Abstract
In response to DNA damage, p53 undergoes post-translational modifications (including acetylation) that are critical for its transcriptional activity. However, the mechanism by which p53 acetylation is regulated is still unclear. Here, we describe an essential role for HLA-B-associated transcript 3 (Bat3)/Scythe in controlling the acetylation of p53 required for DNA damage responses. Depletion of Bat3 from human and mouse cells markedly impairs p53-mediated transactivation of its target genes Puma and p21. Although DNA damage-induced phosphorylation, stabilization, and nuclear accumulation of p53 are not significantly affected by Bat3 depletion, p53 acetylation is almost completely abolished. Bat3 forms a complex with p300, and an increased amount of Bat3 enhances the recruitment of p53 to p300 and facilitates subsequent p53 acetylation. In contrast, Bat3-depleted cells show reduced p53–p300 complex formation and decreased p53 acetylation. Furthermore, consistent with our in vitro findings, thymocytes from Bat3-deficient mice exhibit reduced induction of puma and p21, and are resistant to DNA damage-induced apoptosis in vivo. Our data indicate that Bat3 is a novel and essential regulator of p53-mediated responses to genotoxic stress, and that Bat3 controls DNA damage-induced acetylation of p53.
Keywords: Bat3/Scythe, p53, p300, acetylation, apoptosis, gene targeting
Bat3 (HLA-B-associated transcript 3) was originally identified as a gene within the class III region of the human major histocompatibility complex on chromosome 6 (Spies et al. 1989; Banerji et al. 1990). The biological function of Bat3 is currently unknown. The Xenopus homolog of Bat3, Scythe, was recently shown to interact with Reaper (Rpr), a crucial regulator of apoptosis in Drosophila melanogaster (Thress et al. 1998). The Rpr– Scythe interaction is essential for Rpr-induced apoptosis in a Xenopus egg cell-free system and results in features typical of apoptosis, such as mitochondrial cytochrome c release, caspase activation, and nuclear fragmentation (Thress et al. 1999). Structurally, Bat3/Scythe contains an N-terminal ubiquitin homology region, short tracts of polyproline and polyglycine, and a zinc finger-like domain; however, the functional implications of these domains are unknown (Banerji et al. 1990). Bat3/Scythe also contains a nuclear localization signal (NLS) at its C terminus (CT) (Manchen and Hubberstey 2001). However, a recent report has shown that the majority of endogenous Bat3 can be found in the cytosolic fraction in multiple mouse primary tissues (Kislinger et al. 2006). Certain sequence domains in Bat3/Scythe also appear in the BAG family of proteins that negatively regulate the chaperone function of the Hsp70 family. Thus, Bat3/Scythe may be important for the folding and activity of signaling molecules regulating apoptosis (Thress et al. 2001). Bat3-deficient mice are embryonic lethal with defects in the development of the brain, lung, and kidney (Desmots et al. 2005), and primary neuronal cells deficient for Bat3 are resistant to in vitro apoptosis induced by calcium overloading in the endoplasmic reticulum (ER) (Desmots et al. 2005). Taken together, these data suggest that Bat3 may regulate both proliferation and cell death.
The tumor suppressor protein p53 is a key guardian of genomic stability (Lane 1992). In response to a variety of cellular stresses, p53 governs critical cell fate decisions resulting in cell cycle arrest, DNA repair, senescence, or apoptosis (Levine 1997; Vogelstein et al. 2000; Vousden and Lu 2002). The ability of p53 to inhibit the cell cycle or promote apoptosis depends largely on p53’s ability to control the transcription of target genes regulating cell proliferation and apoptosis. Such genes include p21/WAF1, Gadd45, Noxa, Puma, Bax, p53AIP1, and survivin (Levine 1997; Prives and Hall 1999; Hoffman et al. 2002; Vousden and Lu 2002). The importance of p53’s transcriptional activity to its tumor suppressor function is underscored by the fact that >97% of mutations in human cancers are located within the p53 DNA-binding domain (Olivier et al. 2002).
In healthy cells, p53 is maintained at a low level through interaction with proteins such as Mdm2/Hdm2 (human Mdm2) (Iwakuma and Lozano 2003), COP1 (Dornan et al. 2004), Pirh2 (Leng et al. 2003), and ARF-BP1 (Chen et al. 2005) because these molecules promote ubiquitination and proteasome-mediated degradation of p53 (Haupt et al. 1997; Honda et al. 1997; Kubbutat et al. 1997). In response to stress, p53 becomes stabilized through post-translational modifications, including phosphorylation and acetylation (Appella and Anderson 2001; Brooks and Gu 2003), and performs its biological functions. DNA damage-induced phosphorylation by ATM, Chk2, DNA-PK, or ATR of serine and threonine residues within the p53 N terminus prevents Mdm2 binding and subsequent ubiquitin-mediated degradation of p53 (Shieh et al. 1997; Tibbetts et al. 1999; Hirao et al. 2000). Acetylation of the CT of p53 is carried out by histone acetyltransferases (HATs) such as p300/CREB-binding protein (CBP) and p300/CBP-associated factor (PCAF) (Gu and Roeder 1997; Sakaguchi et al. 1998). In addition to stabilizing p53 by diverting it from Mdm2-mediated ubiquitination (Li et al. 2002), there is some evidence that acetylation of p53 regulates its sequence-specific DNA binding and enhances its transcriptional activity (Gu and Roeder 1997; Sakaguchi et al. 1998; Luo et al. 2004), although controversy still exists (Espinosa and Emerson 2001). Others have reported that p53 acetylation promotes transcription by facilitating the recruitment of cofactors to the promoters of p53-responsive genes (Avantaggiati et al. 1997; Gu and Roeder 1997; Lill et al. 1997; Ito et al. 1999; Barlev et al. 2001; Espinosa and Emerson 2001). The complex and dynamic patterns of post-translational p53 modifications have made this issue difficult to resolve. An understanding of how p53 acetylation is regulated will be essential for defining its precise biological function.
In this study, we have identified a critical requirement for Bat3 in regulating p53 transcriptional activity. Ablation of Bat3 results in reduced apoptosis in response to genotoxic stress. In the absence of Bat3, p300 cannot bind to p53, and p300-mediated acetylation of p53 is almost completely abolished. Consequently, the induction of p53 target genes such as p21 and Puma is severely impaired. Our results demonstrate that Bat3 modulates p300-mediated p53 acetylation and thus functions as a novel regulator of the genotoxic stress response.
Results
Bat3 is necessary for DNA damage-induced apoptosis
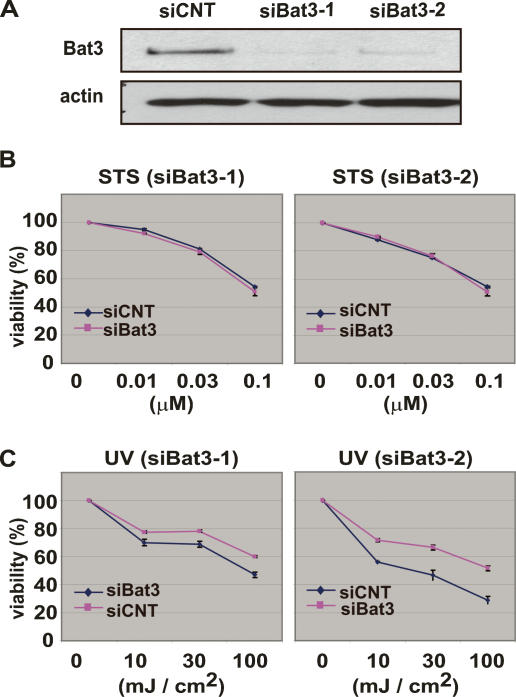
Because homologs of Bat3 have been implicated in apoptotic regulation in other organisms, we determined whether Bat3 deficiency affected the in vitro apoptosis of mammalian cells using “knockdown” (KD) technology. Two independent small interfering RNA (siRNA) oligoduplexes specific for human Bat3 (siBat3-1 and siBat3-2) and a control (siCNT) were introduced into the human osteosacroma cell line U2OS to verify their efficiency. After 48 h, the level of Bat3 protein was reduced to <10% of the control in U2OS cells transfected with siBat3-1 or Bat3-2, whereas introduction of siCNT had no effect on Bat3 protein levels (Fig. 1A). We then transfected siBat3 and siCNT into U2OS cells and induced apoptosis 72 h later by treating the cells with staurosporine (STS; 0, 0.01, 0.03, or 0.1 μM) or UV (0, 10, 30, or 100 mJ/cm2). At 16 h post-treatment, annexin V-FITC/PI staining showed that STS treatment induced apoptosis in siCNT-, siBat3-1-, and siBat3-2-transfected cells to the same extent (Fig. 1B). In contrast, there was a 10%–20% reduction in UV-induced apoptosis in siBat3-1- and siBat3-2-transfected cells compared with siCNT-transfected cells (Fig. 1C). These data suggest that Bat3 plays a regulatory role in DNA damage-induced apoptosis.
Figure 1.
Bat3 is necessary for DNA damage response. (A) Efficient Bat3 KD by siRNA oligo-duplexes. U2OS cells were transfected with control (siCNT) or Bat3 (siBat3-1 and siBat3-2) RNA oligo-duplexes, and Bat3 protein levels were assessed 48 h later by immunoblotting. (actin) Loading control. (B,C) Reduced DNA damage-induced apoptosis. The cells in A were treated with staurosporine (STS) (B) or UV (C) as indicated. Percentage of cell viability was assayed at 16 h post-treatment by annexin-V staining. Data shown are one trial representative of four independent experiments.
Bat3 is required for p53 transcriptional activity
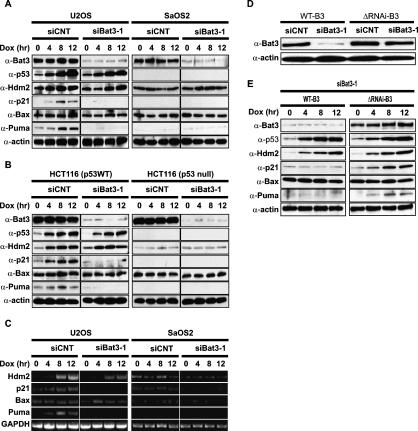
Given the central role played by p53 in DNA damage-induced apoptosis, we next investigated the relationship between Bat3 and the p53 signaling pathway. We first examined the protein levels of p53 and its transcriptional target genes in doxorubicin (Dox)-treated U2OS (p53 wild-type) and SaOS2 (p53-null) cells that were transfected with siCNT or siBat3-1. In U2OS cells transfected with siCNT or siBat3-1, p53 was stabilized such that its protein levels were increased (Fig. 2A, left). However, among the p53 target genes tested, the induction of the p21 and Puma proteins was markedly impaired in siBat3-transfected U2OS cells compared with the normal induction observed in siCNT-transfected U2OS cells (Fig. 2A, left). In contrast, protein levels of the p53 target gene Hdm2 and Bax were comparable in siCNT- and siBat3-transfected U2OS cells (Fig. 2A, left). The induction of p21, Puma, and Hdm2 proteins observed in U2OS cells was p53-dependent since no induction of these proteins was observed in siCNT- or siBat3-transfected SaOS2 cells (Fig. 2A, right). We confirmed this finding in another set of cell lines, p53 wild-type and p53-null HCT116 cells (Bunz et al. 1998). We depleted Bat3 and treated the cells with Dox. The induction of p21 and Puma was severely impaired in HCT116 cells depleted for Bat3 in a p53-dependent manner (Fig. 2B). The protein levels of Hdm2 and Bax were comparable in siCNT- and siBat3-transfected HCT116 cells (Fig. 2B). These data are consistent with our observations in U2OS and SaOS2 cells.
