Abstract
Environmental effects on phenotype can be mediated by epigenetic modifications. The epigenetic state of the murine Avy allele is highly variable, and determines phenotypic effects that vary in a mosaic spectrum that can be shifted by in utero exposure to methyl donor supplementation. We have asked if methyl donor supplementation affects the germ-line epigenetic state of the Avy allele. We find that the somatic epigenetic state of Avy is affected by in utero methyl donor supplementation only when the allele is paternally contributed. Exposure to methyl donor supplementation during midgestation shifts Avy phenotypes not only in the mice exposed as fetuses, but in their offspring. This finding indicates that methyl donors can change the epigenetic state of the Avy allele in the germ line, and that the altered state is retained through the epigenetic resetting that takes place in gametogenesis and embryogenesis. Thus a mother's diet may have an enduring influence on succeeding generations, independent of later changes in diet. Although other reports have suggested such heritable epigenetic changes, this study demonstrates that a specific mammalian gene can be subjected to germ-line epigenetic change.
Keywords: agouti, inheritance
Higher eukaryotes use epigenetic modifications to reversibly suppress transcription of genes and repeat elements, often by stable silencing (1). Epigenetic marks are retained through mitosis, allowing maintenance of characteristic cell types, but they may also be transmitted from one generation to the next (2). By their nature, epigenetic modifications are susceptible to environmental influence: the interposition of epigenetic modifications between genes and the environment provides a way in which the environment can exert heritable influences on phenotype (3–5).
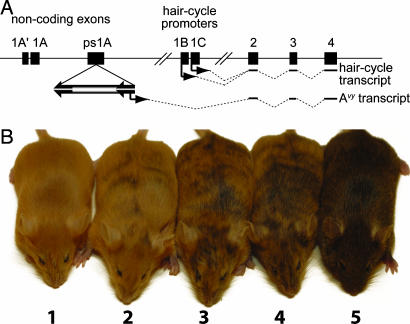
Mice carrying the viable yellow allele of agouti (Avy) are a model of epigenetic variation and inheritance. The Avy allele carries an insertion of an intracisternal A-particle (IAP) retrotransposon into pseudoexon 1A of the agouti locus, upstream of the transcribed region coding for agouti signaling protein (ASP) (Fig. 1A) (3, 4). When the inserted IAP is active, a cryptic promoter in its LTR usurps transcriptional control of agouti and drives ectopic expression of ASP (3, 6). Pancellular expression of ASP gives a neomorphic phenotype of yellow fur, obesity, type II diabetes, and predisposition to tumors; when the IAP is silent agouti is expressed in its normal pattern (6). Avy is dominant, so that when heterozygous with a (nonagouti, a loss-of-function allele of agouti), the epigenetic state of the allele is readily apparent (Fig. 1B). The obese yellow phenotype in Avy mice displays extremely variable expressivity in an isogenic background (7). The activity state of the IAP is typically mosaic and varies widely between isogenic Avy/a mice, whose phenotypes (Fig. 1B) range from fully yellow and obese, through degrees of mottled yellow/agouti with intermediate body mass, to lean fully agouti (called pseudoagouti) (6, 7).
Fig. 1.
The Avy allele and the spectrum of Avy phenotypes. (A) The Avy allele carries an insertion of an IAP retrotransposon in an antisense direction in agouti pseudoexon 1A (3, 4). The Avy transcript originates from a cryptic promoter in the 5′LTR of the IAP and is spliced to agouti coding exons 2, 3 and 4, which encode ASP (3). When the IAP is silent, agouti is transcribed from hair-cycle-specific promoters in exons 1B and 1C. (B) Avy phenotypes are scored from 1 to 5 based on coat color. Fully yellow mice are scored as 1, and fully agouti mice are scored as 5. Phenotypes of mosaic mice range from mostly yellow (2) to mottled yellow/agouti (3) to mostly agouti (4).
