cell. Biol. 2004;24:6931–6946. doi: 10.1128/MCB.24.16.6931-6946.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taverna S.D., Ilin S., Rogers R.S., Tanny J.C., Lavender H., Li H., Baker L., Boyle J., Blair L.P., Chait B.T., Ilin S., Rogers R.S., Tanny J.C., Lavender H., Li H., Baker L., Boyle J., Blair L.P., Chait B.T., Rogers R.S., Tanny J.C., Lavender H., Li H., Baker L., Boyle J., Blair L.P., Chait B.T., Tanny J.C., Lavender H., Li H., Baker L., Boyle J., Blair L.P., Chait B.T., Lavender H., Li H., Baker L., Boyle J., Blair L.P., Chait B.T., Li H., Baker L., Boyle J., Blair L.P., Chait B.T., Baker L., Boyle J., Blair L.P., Chait B.T., Boyle J., Blair L.P., Chait B.T., Blair L.P., Chait B.T., Chait B.T., et al. Yng1 PHD finger binding to H3 trimethylated at K4 promotes NuA3 HAT activity at K14 of H3 and transcription at a subset of targeted ORFs. Mol. Cell. 2006;24:785–796. doi: 10.1016/j.molcel.2006.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White C.L., Suto R.K., Luger K., Suto R.K., Luger K., Luger K. Structure of the yeast nucleosome core particle reveals fundamental changes in internucleosome interactions. EMBO J. 2001;20:5207–5218. doi: 10.1093/emboj/20.18.5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood A., Krogan N.J., Dover J., Schneider J., Heidt J., Boateng M.A., Dean K., Golshani A., Zhang Y., Greenblatt J.F., Krogan N.J., Dover J., Schneider J., Heidt J., Boateng M.A., Dean K., Golshani A., Zhang Y., Greenblatt J.F., Dover J., Schneider J., Heidt J., Boateng M.A., Dean K., Golshani A., Zhang Y., Greenblatt J.F., Schneider J., Heidt J., Boateng M.A., Dean K., Golshani A., Zhang Y., Greenblatt J.F., Heidt J., Boateng M.A., Dean K., Golshani A., Zhang Y., Greenblatt J.F., Boateng M.A., Dean K., Golshani A., Zhang Y., Greenblatt J.F., Dean K., Golshani A., Zhang Y., Greenblatt J.F., Golshani A., Zhang Y., Greenblatt J.F., Zhang Y., Greenblatt J.F., Greenblatt J.F., et al. Bre1, an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter. Mol. Cell. 2003;11:267–274. doi: 10.1016/s1097-2765(02)00802-x. [DOI] [PubMed] [Google Scholar]
- Wood A., Schneider J., Dover J., Johnston M., Shilatifard A., Schneider J., Dover J., Johnston M., Shilatifard A., Dover J., Johnston M., Shilatifard A., Johnston M., Shilatifard A., Shilatifard A. The Bur1/Bur2 complex is required for histone H2B monoubiquitination by Rad6/Bre1 and histone methylation by COMPASS. Mol. Cell. 2005;20:589–599. doi: 10.1016/j.molcel.2005.09.010. [DOI] [PubMed] [Google Scholar]
- Workman J.L. Nucleosome displacement in transcription. Genes & Dev. 2006;20:2009–2017. doi: 10.1101/gad.1435706. [DOI] [PubMed] [Google Scholar]
- Xiao T., Kao C.F., Krogan N.J., Sun Z.W., Greenblatt J.F., Osley M.A., Strahl B.D., Kao C.F., Krogan N.J., Sun Z.W., Greenblatt J.F., Osley M.A., Strahl B.D., Krogan N.J., Sun Z.W., Greenblatt J.F., Osley M.A., Strahl B.D., Sun Z.W., Greenblatt J.F., Osley M.A., Strahl B.D., Greenblatt J.F., Osley M.A., Strahl B.D., Osley M.A., Strahl B.D., Strahl B.D. Histone H2B ubiquitylation is associated with elongating RNA polymerase II. Mol. Cell. Biol. 2005;25:637–651. doi: 10.1128/MCB.25.2.637-651.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan G.C., Liu Y.J., Dion M.F., Slack M.D., Wu L.F., Altschuler S.J., Rando O.J., Liu Y.J., Dion M.F., Slack M.D., Wu L.F., Altschuler S.J., Rando O.J., Dion M.F., Slack M.D., Wu L.F., Altschuler S.J., Rando O.J., Slack M.D., Wu L.F., Altschuler S.J., Rando O.J., Wu L.F., Altschuler S.J., Rando O.J., Altschuler S.J., Rando O.J., Rando O.J. Genome-scale identification of nucleosome positions in S. cerevisiae. Science. 2005;309:626–630. doi: 10.1126/science.1112178. [DOI] [PubMed] [Google Scholar]
- Zhang H., Roberts D.N., Cairns B.R., Roberts D.N., Cairns B.R., Cairns B.R. Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell. 2005;123:219–231. doi: 10.1016/j.cell.2005.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Kolaczkowska A., Devaux F., Panwar S.L., Hallstrom T.C., Jacq C., Moye-Rowley W.S., Kolaczkowska A., Devaux F., Panwar S.L., Hallstrom T.C., Jacq C., Moye-Rowley W.S., Devaux F., Panwar S.L., Hallstrom T.C., Jacq C., Moye-Rowley W.S., Panwar S.L., Hallstrom T.C., Jacq C., Moye-Rowley W.S., Hallstrom T.C., Jacq C., Moye-Rowley W.S., Jacq C., Moye-Rowley W.S., Moye-Rowley W.S. Transcriptional regulation by Lge1p requires a function independent of its role in histone H2B ubiquitination. J. Biol. Chem. 2005;280:2759–2770. doi: 10.1074/jbc.M408333200. [DOI] [PubMed] [Google Scholar]
- Zhu B., Zheng Y., Pham A.D., Mandal S.S., Erdjument-Bromage H., Tempst P., Reinberg D., Zheng Y., Pham A.D., Mandal S.S., Erdjument-Bromage H., Tempst P., Reinberg D., Pham A.D., Mandal S.S., Erdjument-Bromage H., Tempst P., Reinberg D., Mandal S.S., Erdjument-Bromage H., Tempst P., Reinberg D., Erdjument-Bromage H., Tempst P., Reinberg D., Tempst P., Reinberg D., Reinberg D. Monoubiquitination of human histone H2B: The factors involved and their roles in HOX gene regulation. Mol. Cell. 2005;20:601–611. doi: 10.1016/j.molcel.2005.09.025. [DOI] [PubMed] [Google Scholar]
- Zilahi E., Miklos I., Sipiczki M., Miklos I., Sipiczki M., Sipiczki M. The Schizosaccharomyces pombe sep15+ gene encodes a protein homologous to the Med8 subunit of the Saccharomyces cerevisiae transcriptional mediator complex. Curr. Genet. 2000;38:227–232. doi: 10.1007/s002940000158. [DOI] [PubMed] [Google Scholar]