Figure 2.
Impaired induction of p53 target genes in Bat3-depleted cells. (A,B) Impaired protein expression of p53 target genes. U2OS and SaOS2 cells (A) and p53 wild-type and p53-null HCT116 cells (B) were transfected with siCNT or siBat3-1 RNA oligo-duplexes and treated with Dox (0.4 μg/mL) for the indicated times. Protein levels of p53, Hdm2, p21, Bax, and Puma were determined by Western blotting. (C) Impaired p53 target gene transcription. U2OS and SaOS2 cells were transfected and treated as in A, and levels of Hdm2, p21, Bax, and Puma transcripts were determined by semiquantitative RT–PCR analysis. (GAPDH) Loading control. (D) Establishment of a Bat3 mutant resistant to RNAi. U2OS cells subjected to Bat3 KD with siBat3-1 RNA oligo-duplexes were transfected with WT-B3 and ΔRNAi-B3. Protein levels of Bat3 were determined as in A. (E) Rescue of the defective induction of p53 target genes by introducing ΔRNAi-B3. U2OS cells subjected to Bat3 KD with siBat3-1 RNA oligo-duplexes were transfected with WT-B3 and ΔRNAi-B3. The cells were then treated with Dox (0.4 μg/mL) for the indicated times. Protein levels of p53 and its targets were determined as in A.
Next, we examined the transcription of p53 target genes using semiquantitative RT–PCR. We found that levels of p21 and Puma transcripts were severely decreased in siBat3-transfected U2OS cells (Fig. 2C, left), consistent with a defect in protein expression. Induction of Hdm2 transcription was modestly impaired (Fig. 2C, left). However, Bax transcription was not decreased in the absence of Bat3 in this system (Fig. 2C).
To further show that the observed defects in p53 transcriptional activity were indeed caused by specific depletion of Bat3 protein and not by off-target effects, we tested whether the phenotype could be rescued by a Bat3 mutant containing two silent point mutations within the siBat3-1 recognition site (ΔRNAi-B3). We first confirmed that ectopically expressed wild-type Bat3 (WT-B3) was depleted by siBat3-1 transfection, whereas ΔRNAi-B3 was resistant (Fig. 2D). Then, we examined protein levels of p53 and p53 target genes in Dox-treated U2OS cells transfected with siBat3-1 plus either WT-B3 or ΔRNAi-B3. WT-B3 could not rescue defective induction of p21 and Puma (Fig. 2E). In contrast, ΔRNAi-B3 restored the induction of p21 and Puma (Fig. 2E). Taken together, these data suggest that Bat3 influences p53 transactivation and thus is essential for the induction of a certain subset of p53 target genes.
Bat3 plays a role in DNA damage-induced p53 acetylation
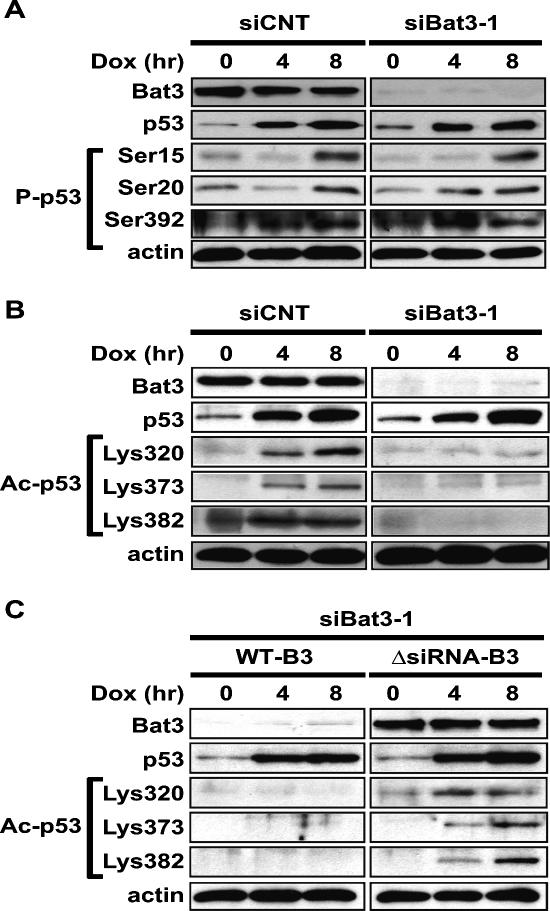
In Dox-treated siBat3-transfected U2OS cells, p53 protein was stabilized (Fig. 2A,B) but p53 transactivation was greatly reduced (Fig. 2C). We therefore speculated that the Bat3 depletion might be affecting the post-translational modification of p53, since most such modifications are known to modulate p53 transcriptional activity (Appella and Anderson 2001; Brooks and Gu 2003). We first examined the phosphorylation of serine (Ser) residues 15, 20, and 392 of human p53, which are known to be phosphorylated in response to DNA damage. U2OS cells transfected with siCNT or siBat3 were treated with Dox for 0, 4, or 8 h, and the phosphorylated forms of Ser15, Ser20, and Ser392 were detected using specific antibodies. Comparable levels of phosphorylation of all three serine residues were detected in siCNT- and siBat3-transfected cells (Fig. 3A), strongly suggesting that Bat3 is not involved in p53 phosphorylation. Next, we examined acetylation of p53. U2OS cells transfected with siCNT or siBat3 were treated with Dox in the presence of the histone deacetylase inhibitor Tricostatin A (TSA), which enhances the proportion of acetylated p53 (Sakaguchi et al. 1998). In siCNT-transfected cells, acetylation of lysine (Lys) residues 320, 373, and 382 of human p53 was readily observed at 4 h post-Dox treatment and had increased by 8 h. In Bat3-depleted cells, acetylation of these lysine residues was severely impaired (Fig. 3B).
Figure 3.
Bat3 is necessary for DNA damage-induced p53 acetylation. (A) Normal p53 phosphorylation. U2OS cells were transfected with siCNT or siBat3 RNA oligo-duplexes and treated 48 h later with Dox (0.4 μg/mL). Protein samples were prepared at the indicated time points post-Dox. Phosphorylation of p53 on Ser15, Ser20, and Ser392 (P-p53) was detected using phospho-specific antibodies. (B) Impaired DNA damage-induced p53 acetylation. U2OS cells were transfected and treated as in A. Extracts were immunoprecipitated with anti-p53 antibody (DO-1) and immunoblotted with antibodies specific for acetylated Lys320, Lys373, and Lys382 (Ac-p53). (C) Rescue of the impaired DNA damage-induced p53 acetylation by the ΔRNAi-B3. U2OS cells were subjected to Bat3 KD and 24 h later they were transfected with WT-B3 and ΔRNAi-B3. The cells were then treated with Dox as in A. Protein levels of acetylated p53 were examined as in B. Data shown are representative of four independent preparations.
Next, to exclude potential off-target effects of siBat3-1, we examined whether the introduction of ΔRNAi-B3 could restore Dox-induced p53 acetylation in the siBat3-1-transfected cells. Introduction of ΔRNAi-B3 restored Dox-induced p53 acetylation (Fig. 3C, right), but WT-B3 did not (Fig. 3C, left). These data indicate that Bat3 is necessary for the acetylation of p53 induced by DNA damage.
Bat3 binds to p300 and enhances p300-mediated p53 acetylation
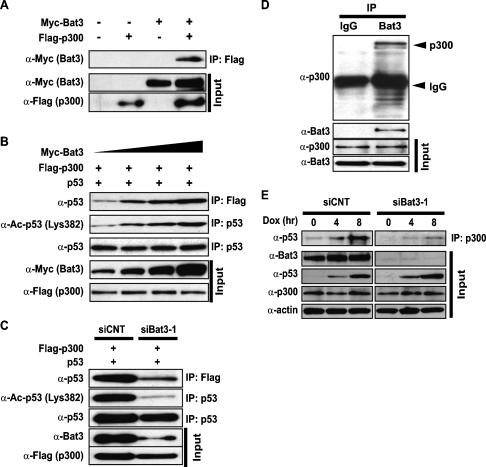
DNA damage-induced acetylation of p53 Lys320 in vitro requires the cofactor PCAF, whereas the acetylation of p53 Lys373/Lys382 depends on p300/CBP (Avantaggiati et al. 1997; Gu and Roeder 1997; Lill et al. 1997; Sakaguchi et al. 1998). To test whether Bat3 interacts with these cofactors, we cotransfected 293T cells with Myc-Bat3 plus Flag-p300, or Myc-Bat3 plus HA-PCAF. Immunoblot analysis revealed an interaction between Myc-Bat3 and Flag-p300 (Fig. 4A), but interaction between Myc-Bat3 and HA-PCAF was not detectable (data not shown). We then investigated whether Bat3 had a positive regulatory effect on p300–p53 interaction or p53 acetylation, or both. We cotransfected H1299 cells with p53, Flag-p300, and increasing amounts of Myc-Bat3. We then immunoprecipitated protein extracts with either anti-Flag or anti-p53 antibodies. Immunoblotting with anti-p53 or anti-Ac-p53 (Lys382) antibodies showed that the amount of p53 associated with Flag-p300 increased with increasing Myc-Bat3, as did the amount of acetylated p53 (Fig. 4B). When H1299 cells were subjected to Bat3 KD followed by cotransfection with p53 and Flag-p300, both the amount of p53 associated with Flag-p300 and p53 acetylation were dramatically decreased (Fig. 4C).
Figure 4.
Bat3 binds to p300 and enhances p300-mediated p53 acetylation. (A) Binding of Bat3 to p300. 293T cells were transfected with empty vector (−), Flag-p300, Myc-Bat3, or Flag-p300 plus Myc-Bat3 as indicated. Extracts prepared at 48 h post-transfection were immunoprecipitated using beads conjugated to anti-Flag antibody. The immunoprecipitated samples were analyzed by Western blotting using anti-Myc antibody. (B) Bat3-mediated enhancement of p53–p300 binding and p53 acetylation. H1299 cells were transfected with expression vectors for p53 (0.3 μg), p300-Flag (0.3 μg), and Bat3-Myc (0.1, 0.2, 0.3, or 0.6 μg). The total amount of DNA transfected was equalized by adding empty vector. At 48 h post-transfection, extracts were immunoprecipitated as in A, followed by immunoblotting with anti-p53 antibody (polyclonal). The extracts were also immunoprecipitated with anti-p53 antibody (DO-1), followed by immunoblotting with anti-acetylated p53 (Lys382). (C) Suppression of p53–p300 binding and p53 acetylation by Bat3 KD. H1299 cells were transfected with control or Bat3-specific siRNA oligo-duplexes and cotransfected 24 h later with Flag-p300 (0.6 μg) and p53 (0.6 μg). After another 24 h, extracts were immunoprecipitated to examine p53–p300 interaction and p53 acetylation as in B. (D) Binding of endogenous Bat3 and p300. Extract prepared from U2OS cells was immunoprecipitated using anti-Bat3 antibody or control IgG. The immunoprecipitated samples were analyzed by Western blotting using anti-p300 antibody. (E) Reduced interaction of endogenous p53 and p300 following Bat3 KD. U2OS cells were transfected with control or Bat3-specific siRNA oligo-duplexes and treated 48 h later with Dox (0.4 μg/mL) for the indicated times. Extracts were immunoprecipitated with anti-p300 antibody followed by immunoblotting with anti-p53 antibody. For A–E, “Input” represents the protein levels of the indicated molecules in the original extracts as determined by conventional Western blotting.