The observed pattern of epigenetic mosaicism of the Avy allele is consistent with a somatically stable epigenetic state (either on or off) established in early embryogenesis. Phenotypic variation is a direct result of variation in the epigenotype of the IAP retrotransposon from which the aberrant Avy transcript originates; thus the syndrome represents a case of a retrotransposon acting as a controlling element (2). The IAP's epigenotype correlates closely with cytosine methylation of its 5′ LTR: in pseudoagouti mice the LTR is heavily methylated, in yellow mice it is unmethylated, and in mottled mice methylation is intermediate (4, 8, 9). Whereas other retrotransposons are maintained in a state of stable epigenetic silence, the behavior of the IAP that controls the Avy allele is highly unusual: it exhibits a strong tendency for reversion in the germ line from the active to the silent state (or vice versa), whereas its somatic state (active or silent) is stable. The mechanistic basis of epigenetic variation in the Avy allele is not known.
Somatic activity of the Avy allele can be affected by maternal nutrition. When pregnant dams receive a diet supplemented with methyl donors (folate, choline, betaine, and vitamin B12), the spectrum of phenotypes in Avy offspring is shifted toward the epigenetically suppressed state that is termed pseudoagouti (4, 8, 10). This change correlates with an increase in cytosine methylation of the Avy allele (4, 8), indicating that the epigenetic state of the allele can be influenced by environmental factors. Methyl donors may influence the pool of S-adenosylmethionine, which donates methyl groups to cytosine and many proteins (10); the epigenetic effects of dietary methyl donors could be direct (methylation of cytosine) or indirect.
The somatic epigenetic changes in response to methyl donor supplementation raise the question of whether methyl donor supplementation also affects the germ line, and whether any changes could be maintained into the next generation. Epigenetic marks at genes and repeat elements are usually removed and reset soon after fertilization (11), but some mammalian genes, and the Avy IAP, can at least partially retain epigenetic marks so that their epigenotype is inherited (9, 12). Reports of heritable effects mediated by environmental factors (13–15), sometimes involving methylation changes (13), have not identified specific genes.
We have assessed the ability of methyl donor supplementation to alter the epigenetic state of the Avy allele in the germ line. We find that the Avy allele is responsive to methyl donor supplementation only when it is contributed by the sire. Supplementation of the maternal diet for a period during midgestation produces changes in the offspring exposed in utero, but also in pups born in the subsequent generation that was not exposed to higher levels of methyl donors. Taken together, these results demonstrate that environmental influences on phenotype may act by inducing epigenetic changes, and that such changes may be heritable and allele-specific.
Results
The Avy allele is dominant: when heterozygous with a, the epigenetic state of the Avy allele is readily apparent by inspection of the coat (Fig. 1) (6, 7). Coat color (yellow, mosaic yellow/agouti, or pseudoagouti) is closely linked to other manifestations of the viable yellow phenotype (obesity, type II diabetes, and tumor incidence) and to methylation of the IAP retrotransposon that drives ectopic expression of ASP (4, 6–9). Because in utero methyl donor supplementation subtly shifts the spectrum of phenotypes displayed by offspring, detection of any effect requires breeding of a sufficiently large number of mice to establish its significance. In our experiments, offspring in each treatment or control group were considered as independent variables for statistical analysis (see Methods), so that all offspring in each treatment group were compared with all offspring in the corresponding control group, without reference to their respective dams.
The Avy Allele Is Affected by Dietary Methyl Donors Only When It Is Paternally Contributed.
Our experiments aimed to assess the heritability of effects on the Avy allele created by supplementation of the maternal diet with methyl donors; that is, the ability of methyl donors to alter the epigenotype of the Avy allele in the germ line. The epigenotype of the Avy allele is partially stable in the female germ line, leading to weak inheritance of the maternal phenotype, but paternal Avy epigenotype does not influence the phenotype of offspring (9, 16). Thus, the spectrum of phenotypes in offspring of Avy/a females is skewed toward the maternal phenotype, whereas the spectrum of phenotypes in offspring of Avy/a males is the same regardless of paternal phenotype (9, 16). We thus reasoned that methyl donors would be more likely to stably alter the epigenotype of Avy in the female germ line. For this reason we began by providing methyl donor supplementation to Avy females mated to congenic a/a males.