We next confirmed this finding for endogenous p53 and p300. Immunoprecipitation of protein extracts with anti-Bat3 antibody and subsequent immunoblot analysis with anti-p300 antibody revealed the interaction of the two endogenous proteins (Fig. 4D). Then, U2OS cells were subjected to Bat3 KD followed by Dox treatment. Immunoblotting of protein extracts at 0, 4, or 8 h post-Dox using anti-p300 antibody showed that the amount of p53 interacting with p300 was significantly reduced in siBat3-transfected cells (Fig. 4E). These data indicate that, in response to DNA damage, Bat3 positively regulates p53–p300 interaction and the subsequent acetylation of p53.
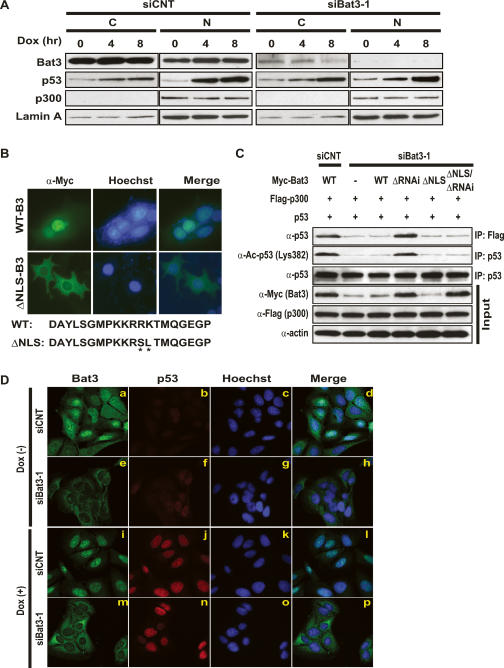
Nuclear Bat3 is necessary for p53 acetylation
Because p300 is a nuclear protein and endogenous Bat3 in primary tissues has been reported to be in the cytosol (Kislinger et al. 2006), we investigated whether nuclear localization of Bat3 might be essential for its interaction with p300 and its effect on p53 acetylation. We prepared nuclear and cytosolic fractions from U2OS cells that had been transfected with either siCNT or siBat3 and treated with Dox. We found that p53 was stabilized in the presence of Dox, and that the majority of the p53 protein was localized in the nucleus in both siCNT- and siBat3-transfected cells (Fig. 5A). In untreated siCNT-transfected cells (time 0 h), the majority of Bat3 protein was present in the cytosol, and a smaller amount was detected in the nucleus. However, after Dox treatment, Bat3 levels were slightly increased in the nucleus (Fig. 5A, left). In contrast, in Bat3-depleted cells, Bat3 protein was markedly reduced in the cytosol and undetectable in the nucleus (Fig. 5A, right). In both siCNT- and siBat3-transfected cells, p300 was detectable only in the nucleus and p300 protein levels did not change significantly after Dox treatment (Fig. 5A). These data are consistent with the idea that nuclear Bat3 is necessary for p300-mediated p53 acetylation.
Figure 5.
Nuclear localization of Bat3 is necessary for its activity. (A) Biochemical demonstration. U2OS cells transfected with control or Bat3-specific siRNA oligo-duplexes were subjected to Dox (0.4 μg/mL) treatment. After 0, 4, or 8 h, cytosolic (C) and nuclear (N) fractions were prepared from extracts. Protein levels of Bat3, p53, and p300 were determined by immunoblotting. (Lamin A) Loading control for nuclear samples. (B) Establishment of a nuclear-localization-defective mutant of Bat3. U2OS cells transfected with Myc-tagged WT-B3 and ΔNLS-B3 were stained with anti-myc-FITC antibody. Nuclei were visualized using Hoechst 33258 (blue), merge of myc and Hoechst 33258 staining. The point mutations introduced in ΔNLS-B3 are indicated by asterisk. (C) Failure of the enhancement of p53–p300 binding and p53 acetylation by ΔNLS-B3. H1299 cells were transfected with control or Bat3-specific siRNA oligo-duplexes and cotransfected 24 h later with WT-B3, ΔRNAi-B3, ΔNLS-B3, or ΔNLS/ΔRNAi-B3. At 48 h post-transfection, p53–p300 binding and p53 acetylation were examined as in Figure 3. (D) Cellular demonstration. U2OS cells were transfected as in A and treated with Dox for 0 or 8 h, at which times the cells were fixed, permeabilized, and stained with anti-Bat3 (panels a,e,i,m) or anti-p53 (DO-1; panels b,f,j,n) antibodies, followed by anti-rabbit IgG (green) or anti-mouse IgG (red). Nuclei were visualized using Hoechst 33258 (blue; panels c,g,k,o). (Panels d,h,l,p) Merge of Bat3 and Hoechst 33258 staining. Data shown are representative of four independent preparations.
To confirm the importance of nuclear Bat3 for its positive regulatory effects on p300–p53 interaction and p53 acetylation, we have created two additional Bat3 mutants: ΔNLS-B3, which has two point mutations within NLS, and ΔNLS/ΔRNAi-B3, which contains both ΔNLS and ΔRNAi mutations. ΔNLS-B3 localized only in the cytosol (Fig. 5B), whereas WT-B3 localized mostly in the nucleus (Fig. 5B). We then investigated whether WT-B3, ΔRNAi-B3, ΔNLS-B3, and ΔNLS/ΔRNAi-B3 had a positive regulatory effect on p300–p53 interaction, p53 acetylation, or both. We transfected H1299 cells with siBat3-1, followed by cotransfection of p53 and Flag-p300 plus either Myc-WT-B3, Myc-ΔRNAi-B3, Myc-ΔNLS-B3, or Myc-ΔNLS/ΔRNAi-B3. Myc-ΔRNAi-B3 and Myc-ΔNLS/ΔRNAi-B3 were resistant to siBat3-1 and expressed comparable levels of protein (Fig. 5C). We then immunoprecipitated protein extracts with either anti-Flag or anti-p53 antibodies. Immunoblotting with anti-p53 or anti-Ac-p53 (Lys382) antibodies showed that Myc-ΔNLS/ΔRNAi-B3 failed to increase the amount of acetylated p53 and p53 associating with p300 (Fig. 5C). In contrast, Myc-ΔRNAi-B3 enhanced p53–p300 association and p53 acetylation (Fig. 5C). These data provide further support that nuclear Bat3 is required for its positive regulatory effect on p300-mediated p53 acetylation.
To confirm these biochemical findings at the cellular level, we investigated the subcellular localization of p53 and Bat3 in U2OS cells. We transfected U2OS cells with siCNT and siBat3, cultured them for 48 h, and treated them with Dox for 8 h. The cells were then fixed and stained with anti-p53 and anti-Bat3 antibodies, as well as with Hoechst 33258 (to visualize nuclei). In the absence of Dox, there was only trace staining of p53 in both control and Bat3-depleted cells (Fig. 5D, panels b,f). In siCNT-transfected cells, Bat3 was localized in both the cytosol and nucleus (Fig. 5D, panel a). However, in siBat3-transfected cells, Bat3 was undetectable in the nucleus and stained only very weakly in the cytosol (Fig. 5D, panel e). In the presence of Dox, nuclear staining of Bat3 became stronger in siCNT-transfected cells (Fig. 5D, panel i). In siBat3-transfected cells, Bat3 was stained only in the cytosol even after Dox treatment (Fig. 5D, panel m). Dox treatment also induced p53, which accumulated in the nucleus in both siCNT- and siBat3-transfected cells (Fig. 5D, panel j,n). These observations are consistent with our biochemical assays and further suggest that nuclear localization of Bat3 is necessary for p300-mediated p53 acetylation.
Bat3 enhances p300-dependent p53 transcriptional activity
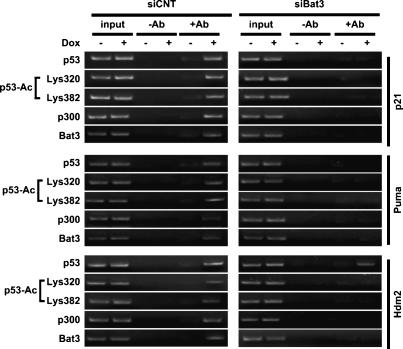
We next examined the effect of Bat3 on the induction of p53 target genes in a physiological setting. We used a chromatin immunoprecipitation (ChIP) assay to determine whether p300, Bat3, acetylated p53, and nonacetylated p53 could bind to the promoters of the endogenous p21, Puma, and Hdm2 genes in U2OS cells transfected with siCNT or siBat3 and treated with Dox. Chromatin was immunoprecipitated with anti-p53, anti-acetyl-p53 (Lys320/Lys380), anti-p300, or anti-Bat3 antibodies and analyzed by PCR using primer pairs specific for the p53-binding region of the p21, Puma, and Hdm2 promoters. In Dox-treated siCNT-transfected cells, p53, acetylated p53, p300, and Bat3 were recruited to the p53-binding site in the p21, Puma, and Hdm2 promoters (Fig. 6, left). In contrast, in Dox-treated Bat3-depleted cells, no antibody was able to precipitate the p53-binding region of p21 and Puma (Fig. 6, right). Interestingly, p53 binding to the Hdm2 promoter occurred in Dox-treated Bat3-depleted cells, although to a lesser extent than in the control (Fig. 6, right). On the other hand, recruitment of acetylated p53 or p300 to the Hdm2 promoter was severely impaired in Bat3-depleted cells (Fig. 6, right). These data are consistent with previous reports describing a functional and physical interaction between p53 acetylation and p300 recruitment that is necessary for binding to p53-responsive elements (p53REs) in p53 target gene promoters (Avantaggiati et al. 1997; Lill et al. 1997; Barlev et al. 2001).
Figure 6.
Bat3 is required for p53-mediated transcription. Impaired recruitment of p53 to the p21, Puma, and Hdm2 promoters in the absence of Bat3. U2OS cells transfected with control or Bat3-specific siRNA oligo-duplexes were treated with Dox (0.4 μg/mL) for 8 h, and extracts were immunoprecipitated with anti-p53, anti-acetylated p53 (Lys382 and 320), anti-p300, or Bat3 antibodies. The p53-binding regions of the p21, Puma, and Hdm2 promoters were amplified by PCR. Data shown are representative of four independent preparations.
Bat3 deficiency impairs genotoxic stress responses in a mouse model
Our in vitro data indicated that Bat3 was involved in p53-mediated responses to genotoxic stress through the regulation of p300-mediated p53 acetylation. To confirm this finding in a mouse model, we took advantage of Bat3 knockout mice independently established in our laboratory (Supplementary Fig. 1). Although an initial report indicated that Bat3-deficient mice are embryonic lethal (Desmots et al. 2005), we found that the lethal phenotype can be partially rescued in the ICR genetic background (Supplementary Table 1).
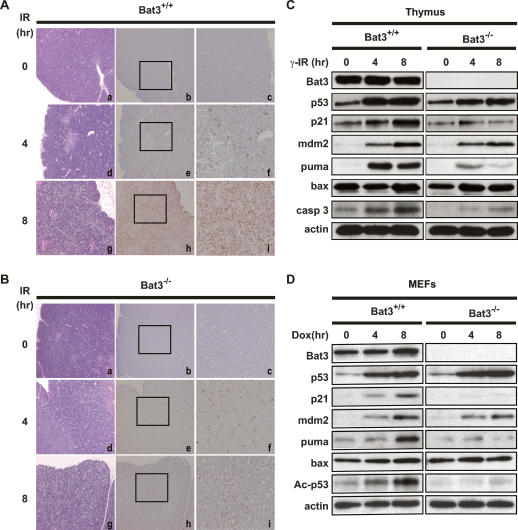
Since puma deficiency is sufficient to suppress DNA damage-induced apoptosis in thymocytes (Jeffers et al. 2003; Villunger et al. 2003), and the induction of Puma was markedly impaired in our Bat3-depleted cells (Fig. 2A–C), we chose the thymus in which to investigate whether Bat3 regulates apoptosis in vivo. Bat3+/+ and Bat3−/− littermate mice were subjected to whole-body γ-irradiation (γ-IR; 10 Gy) and the thymi were removed at 0, 4, or 8 h post γ-IR. Each thymus was cut in half, with one half used for histological analysis and the other half used for biochemical analysis. Bat3−/− mice were slightly smaller in size than their wild-type counterparts (∼80% of wild-type weight at 6–8 wk of age). However, there was no significant difference in total thymocyte number or in subpopulations of CD4/CD8-expressing cells (Supplementary Fig. 2). Bat3 deficiency itself did not induce spontaneous apoptosis or histological abnormalities in the thymus (Fig. 7A [panels a–c], B [panels a–c]). In the wild type, low levels of apoptosis were detected at 4 h post-γ-IR that increased dramatically by 8 h post-γ-IR, consistent with previous reports (Lowe et al. 1993; Erster et al. 2004). Bat3−/− mice showed significantly fewer apoptotic cells at 4 h post-γ-IR than did the wild type (Fig. 7A [panels e, f], B [panels e, f]), and the impairment of γ-IR-induced apoptosis at 8 h post-γ-IR was much more prominent in Bat3-deficient thymus than in the wild type (Fig. 7A [panels h, i], B [panels h, i]).
Figure 7.
Impaired γ-IR-induced apoptosis in Bat3-deficient thymus. (A,B) Histological determination of impaired apoptosis. Six-week-old Bat3+/+ (A) and Bat3−/− (B) littermate mice were left untreated or subjected to 10 Gy whole-body γ-IR. After 4 or 8 h, the thymus was removed and cut in half. One half was fixed and subjected to histological analysis. The other half was used for biochemical analysis, as shown in C. Serial sections of paraffin-embedded thymus were stained with H&E (A-,B, panels -a,d,g) and subjected to in situ end-labeling (IEL) of fragmented DNA (A-,B, panels b,c,e,f,h,i). For each IEL preparation, two magnifications are shown (A-,B, panels b,e,h: 10 × 10; A-,B, panels c,f,i: 10 × 25). Data shown are representative of three independent preparations. (C) Impaired induction of p53 target genes and apoptosis. Half of the thymic lysates prepared in A were analyzed by Western blotting for protein levels of the indicated proteins. (casp 3) Activated caspase-3. (D) Impaired induction of p53 target genes and p53 acetylation in Dox-treated Bat3-deficient MEFs. Protein lysates prepared from Dox-treated Bat3+/+ and Bat3−/− MEFs were analyzed by Western blotting for protein levels of the indicated proteins. Data shown are representative of three independent preparations.
To confirm that this impaired apoptosis was due to a defect in the p53 pathway, we examined the protein levels of p53 and several p53 target genes in the thymi of γ-IR-treated Bat3+/+ and Bat3−/− mice. Consistent with our in vitro findings, p53 was stabilized in both wild-type and Bat3-deficient thymi. However, the induction of puma and p21 protein expression was greatly inhibited in the absence of Bat3 (Fig. 7C). A much less striking difference occurred in the induction of the bax and mdm2 proteins. We also examined the level of activated caspase-3, a conventional marker for apoptosis induction, to confirm our histological findings. Activated caspase-3 was significantly reduced in Bat3-deficient thymus. We also examined the protein levels of p53, acetylated p53 Lys370 (corresponding to human Lys373), and its target genes in Dox-treated Bat3+/+ and Bat3−/− mouse embryonic fibroblasts (MEFs). Consistent with our previous findings, the induction of puma and p21 protein expression as well as p53 acetylation was markedly impaired in Bat3-deficient MEFs (Fig. 7D).
These findings in multiple tissues of the mouse model further support our hypothesis that Bat3 is a critical regulator of p53 function induced by genotoxic stress.
Discussion
In this study, we have defined the positive regulatory role of Bat3 in DNA damage-induced p53 transactivation and have started to unravel the molecular mechanisms underlying its effects. Using biochemical and genetic approaches, we have shown that Bat3 is required for the acetylation of p53 in response to DNA damage.
Bat3 is critical for p300-mediated acetylation of p53
It is well established that p53 is a node of genotoxic stress signaling pathways. However, the precise mechanism of action of p53 has been difficult to ascertain, partly because of the complex and dynamic post-translational modifications undergone by p53. This study demonstrates that Bat3 plays a fundamental role in p300-mediated acetylation of p53 and ultimately regulates DNA damage-induced stress responses in vitro and in vivo (Figs. 1, 7). Bat3 can form a complex with p300 and p53 (Fig. 4; Supplementary Fig. 4), and is recruited to p53RE (Fig. 6). Thus, Bat3 may act as a cofactor to enhance p53 transactivation. In the absence of Bat3, p53 is stabilized (Figs. 2, 7), phosphorylated (Fig. 3), localizes in the nucleus (Fig. 5), and fails to become acetylated on Lys320, Lys373, and Lys382 (Figs. 3, 7). Nonacetylated p53 cannot bind to the p53REs in the promoters of p53 target genes such as p21 and Puma (Fig. 6), and, as a consequence, p21 and Puma cannot be induced (Figs. 2, 7). In addition, binding of nuclear Bat3 to p300 positively regulates p300–p53 binding and p53 acetylation (Figs. 4, 5). Furthermore, since Lys320 and Lys378/Lys382 acetylation are mediated by PCAF and p300/CBP in vitro, respectively, our data suggest that p300 function may be necessary for PCAF-mediated Lys320 acetylation in vivo. Since p300 physically interacts with PCAF (Yang et al. 1996), the recruitment of PCAF by p300 might be required for its activity in vivo. The other possibility is that conformational changes of the p53 CT by p300-mediated acetylation may allow PCAF to access Lys320. Indeed, Lys320 acetylation was significantly impaired when p300 was ablated in U2OS cells (Supplementary Fig. 3). These lines of evidence further support our hypothesis of an important Bat3–p300 connection.
Effects of CT modification on p53-inducible genes
To date, three major mechanisms have been proposed to explain how modifications of the CT of p53 (including phosphorylation, acetylation, CT-specific antibody treatment, and deletion) regulate p53 activity. First, p53 CT modifications may enhance sequence-specific binding to p53REs. Second, p53 CT modifications may accelerate the recruitment of cofactors and alter chromatin structure, thereby promoting transcription. Third, p53 CT modifications may stabilize the p53 protein by inhibiting Hdm2/Mdm2-mediated p53 ubiquitination.
There has been extended debate on how CT modification might regulate sequence-specific DNA binding by p53. In the allosteric model, the unmodified CT is a negative regulator of the p53 DNA-binding domain and maintains the inactive conformation until modification occurs. The CT modification releases the negative effect and allows active p53 to bind to DNA. Consistent with this model, the acetylation of the p53 CT is induced by numerous stress stimuli and significantly increases p53 transcriptional activity (Gu and Roeder 1997; Sakaguchi et al. 1998; Ahn and Prives 2001; Luo et al. 2004). On the other hand, several reports weigh against this model. Since CT-deleted p53 (ΔCp53) is transcriptionally inactive in an artificially reconstituted chromatin assay, the p53 CT may be a positive regulator of p53 DNA binding and transactivation (Espinosa and Emerson 2001). In addition, NMR analysis has shown that wild-type and ΔCp53 are identical in conformation and that the CT does not interact with other p53 domains (Ayed et al. 2001). In addition to this controversy, recent reports have supported the importance of the p53 CT for p53 binding to nonlinear DNA (McKinney and Prives 2002; Fojta et al. 2004; McKinney et al. 2004; Palecek et al. 2004). Finally, CT modification is necessary for efficient promoter activation mediated by p53 (Liu et al. 2004; McKinney et al. 2004). While our model cannot offer a reconciliation of these discrepancies, we propose that CT modifications may regulate both p53 DNA binding and transactivation by modifying either p53 DNA binding or stability of p53–DNA complex, or both.
An inhibition of CT modification would not necessarily have an “all or none” effect on the induction of p53 target genes. The sequence and structure of p53REs, as well as the chromatin structure of the promoters of target genes, would also likely have a great influence. In our model, levels of p21 and Puma induction were severely impaired in the absence of Bat3, but Hdm2/Mdm2 induction was only modestly affected (Figs. 2, 7). These results suggest that at least two sets of p53 target genes may exist: one (including genes such as Puma and p21) that requires p53 acetylation for efficient induction, and the other (including genes like Hdm2/Mdm2) that can be induced to a limited extent by nonacetylated p53. Several studies support this hypothesis. Firstly, ΔCp53 is defective in its induction of p21 and apoptosis (Chen et al. 1996). Secondly, CT modification is necessary for efficient p53 binding to the p21 promoter but not for binding to the Mdm2 promoter, presumably because these two genes have structurally different p53REs (Kaku et al. 2001). Thirdly, ΔCp53 is much less efficient in binding to the p21 promoter than to the Hdm2 promoter, in a way that closely correlates with decreased induction of the target gene (McKinney et al. 2004). Taken together, our findings and these reports suggest that a p53 protein that cannot sustain CT acetylation will bind moderately well to the Hdm2/Mdm2 promoter but hardly at all to the Puma and p21 promoters.
It is also possible that p53 acetylation affects the recruitment of cofactors like TRAP and p300/CBP. TRAP is required for the formation of preinitiation complexes (Ito et al. 1999), while p300/CBP has HATs activity required for the remodeling of chromatin structure (Avantaggiati et al. 1997; Barlev et al. 2001; Espinosa and Emerson 2001). We have shown that, although p300 recruitment to the p21 and Hdm2 promoters was impaired in Bat3-depleted cells, a small amount of p53 binding to the Hdm2 promoter did occur (Fig. 6). Thus, unacetylated p53 appears to be able to induce Hdm2. It also has been reported that the patterns of p53-dependent histone acetylation of the p21, Puma, and Hdm2 promoters differ in H3 and H4. The histone acetylation is increased in both the p21 and Puma promoters in response to p53 activation, whereas there is only a small increase in H4 acetylation and no increase in H3 acetylation in the Hdm2 promoter (Kaeser and Iggo 2004). Thus, the Hdm2 promoter may be activated without the acetylated p53-mediated recruitment of cofactors and HATs and the histone modifications that are essential for p21 and Puma induction. A more precise correlation between p53 target gene promoter sequences and patterns of p53 post-translational modifications is necessary to clarify this issue.
The last major mechanism proposes that acetylation of the p53 CT influences p53 stability, since the CT lysine residues that are targets for acetylation overlap those ubiquitinated by Mdm2 (Rodriguez et al. 2000; Ito et al. 2001). We have shown that impairment of p53 acetylation does not significantly affect the stability of p53 in vitro or in vivo (Figs. 2, 7), implying that the phosphorylation of the p53 N terminus that occurs in response to DNA damage has a dominant inhibitory effect on Hdm2/Mdm2-mediated p53 degradation. Consistent with this notion, a mutant form of p53 in which all CT lysines are substituted with arginines shows virtually normal protein stability in a mouse model (Feng et al. 2005; Krummel et al. 2005).
Bat3 modulates the function of apoptotic regulators
In cell-free extracts prepared from Xenopus eggs, the Bat3 homolog, Scythe, is essential for Rpr-induced apoptosis in that immunodepletion of Scythe impedes Rpr-induced caspase activation. Paradoxically, an excess recombinant Scythe in this system also suppresses Rpr-induced apoptosis (Thress et al. 1999). To explain this, it has been suggested that Scythe sequesters a proapoptotic factor required for mitochondrial cytochrome c release and that Scythe is necessary for the Hsp70-mediated folding and activation of this putative factor. These data are consistent with Scythe being a member of the BAG family of proteins that modulate Hsp70-mediated protein folding. Indeed, although no mammalian homolog of Rpr has been identified to date, Drosophila Rpr and its associated proapoptotic molecules Hid and Grim can induce apoptosis in mammalian cells (McCarthy and Dixit 1998; Goyal et al. 2000). In addition, viral proteins with homology with Rpr have been shown to bind Bat3 and trigger apoptosis (Colon-Ramos et al. 2003). Thus, functional homologs of Rpr (Hid and Grim), a parallel signaling pathway, or both may exist in mammalian systems. We have shown that, in response to DNA damage, a proapoptotic molecule, p53, is activated in a Bat3-dependent manner and induces apoptosis. Although it is not clear how our findings relate Scythe’s role in Rpr-induced apoptosis, it may be that chaperone function of Bat3/Scythe is important for its effects on p53. Nonetheless, we have clearly established that proapoptotic molecules exist as downstream targets of the Bat3 signaling pathway in mammals. Moreover, an absence of Bat3 results in impaired apoptosis induction in vitro and in vivo (Figs. 1, 7).
Bat3 may play a role in apoptosis induced by stimuli other than DNA damage. Ricin, a type II ribosome-inactivating protein, uses its RNA N-glycosidase activity to inhibit protein biosynthesis (Wu et al. 2004). In addition, ricin induces apoptosis by a mechanism that requires caspase-3-mediated cleavage and release of the CT of Bat3 (Wu et al. 2004), suggesting that the Bat3 CT retains proapoptotic activity. This finding is quite intriguing because the 312 CT amino acids of Scythe (C312) also exhibit an autonomous Rpr-independent ability to activate caspases in the egg extract (Thress et al. 1998). However, the precise role of the CT product in the regulation of Bat3-mediated apoptosis is not clear because the cleavage of the CT appears to be stimulus-dependent. For instance, staurosporine treatment does not result in the release of the Bat3 CT (Manchen and Hubberstey 2001). We also could not detect a significant amount of Bat3 cleavage product upon DNA damage (data not shown). However, our results indicate that nuclear Bat3 is essential for the p300–p53 interaction (Fig. 4), and the Bat3/Scythe CT is known to contain a NLS (Manchen and Hubberstey 2001), which is required for nuclear localization of Bat3 (Fig. 5). Thus, the modification of Bat3 CT by caspase-3-mediated cleavage and other mechanisms may have a regulatory effect on Bat3/Scythe function.
Most Bat3-deficient embryos exhibit impaired development of multiple tissues due to a mixture of cell death and proliferation defects (Desmots et al. 2005). This complex phenotype clearly indicates that Bat3’s role in vivo is not straightforward and that regulation of p53 may be only part of Bat3’s functions. Our findings, coupled with previous reports, suggest that the contribution of Bat3/Scythe to apoptotic regulation is highly context-specific (Manchen and Hubberstey 2001; Kumar et al. 2004; Wu et al. 2004; Desmots et al. 2005). For instance, Bat3-deficient primary neuronal cells are resistant to ER stress-induced apoptosis (Desmots et al. 2005), a form of cell death in which p53 acetylation may not be a major factor. Thus, more precise biochemical and genetic analyses to determine tissue-specific functions of Bat3/Scythe will be necessary to fully understand the contribution of this molecule to the regulation of apoptosis and cell proliferation.
In conclusion, our work demonstrates that Bat3 is necessary for p300-mediated p53 acetylation, and functions as a novel positive regulator of p53-mediated apoptosis induced by genotoxic stress in vivo and in vitro. Since dysregulation of the p300–p53 pathway has been observed in multiple cancers (Goodman and Smolik 2000) and a biallelic inactivating mutation of Bat3 has been found in a colon cancer cell line (Ivanov et al. 2006), it will be important to investigate whether mutations of Bat3 are associated with the human diseases.
Materials and methods
Cell lines, transfections, and RNA interference (RNAi)
U2OS (p53 wild-type), SaOS2 (p53-null), and H1299 (p53-null) cells were all originally obtained from the American Tissue Culture Collection (ATCC). HCT116 cells (p53 wild-type and p53-null) were a generous gift from Dr. Bert Vogelstein (Johns Hopkins Kimmel Cancer Center, Baltimore, MD). Synthetic siRNA oligo-duplexes for human Bat3 and human p300 were purchased from Dharmacon and Santa Cruz Biotechnology, respectively (sequences for siRNA oligo-duplexes are in the Supplemental Material). Cells were cultured to 60% confluency in Opti-MEM medium (Invitrogen) and transfected with siRNA oligo-duplexes (final concentration, 40 nM) plus 3 μL of Lipofectamine 2000 (Invitrogen). At 6 h post-transfection, the medium was changed to complete medium. Transfected cells were used for assays at 48 h post-transfection unless otherwise indicated. All control samples were transfected with control siRNA oligo-duplex (Dharmacon).
Apoptosis
U2OS cells transfected with Bat3-specific (siBat3-1 and siBat3-2) or control siRNA oligo-duplexes were plated on six-well plates. The cells were washed twice with prewarmed PBS (Mg- and Ca-free) and treated with UV (0, 10, 30, or 100 mJ/cm2; Strata Cross Linker) or staurosporine (0, 0.01, 0.03, or 1 μM; Sigma-Aldrich). Treated cells were trypsinized after 16 h, washed in PBS, and resuspended in 100 μL of 1× binding buffer (10 mM HEPES, 140 mM NaCl, 2.5 mM CaCl2 at pH 7.4) containing annexin V-FITC (final concentration, 25 μg/mL; Invitrogen) plus propidium iodide (PI; final concentration, 10 μg/mL; Sigma-Aldrich). Samples were incubated for 15 min at room temperature with gentle agitation in the dark. An additional 300 μL of 1× binding buffer were added to each sample prior to flow cytometric analysis using a FACScalibur (Becton Dickinson) as previously described (Okada et al. 2002). Data were analyzed using CellQuest software.
Antibody production
For production of anti-human Bat3 antibody (Ab), human Bat3 DNA was subcloned into the pFastBac-HT vector (Invitrogen). The 6× His-tagged Bat3 fusion protein was expressed in Spodoptera frugiperda Sf-9 cells using the BAC-TO BAC Baculovirus Expression System according to the manufacturer’s instructions (Invitrogen) and purified over a Ni-NTA column (Qiagen).
Polyclonal anti-mouse Bat3 Ab was generated by a custom antiserum production service (SigmaGenosys) using a 6× His-tagged recombinant protein corresponding to amino acids 1–160 of mouse Bat3. The bacterially produced recombinant protein was purified using a Ni-NTA column (Qiagen) according to the manufacturer’s instructions. Rabbits were immunized with the recombinant proteins.
Western blot analysis
Protein samples were prepared in lysis buffer (0.1% SDS, 0.1% DOC, 1% Triton X-100, 20 mM Tris-HCl at pH 7.5, 150 mM NaCl, 1 mM EDTA, 10 mM NaF, 1 mM NaVO4, protease inhibitor cocktail [Roche Diagnostics]) and 1 μM tricostatin A (Sigma-Aldrich), and 20 μg were immunoblotted according to standard procedures. Abs recognizing the following proteins were used in this study: p53 (DO-1), p300 (N-15, C-20), MDM2 (C-18), Bax (N-20), and Lamin A, all from SantaCruz Biotechnology; phospho-p53 (Ser15, Ser20, and Ser392) and p53 polyclonal Ab, both from Cell Signaling Technology; acetylated p53 (Lys320 and Lys373) from Upstate Biotechnology; acetylated p53 (Lys382) from Cell Signaling; PUMA from ProSci; p21 and active caspase-3 from BD Pharmingen; Flag (M2) and actin, both from Sigma. Subcellular fractionation was performed as previously described (Andrews and Faller 1991).
Immunofluorescent microscopy
Cells were fixed in cold methanol for 15 min and blocked in 2.0% BSA, 0.2% Triton X-100, and PBS for 1 h. Slides were incubated with mouse anti-p53 and rabbit anti-Bat3 as primary antibodies followed by AlexaFluor488 goat anti-rabbit IgG and AlexaFluor594 goat anti-mouse IgG (Molecular Probes) as secondary antibodies. Hoechst 33258 (Molecular Probes) was used to stain DNA. Images were acquired on a Zeiss LSM510 confocal microscope (Jena) using a 63×/1.2 NA water immersion objective. The three fluorophores DAPI, Alexa 488, and Alexa 594 were imaged sequentially using 351-, 488-, and 543-nm laser excitation and appropriate band-pass emission filters.
Imunoprecipitations
293T cells were transfected with empty vector (pcDNA3; Invitrogen), Flag-300 pcDNA3, or Myc-Bat3 pcDNA3. Cells were lysed in lysis buffer (50 mM Tris HCl, 150 mM NaCl, 1% NP40) supplemented with protease inhibitor cocktail (Roche Diagnostics) for 30 min on ice and cleared by centrifugation. Extracts were incubated with the appropriate antibodies for 16 h at 4°C with rotation and incubated for an additional 2–3 h with protein A/G agarose beads (SantaCruz Biotechnology). In some experiments, anti-Myc agarose beads (SantaCruz Biotechnology) were used to pull down Myc-tagged proteins. The samples were fractionated by SDS-PAGE and subjected to immunoblotting as described above.
RT–PCR analysis
Total RNA was extracted from cells using TRIzol (Life Technologies). First-strand DNA was synthesized using the SuperScript First-Strand Synthesis System (Invitrogen) followed by PCR specific for the target genes. Primer sequences are in the Supplemental Material.
ChIP
Cells were cross-linked with 1% (v/v) formaldehyde, and ChIP was performed according to the manufacturer’s protocol (CHIP-IT Enzymatic Kit; Active Motif). The primers for p21 and Hdm2 have been previously described (McKinney et al. 2004; Wei et al. 2005). Primer sequences are in the Supplemental Material.
Bat3 mutants
To obtain a human Bat3 mutant resistant to RNAi by siBat3-1 (ΔRNAi-B3), two silent point mutations were introduced within the siBat3-1 recognition site using the oligonucleotide 5′-GGA CAAATTTGGAATTTCTCCAA-3′. To create a NLS mutant (ΔNLS-B3), we introduced two point mutations within the NLS consensus sequence as previously reported (Manchen and Hubberstey 2001). We also established a mutant containing both mutations, ΔNLS/ΔRNAi-B3. The mutations were introduced by the QuikChange Mutagenesis Kit (Stratagene) according to the manufacturer’s protocol. All of the mutations were verified by sequencing.
Histology
To analyze apoptosis in the thymus in vivo, mice (6- to 8-wk-old) were irradiated with 10 Gy from a 137Cs source (Gammacell 40) at a dose of 1.0 Gy/min. The thymus was removed either immediately (0 h) or at 4 or 8 h post-γ-IR and fixed in 10% buffered formalin overnight at 4°C. Tissue samples were embedded in wax according to standard procedures and sectioned at a thickness of 4 μm. Apoptosis in tissue sections was analyzed by in situ end-labeling of fragmented DNA as previously described (Wijsman et al. 1993). Hematoxylin-eosin (H&E) staining was performed according to a standard protocol.
Acknowledgments
We thank Kohei Miyazono, Takeshi Imamura, and Suk-Chul Bae for p300; Melanie Ott for HA-PCAF; Bert Vogelstein for HCT116 cell lines; Samuel Benchimol for p53, critical reading of the manuscript, and helpful discussion; the staff of the Advanced Optical Microscopy Facility (AOMF) at University Health Network for assistance with imaging; and Pathology Research Program (PRP) for histology sample preparation. We are grateful to Mary Saunders for scientific editing. This work was supported by the Start-up Research Grant from CFIBCR/OCI to H.O. S.K. is supported by NIH grant (RO1 GM61919). T.W.M. is supported by NCIC.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1534107
References
- Ahn J., Prives C., Prives C. The C-terminus of p53: The more you learn the less you know. Nat. Struct. Biol. 2001;8:730–732. doi: 10.1038/nsb0901-730. [DOI] [PubMed] [Google Scholar]
- Andrews N.C., Faller D.V., Faller D.V. A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res. 1991;19:2499. doi: 10.1093/nar/19.9.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appella E., Anderson C.W., Anderson C.W. Post-translational modifications and activation of p53 by genotoxic stresses. Eur. J. Biochem. 2001;268:2764–2772. doi: 10.1046/j.1432-1327.2001.02225.x. [DOI] [PubMed] [Google Scholar]
- Avantaggiati M.L., Ogryzko V., Gardner K., Giordano A., Levine A.S., Kelly K., Ogryzko V., Gardner K., Giordano A., Levine A.S., Kelly K., Gardner K., Giordano A., Levine A.S., Kelly K., Giordano A., Levine A.S., Kelly K., Levine A.S., Kelly K., Kelly K. Recruitment of p300/CBP in p53-dependent signal pathways. Cell. 1997;89:1175–1184. doi: 10.1016/s0092-8674(00)80304-9. [DOI] [PubMed] [Google Scholar]
- Ayed A., Mulder F.A., Yi G.S., Lu Y., Kay L.E., Arrowsmith C.H., Mulder F.A., Yi G.S., Lu Y., Kay L.E., Arrowsmith C.H., Yi G.S., Lu Y., Kay L.E., Arrowsmith C.H., Lu Y., Kay L.E., Arrowsmith C.H., Kay L.E., Arrowsmith C.H., Arrowsmith C.H. Latent and active p53 are identical in conformation. Nat. Struct. Biol. 2001;8:756–760. doi: 10.1038/nsb0901-756. [DOI] [PubMed] [Google Scholar]
- Banerji J., Sands J., Strominger J.L., Spies T., Sands J., Strominger J.L., Spies T., Strominger J.L., Spies T., Spies T. A gene pair from the human major histocompatibility complex encodes large proline-rich proteins with multiple repeated motifs and a single ubiquitin-like domain. Proc. Natl. Acad. Sci. 1990;87:2374–2378. doi: 10.1073/pnas.87.6.2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlev N.A., Liu L., Chehab N.H., Mansfield K., Harris K.G., Halazonetis T.D., Berger S.L., Liu L., Chehab N.H., Mansfield K., Harris K.G., Halazonetis T.D., Berger S.L., Chehab N.H., Mansfield K., Harris K.G., Halazonetis T.D., Berger S.L., Mansfield K., Harris K.G., Halazonetis T.D., Berger S.L., Harris K.G., Halazonetis T.D., Berger S.L., Halazonetis T.D., Berger S.L., Berger S.L. Acetylation of p53 activates transcription through recruitment of coactivators/histone acetyltransferases. Mol. Cell. 2001;8:1243–1254. doi: 10.1016/s1097-2765(01)00414-2. [DOI] [PubMed] [Google Scholar]
- Brooks C.L., Gu W., Gu W. Ubiquitination, phosphorylation and acetylation: The molecular basis for p53 regulation. Curr. Opin. Cell Biol. 2003;15:164–171. doi: 10.1016/s0955-0674(03)00003-6. [DOI] [PubMed] [Google Scholar]
- Bunz F., Dutriaux A., Lengauer C., Waldman T., Zhou S., Brown J.P., Sedivy J.M., Kinzler K.W., Vogelstein B., Dutriaux A., Lengauer C., Waldman T., Zhou S., Brown J.P., Sedivy J.M., Kinzler K.W., Vogelstein B., Lengauer C., Waldman T., Zhou S., Brown J.P., Sedivy J.M., Kinzler K.W., Vogelstein B., Waldman T., Zhou S., Brown J.P., Sedivy J.M., Kinzler K.W., Vogelstein B., Zhou S., Brown J.P., Sedivy J.M., Kinzler K.W., Vogelstein B., Brown J.P., Sedivy J.M., Kinzler K.W., Vogelstein B., Sedivy J.M., Kinzler K.W., Vogelstein B., Kinzler K.W., Vogelstein B., Vogelstein B. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998;282:1497–1501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- Chen X., Ko L.J., Jayaraman L., Prives C., Ko L.J., Jayaraman L., Prives C., Jayaraman L., Prives C., Prives C. p53 levels, functional domains, and DNA damage determine the extent of the apoptotic response of tumor cells. Genes & Dev. 1996;10:2438–2451. doi: 10.1101/gad.10.19.2438. [DOI] [PubMed] [Google Scholar]
- Chen D., Kon N., Li M., Zhang W., Qin J., Gu W., Kon N., Li M., Zhang W., Qin J., Gu W., Li M., Zhang W., Qin J., Gu W., Zhang W., Qin J., Gu W., Qin J., Gu W., Gu W. ARF-BP1/Mule is a critical mediator of the ARF tumor suppressor. Cell. 2005;121:1071–1083. doi: 10.1016/j.cell.2005.03.037. [DOI] [PubMed] [Google Scholar]
- Colon-Ramos D.A., Irusta P.M., Gan E.C., Olson M.R., Song J., Morimoto R.I., Elliott R.M., Lombard M., Hollingsworth R., Hardwick J.M., Irusta P.M., Gan E.C., Olson M.R., Song J., Morimoto R.I., Elliott R.M., Lombard M., Hollingsworth R., Hardwick J.M., Gan E.C., Olson M.R., Song J., Morimoto R.I., Elliott R.M., Lombard M., Hollingsworth R., Hardwick J.M., Olson M.R., Song J., Morimoto R.I., Elliott R.M., Lombard M., Hollingsworth R., Hardwick J.M., Song J., Morimoto R.I., Elliott R.M., Lombard M., Hollingsworth R., Hardwick J.M., Morimoto R.I., Elliott R.M., Lombard M., Hollingsworth R., Hardwick J.M., Elliott R.M., Lombard M., Hollingsworth R., Hardwick J.M., Lombard M., Hollingsworth R., Hardwick J.M., Hollingsworth R., Hardwick J.M., Hardwick J.M., et al. Inhibition of translation and induction of apoptosis by Bunyaviral nonstructural proteins bearing sequence similarity to reaper. Mol. Biol. Cell. 2003;14:4162–4172. doi: 10.1091/mbc.E03-03-0139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmots F., Russell H.R., Lee Y., Boyd K., McKinnon P.J., Russell H.R., Lee Y., Boyd K., McKinnon P.J., Lee Y., Boyd K., McKinnon P.J., Boyd K., McKinnon P.J., McKinnon P.J. The reaper-binding protein scythe modulates apoptosis and proliferation during mammalian development. Mol. Cell. Biol. 2005;25:10329–10337. doi: 10.1128/MCB.25.23.10329-10337.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dornan D., Wertz I., Shimizu H., Arnott D., Frantz G.D., Dowd P., O’Rourke K., Koeppen H., Dixit V.M., Wertz I., Shimizu H., Arnott D., Frantz G.D., Dowd P., O’Rourke K., Koeppen H., Dixit V.M., Shimizu H., Arnott D., Frantz G.D., Dowd P., O’Rourke K., Koeppen H., Dixit V.M., Arnott D., Frantz G.D., Dowd P., O’Rourke K., Koeppen H., Dixit V.M., Frantz G.D., Dowd P., O’Rourke K., Koeppen H., Dixit V.M., Dowd P., O’Rourke K., Koeppen H., Dixit V.M., O’Rourke K., Koeppen H., Dixit V.M., Koeppen H., Dixit V.M., Dixit V.M. The ubiquitin ligase COP1 is a critical negative regulator of p53. Nature. 2004;429:86–92. doi: 10.1038/nature02514. [DOI] [PubMed] [Google Scholar]
- Erster S., Mihara M., Kim R.H., Petrenko O., Moll U.M., Mihara M., Kim R.H., Petrenko O., Moll U.M., Kim R.H., Petrenko O., Moll U.M., Petrenko O., Moll U.M., Moll U.M. In vivo mitochondrial p53 translocation triggers a rapid first wave of cell death in response to DNA damage that can precede p53 target gene activation. Mol. Cell. Biol. 2004;24:6728–6741. doi: 10.1128/MCB.24.15.6728-6741.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinosa J.M., Emerson B.M., Emerson B.M. Transcriptional regulation by p53 through intrinsic DNA/chromatin binding and site-directed cofactor recruitment. Mol. Cell. 2001;8:57–69. doi: 10.1016/s1097-2765(01)00283-0. [DOI] [PubMed] [Google Scholar]
- Feng L., Lin T., Uranishi H., Gu W., Xu Y., Lin T., Uranishi H., Gu W., Xu Y., Uranishi H., Gu W., Xu Y., Gu W., Xu Y., Xu Y. Functional analysis of the roles of posttranslational modifications at the p53 C terminus in regulating p53 stability and activity. Mol. Cell. Biol. 2005;25:5389–5395. doi: 10.1128/MCB.25.13.5389-5395.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fojta M., Pivonkova H., Brazdova M., Nemcova K., Palecek J., Vojtesek B., Pivonkova H., Brazdova M., Nemcova K., Palecek J., Vojtesek B., Brazdova M., Nemcova K., Palecek J., Vojtesek B., Nemcova K., Palecek J., Vojtesek B., Palecek J., Vojtesek B., Vojtesek B. Investigations of the supercoil-selective DNA binding of wild type p53 suggest a novel mechanism for controlling p53 function. Eur. J. Biochem. 2004;271:3865–3876. doi: 10.1111/j.1432-1033.2004.04323.x. [DOI] [PubMed] [Google Scholar]
- Goodman R.H., Smolik S., Smolik S. CBP/p300 in cell growth, transformation, and development. Genes & Dev. 2000;14:1553–1577. [PubMed] [Google Scholar]
- Goyal L., McCall K., Agapite J., Hartwieg E., Steller H., McCall K., Agapite J., Hartwieg E., Steller H., Agapite J., Hartwieg E., Steller H., Hartwieg E., Steller H., Steller H. Induction of apoptosis by Drosophila reaper, hid and grim through inhibition of IAP function. EMBO J. 2000;19:589–597. doi: 10.1093/emboj/19.4.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W., Roeder R.G., Roeder R.G. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- Haupt Y., Maya R., Kazaz A., Oren M., Maya R., Kazaz A., Oren M., Kazaz A., Oren M., Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- Hirao A., Kong Y.Y., Matsuoka S., Wakeham A., Ruland J., Yoshida H., Liu D., Elledge S.J., Mak T.W., Kong Y.Y., Matsuoka S., Wakeham A., Ruland J., Yoshida H., Liu D., Elledge S.J., Mak T.W., Matsuoka S., Wakeham A., Ruland J., Yoshida H., Liu D., Elledge S.J., Mak T.W., Wakeham A., Ruland J., Yoshida H., Liu D., Elledge S.J., Mak T.W., Ruland J., Yoshida H., Liu D., Elledge S.J., Mak T.W., Yoshida H., Liu D., Elledge S.J., Mak T.W., Liu D., Elledge S.J., Mak T.W., Elledge S.J., Mak T.W., Mak T.W. DNA damage-induced activation of p53 by the checkpoint kinase Chk2. Science. 2000;287:1824–1827. doi: 10.1126/science.287.5459.1824. [DOI] [PubMed] [Google Scholar]
- Hoffman W.H., Biade S., Zilfou J.T., Chen J., Murphy M., Biade S., Zilfou J.T., Chen J., Murphy M., Zilfou J.T., Chen J., Murphy M., Chen J., Murphy M., Murphy M. Transcriptional repression of the anti-apoptotic survivin gene by wild type p53. J. Biol. Chem. 2002;277:3247–3257. doi: 10.1074/jbc.M106643200. [DOI] [PubMed] [Google Scholar]
- Honda R., Tanaka H., Yasuda H., Tanaka H., Yasuda H., Yasuda H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 1997;420:25–27. doi: 10.1016/s0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- Ito M., Yuan C.X., Malik S., Gu W., Fondell J.D., Yamamura S., Fu Z.Y., Zhang X., Qin J., Roeder R.G., Yuan C.X., Malik S., Gu W., Fondell J.D., Yamamura S., Fu Z.Y., Zhang X., Qin J., Roeder R.G., Malik S., Gu W., Fondell J.D., Yamamura S., Fu Z.Y., Zhang X., Qin J., Roeder R.G., Gu W., Fondell J.D., Yamamura S., Fu Z.Y., Zhang X., Qin J., Roeder R.G., Fondell J.D., Yamamura S., Fu Z.Y., Zhang X., Qin J., Roeder R.G., Yamamura S., Fu Z.Y., Zhang X., Qin J., Roeder R.G., Fu Z.Y., Zhang X., Qin J., Roeder R.G., Zhang X., Qin J., Roeder R.G., Qin J., Roeder R.G., Roeder R.G. Identity between TRAP and SMCC complexes indicates novel pathways for the function of nuclear receptors and diverse mammalian activators. Mol. Cell. 1999;3:361–370. doi: 10.1016/s1097-2765(00)80463-3. [DOI] [PubMed] [Google Scholar]
- Ito A., Lai C.H., Zhao X., Saito S., Hamilton M.H., Appella E., Yao T.P., Lai C.H., Zhao X., Saito S., Hamilton M.H., Appella E., Yao T.P., Zhao X., Saito S., Hamilton M.H., Appella E., Yao T.P., Saito S., Hamilton M.H., Appella E., Yao T.P., Hamilton M.H., Appella E., Yao T.P., Appella E., Yao T.P., Yao T.P. p300/CBP-mediated p53 acetylation is commonly induced by p53-activating agents and inhibited by MDM2. EMBO J. 2001;20:1331–1340. doi: 10.1093/emboj/20.6.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov I., Lo K.C., Hawthorn L., Cowell J.K., Ionov Y., Lo K.C., Hawthorn L., Cowell J.K., Ionov Y., Hawthorn L., Cowell J.K., Ionov Y., Cowell J.K., Ionov Y., Ionov Y. Identifying candidate colon cancer tumor suppressor genes using inhibition of nonsense-mediated mRNA decay in colon cancer cells. Oncogene. 2006 doi: 10.1038/sj.onc.1210098. Epub November 6, 2006. DOI: 10.1038/sj.onc.1210098. [DOI] [PubMed] [Google Scholar]
- Iwakuma T., Lozano G., Lozano G. MDM2, an introduction. Mol. Cancer Res. 2003;1:993–1000. [PubMed] [Google Scholar]
- Jeffers J.R., Parganas E., Lee Y., Yang C., Wang J., Brennan J., MacLean K.H., Han J., Chittenden T., Ihle J.N., Parganas E., Lee Y., Yang C., Wang J., Brennan J., MacLean K.H., Han J., Chittenden T., Ihle J.N., Lee Y., Yang C., Wang J., Brennan J., MacLean K.H., Han J., Chittenden T., Ihle J.N., Yang C., Wang J., Brennan J., MacLean K.H., Han J., Chittenden T., Ihle J.N., Wang J., Brennan J., MacLean K.H., Han J., Chittenden T., Ihle J.N., Brennan J., MacLean K.H., Han J., Chittenden T., Ihle J.N., MacLean K.H., Han J., Chittenden T., Ihle J.N., Han J., Chittenden T., Ihle J.N., Chittenden T., Ihle J.N., Ihle J.N., et al. Puma is an essential mediator of p53-dependent and -independent apoptotic pathways. Cancer Cell. 2003;4:321–328. doi: 10.1016/s1535-6108(03)00244-7. [DOI] [PubMed] [Google Scholar]
- Kaeser M.D., Iggo R.D., Iggo R.D. Promoter-specific p53-dependent histone acetylation following DNA damage. Oncogene. 2004;23:4007–4013. doi: 10.1038/sj.onc.1207536. [DOI] [PubMed] [Google Scholar]
- Kaku S., Iwahashi Y., Kuraishi A., Albor A., Yamagishi T., Nakaike S., Kulesz-Martin M., Iwahashi Y., Kuraishi A., Albor A., Yamagishi T., Nakaike S., Kulesz-Martin M., Kuraishi A., Albor A., Yamagishi T., Nakaike S., Kulesz-Martin M., Albor A., Yamagishi T., Nakaike S., Kulesz-Martin M., Yamagishi T., Nakaike S., Kulesz-Martin M., Nakaike S., Kulesz-Martin M., Kulesz-Martin M. Binding to the naturally occurring double p53 binding site of the Mdm2 promoter alleviates the requirement for p53 C-terminal activation. Nucleic Acids Res. 2001;29:1989–1993. doi: 10.1093/nar/29.9.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kislinger T., Cox B., Kannan A., Chung C., Hu P., Ignatchenko A., Scott M.S., Gramolini A.O., Morris Q., Hallett M.T., Cox B., Kannan A., Chung C., Hu P., Ignatchenko A., Scott M.S., Gramolini A.O., Morris Q., Hallett M.T., Kannan A., Chung C., Hu P., Ignatchenko A., Scott M.S., Gramolini A.O., Morris Q., Hallett M.T., Chung C., Hu P., Ignatchenko A., Scott M.S., Gramolini A.O., Morris Q., Hallett M.T., Hu P., Ignatchenko A., Scott M.S., Gramolini A.O., Morris Q., Hallett M.T., Ignatchenko A., Scott M.S., Gramolini A.O., Morris Q., Hallett M.T., Scott M.S., Gramolini A.O., Morris Q., Hallett M.T., Gramolini A.O., Morris Q., Hallett M.T., Morris Q., Hallett M.T., Hallett M.T., et al. Global survey of organ and organelle protein expression in mouse: Combined proteomic and transcriptomic profiling. Cell. 2006;125:173–186. doi: 10.1016/j.cell.2006.01.044. [DOI] [PubMed] [Google Scholar]
- Krummel K.A., Lee C.J., Toledo F., Wahl G.M., Lee C.J., Toledo F., Wahl G.M., Toledo F., Wahl G.M., Wahl G.M. The C-terminal lysines fine-tune P53 stress responses in a mouse model but are not required for stability control or transactivation. Proc. Natl. Acad. Sci. 2005;102:10188–10193. doi: 10.1073/pnas.0503068102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubbutat M.H., Jones S.N., Vousden K.H., Jones S.N., Vousden K.H., Vousden K.H. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- Kumar R., Lutz W., Frank E., Im H.J., Lutz W., Frank E., Im H.J., Frank E., Im H.J., Im H.J. Immediate early gene X-1 interacts with proteins that modulate apoptosis. Biochem. Biophys. Res. Commun. 2004;323:1293–1298. doi: 10.1016/j.bbrc.2004.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane D.P. Cancer. p53, guardian of the genome. Nature. 1992;358:15–16. doi: 10.1038/358015a0. [DOI] [PubMed] [Google Scholar]
- Leng R.P., Lin Y., Ma W., Wu H., Lemmers B., Chung S., Parant J.M., Lozano G., Hakem R., Benchimol S., Lin Y., Ma W., Wu H., Lemmers B., Chung S., Parant J.M., Lozano G., Hakem R., Benchimol S., Ma W., Wu H., Lemmers B., Chung S., Parant J.M., Lozano G., Hakem R., Benchimol S., Wu H., Lemmers B., Chung S., Parant J.M., Lozano G., Hakem R., Benchimol S., Lemmers B., Chung S., Parant J.M., Lozano G., Hakem R., Benchimol S., Chung S., Parant J.M., Lozano G., Hakem R., Benchimol S., Parant J.M., Lozano G., Hakem R., Benchimol S., Lozano G., Hakem R., Benchimol S., Hakem R., Benchimol S., Benchimol S. Pirh2, a p53-induced ubiquitin-protein ligase, promotes p53 degradation. Cell. 2003;112:779–791. doi: 10.1016/s0092-8674(03)00193-4. [DOI] [PubMed] [Google Scholar]
- Levine A.J. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- Li M., Luo J., Brooks C.L., Gu W., Luo J., Brooks C.L., Gu W., Brooks C.L., Gu W., Gu W. Acetylation of p53 inhibits its ubiquitination by Mdm2. J. Biol. Chem. 2002;277:50607–50611. doi: 10.1074/jbc.C200578200. [DOI] [PubMed] [Google Scholar]
- Lill N.L., Grossman S.R., Ginsberg D., DeCaprio J., Livingston D.M., Grossman S.R., Ginsberg D., DeCaprio J., Livingston D.M., Ginsberg D., DeCaprio J., Livingston D.M., DeCaprio J., Livingston D.M., Livingston D.M. Binding and modulation of p53 by p300/CBP coactivators. Nature. 1997;387:823–827. doi: 10.1038/42981. [DOI] [PubMed] [Google Scholar]
- Liu Y., Lagowski J.P., Vanderbeek G.E., Kulesz-Martin M.F., Lagowski J.P., Vanderbeek G.E., Kulesz-Martin M.F., Vanderbeek G.E., Kulesz-Martin M.F., Kulesz-Martin M.F. Facilitated search for specific genomic targets by p53 C-terminal basic DNA binding domain. Cancer Biol. Ther. 2004;3:1102–1108. doi: 10.4161/cbt.3.11.1189. [DOI] [PubMed] [Google Scholar]
- Lowe S.W., Schmitt E.M., Smith S.W., Osborne B.A., Jacks T., Schmitt E.M., Smith S.W., Osborne B.A., Jacks T., Smith S.W., Osborne B.A., Jacks T., Osborne B.A., Jacks T., Jacks T. p53 is required for radiation-induced apoptosis in mouse thymocytes. Nature. 1993;362:847–849. doi: 10.1038/362847a0. [DOI] [PubMed] [Google Scholar]
- Luo J., Li M., Tang Y., Laszkowska M., Roeder R.G., Gu W., Li M., Tang Y., Laszkowska M., Roeder R.G., Gu W., Tang Y., Laszkowska M., Roeder R.G., Gu W., Laszkowska M., Roeder R.G., Gu W., Roeder R.G., Gu W., Gu W. Acetylation of p53 augments its site-specific DNA binding both in vitro and in vivo. Proc. Natl. Acad. Sci. 2004;101:2259–2264. doi: 10.1073/pnas.0308762101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manchen S.T., Hubberstey A.V., Hubberstey A.V. Human Scythe contains a functional nuclear localization sequence and remains in the nucleus during staurosporine-induced apoptosis. Biochem. Biophys. Res. Commun. 2001;287:1075–1082. doi: 10.1006/bbrc.2001.5701. [DOI] [PubMed] [Google Scholar]
- McCarthy J.V., Dixit V.M., Dixit V.M. Apoptosis induced by Drosophila reaper and grim in a human system. Attenuation by inhibitor of apoptosis proteins (cIAPs) J. Biol. Chem. 1998;273:24009–24015. doi: 10.1074/jbc.273.37.24009. [DOI] [PubMed] [Google Scholar]
- McKinney K., Prives C., Prives C. Efficient specific DNA binding by p53 requires both its central and C-terminal domains as revealed by studies with high-mobility group 1 protein. Mol. Cell. Biol. 2002;22:6797–6808. doi: 10.1128/MCB.22.19.6797-6808.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney K., Mattia M., Gottifredi V., Prives C., Mattia M., Gottifredi V., Prives C., Gottifredi V., Prives C., Prives C. p53 linear diffusion along DNA requires its C terminus. Mol. Cell. 2004;16:413–424. doi: 10.1016/j.molcel.2004.09.032. [DOI] [PubMed] [Google Scholar]
- Okada H., Suh W.K., Jin J., Woo M., Du C., Elia A., Duncan G.S., Wakeham A., Itie A., Lowe S.W., Suh W.K., Jin J., Woo M., Du C., Elia A., Duncan G.S., Wakeham A., Itie A., Lowe S.W., Jin J., Woo M., Du C., Elia A., Duncan G.S., Wakeham A., Itie A., Lowe S.W., Woo M., Du C., Elia A., Duncan G.S., Wakeham A., Itie A., Lowe S.W., Du C., Elia A., Duncan G.S., Wakeham A., Itie A., Lowe S.W., Elia A., Duncan G.S., Wakeham A., Itie A., Lowe S.W., Duncan G.S., Wakeham A., Itie A., Lowe S.W., Wakeham A., Itie A., Lowe S.W., Itie A., Lowe S.W., Lowe S.W., et al. Generation and characterization of Smac/DIABLO-deficient mice. Mol. Cell. Biol. 2002;22:3509–3517. doi: 10.1128/MCB.22.10.3509-3517.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivier M., Eeles R., Hollstein M., Khan M.A., Harris C.C., Hainaut P., Eeles R., Hollstein M., Khan M.A., Harris C.C., Hainaut P., Hollstein M., Khan M.A., Harris C.C., Hainaut P., Khan M.A., Harris C.C., Hainaut P., Harris C.C., Hainaut P., Hainaut P. The IARC TP53 database: New online mutation analysis and recommendations to users. Hum. Mutat. 2002;19:607–614. doi: 10.1002/humu.10081. [DOI] [PubMed] [Google Scholar]
- Palecek E., Brazda V., Jagelska E., Pecinka P., Karlovska L., Brazdova M., Brazda V., Jagelska E., Pecinka P., Karlovska L., Brazdova M., Jagelska E., Pecinka P., Karlovska L., Brazdova M., Pecinka P., Karlovska L., Brazdova M., Karlovska L., Brazdova M., Brazdova M. Enhancement of p53 sequence-specific binding by DNA supercoiling. Oncogene. 2004;23:2119–2127. doi: 10.1038/sj.onc.1207324. [DOI] [PubMed] [Google Scholar]
- Prives C., Hall P.A., Hall P.A. The p53 pathway. J. Pathol. 1999;187:112–126. doi: 10.1002/(SICI)1096-9896(199901)187:1<112::AID-PATH250>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Rodriguez M.S., Desterro J.M., Lain S., Lane D.P., Hay R.T., Desterro J.M., Lain S., Lane D.P., Hay R.T., Lain S., Lane D.P., Hay R.T., Lane D.P., Hay R.T., Hay R.T. Multiple C-terminal lysine residues target p53 for ubiquitin-proteasome-mediated degradation. Mol. Cell. Biol. 2000;20:8458–8467. doi: 10.1128/mcb.20.22.8458-8467.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi K., Herrera J.E., Saito S., Miki T., Bustin M., Vassilev A., Anderson C.W., Appella E., Herrera J.E., Saito S., Miki T., Bustin M., Vassilev A., Anderson C.W., Appella E., Saito S., Miki T., Bustin M., Vassilev A., Anderson C.W., Appella E., Miki T., Bustin M., Vassilev A., Anderson C.W., Appella E., Bustin M., Vassilev A., Anderson C.W., Appella E., Vassilev A., Anderson C.W., Appella E., Anderson C.W., Appella E., Appella E. DNA damage activates p53 through a phosphorylation–acetylation cascade. Genes & Dev. 1998;12:2831–2841. doi: 10.1101/gad.12.18.2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shieh S.Y., Ikeda M., Taya Y., Prives C., Ikeda M., Taya Y., Prives C., Taya Y., Prives C., Prives C. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell. 1997;91:325–334. doi: 10.1016/s0092-8674(00)80416-x. [DOI] [PubMed] [Google Scholar]
- Spies T., Blanck G., Bresnahan M., Sands J., Strominger J.L., Blanck G., Bresnahan M., Sands J., Strominger J.L., Bresnahan M., Sands J., Strominger J.L., Sands J., Strominger J.L., Strominger J.L. A new cluster of genes within the human major histocompatibility complex. Science. 1989;243:214–217. doi: 10.1126/science.2911734. [DOI] [PubMed] [Google Scholar]
- Thress K., Henzel W., Shillinglaw W., Kornbluth S., Henzel W., Shillinglaw W., Kornbluth S., Shillinglaw W., Kornbluth S., Kornbluth S. Scythe: A novel reaper-binding apoptotic regulator. EMBO J. 1998;17:6135–6143. doi: 10.1093/emboj/17.21.6135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thress K., Evans E.K., Kornbluth S., Evans E.K., Kornbluth S., Kornbluth S. Reaper-induced dissociation of a Scythe-sequestered cytochrome c-releasing activity. EMBO J. 1999;18:5486–5493. doi: 10.1093/emboj/18.20.5486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thress K., Song J., Morimoto R.I., Kornbluth S., Song J., Morimoto R.I., Kornbluth S., Morimoto R.I., Kornbluth S., Kornbluth S. Reversible inhibition of Hsp70 chaperone function by Scythe and Reaper. EMBO J. 2001;20:1033–1041. doi: 10.1093/emboj/20.5.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibbetts R.S., Brumbaugh K.M., Williams J.M., Sarkaria J.N., Cliby W.A., Shieh S.Y., Taya Y., Prives C., Abraham R.T., Brumbaugh K.M., Williams J.M., Sarkaria J.N., Cliby W.A., Shieh S.Y., Taya Y., Prives C., Abraham R.T., Williams J.M., Sarkaria J.N., Cliby W.A., Shieh S.Y., Taya Y., Prives C., Abraham R.T., Sarkaria J.N., Cliby W.A., Shieh S.Y., Taya Y., Prives C., Abraham R.T., Cliby W.A., Shieh S.Y., Taya Y., Prives C., Abraham R.T., Shieh S.Y., Taya Y., Prives C., Abraham R.T., Taya Y., Prives C., Abraham R.T., Prives C., Abraham R.T., Abraham R.T. A role for ATR in the DNA damage-induced phosphorylation of p53. Genes & Dev. 1999;13:152–157. doi: 10.1101/gad.13.2.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villunger A., Michalak E.M., Coultas L., Mullauer F., Bock G., Ausserlechner M.J., Adams J.M., Strasser A., Michalak E.M., Coultas L., Mullauer F., Bock G., Ausserlechner M.J., Adams J.M., Strasser A., Coultas L., Mullauer F., Bock G., Ausserlechner M.J., Adams J.M., Strasser A., Mullauer F., Bock G., Ausserlechner M.J., Adams J.M., Strasser A., Bock G., Ausserlechner M.J., Adams J.M., Strasser A., Ausserlechner M.J., Adams J.M., Strasser A., Adams J.M., Strasser A., Strasser A. p53- and drug-induced apoptotic responses mediated by BH3-only proteins puma and noxa. Science. 2003;302:1036–1038. doi: 10.1126/science.1090072. [DOI] [PubMed] [Google Scholar]
- Vogelstein B., Lane D., Levine A.J., Lane D., Levine A.J., Levine A.J. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- Vousden K.H., Lu X., Lu X. Live or let die: The cell’s response to p53. Nat. Rev. Cancer. 2002;2:594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- Wei G., Li A.G., Liu X., Li A.G., Liu X., Liu X. Insights into selective activation of p53 DNA binding by c-Abl. J. Biol. Chem. 2005;280:12271–12278. doi: 10.1074/jbc.M409522200. [DOI] [PubMed] [Google Scholar]
- Wijsman J.H., Jonker R.R., Keijzer R., de van Vel C.J., Cornelisse C.J., van Dierendonck J.H., Jonker R.R., Keijzer R., de van Vel C.J., Cornelisse C.J., van Dierendonck J.H., Keijzer R., de van Vel C.J., Cornelisse C.J., van Dierendonck J.H., de van Vel C.J., Cornelisse C.J., van Dierendonck J.H., Cornelisse C.J., van Dierendonck J.H., van Dierendonck J.H. A new method to detect apoptosis in paraffin sections: In situ end-labeling of fragmented DNA. J. Histochem. Cytochem. 1993;41:7–12. doi: 10.1177/41.1.7678025. [DOI] [PubMed] [Google Scholar]
- Wu Y.H., Shih S.F., Lin J.Y., Shih S.F., Lin J.Y., Lin J.Y. Ricin triggers apoptotic morphological changes through caspase-3 cleavage of BAT3. J. Biol. Chem. 2004;279:19264–19275. doi: 10.1074/jbc.M307049200. [DOI] [PubMed] [Google Scholar]
- Yang X.J., Ogryzko V.V., Nishikawa J., Howard B.H., Nakatani Y., Ogryzko V.V., Nishikawa J., Howard B.H., Nakatani Y., Nishikawa J., Howard B.H., Nakatani Y., Howard B.H., Nakatani Y., Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]