Avy/a dams mated to a/a sires were fed an NIH-31 diet supplemented with folate, choline, betaine, vitamin B12, zinc, and methionine (Specialty Feeds, Glen Forest, WA, Australia) (8) for 2 weeks before mating and during pregnancy and lactation. Offspring were scored for coat color phenotype (Fig. 1B) at weaning.
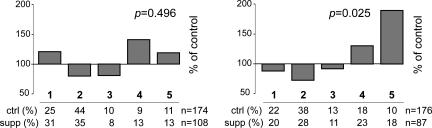
In apparent conflict with previously reported experiments (4, 8, 10) we observed no effect on the phenotypes of Avy/a offspring of supplemented dams when compared with offspring of unsupplemented Avy/a dams (Fig. 2Left). In those experiments, however, the Avy allele had been contributed by the sire (4, 8, 10). We thus repeated the supplementation experiment using Avy/a sires and a/a dams, and obtained a result (Fig. 2 Right) consistent with the previous reports: a significant shift in the spectrum of phenotypes toward pseudoagouti (silent, methylated Avy).
Fig. 2.
Parent-of-origin effect of methyl donor supplementation. Parents were supplemented with methyl donors throughout pregnancy and offspring phenotypes scored as in Fig. 1B. (Left) Maternal transmission. Shown is the maternally derived Avy allele (Avy/a dam, a/a sire). (Right) Paternal transmission. Shown is the paternally derived Avy allele (a/a dam, Avy/a sire). Results are expressed as the percentage of supplemented offspring with the same phenotype as unsupplemented controls; the percentage of offspring of each coat color is shown beneath each graph.
These results suggest that in utero methyl donor supplementation affects the somatic epigenetic state of the Avy allele only when the allele is derived from the sire.
Methyl Donors Induce a Germ-Line Epigenetic Change in the Avy Allele.
We reasoned that if a germ-line change in Avy epigenotype were induced by methyl donors, it could be reflected in phenotypes in a later generation that had not been exposed to methyl donor supplementation. To test this idea, we supplemented pregnant dams and bred their F1 offspring to observe phenotypes in the F2 generation. Because we found that methyl donor supplementation affects only the paternally contributed Avy allele (see above), we mated a/a dams to Avy/a sires to produce the F1 generation. The methyl donor supplemented diet was provided only from embryonic day 8.5 (E8.5) to E15.5. This restricted period of supplementation had two related purposes. First, because it commenced well past the period when somatic Avy epigenotype appears to be set, we supposed that it might have no effect on the phenotypes of F1 mice. Second, because the supplementation encompasses the period when primordial germ cells differentiate and reset epigenetic marks (11), it may be the optimum point to induce an epigenetic change in the germ line.
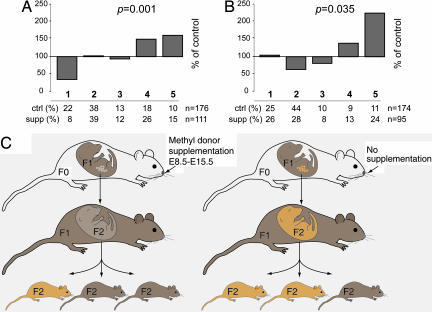
The pattern of mosaicism in Avy mice is consistent with an epigenetic state that is set during early embryogenesis and is stable (in somatic cells) thereafter. This stability resembles that seen with other epigenetic phenomena such as parental imprinting and X-chromosome inactivation. By recording phenotypes of F1 mice in our experiment, we were able to observe any effects of in utero methyl donor supplementation on the epigenotype of the Avy allele. Despite the restricted period of supplementation, the spectrum of phenotypes in F1 mice was shifted significantly (Fig. 3A), indicating that the Avy allele is susceptible to induced epigenetic change even after the early embryonic period when epigenetic resetting takes place.
Fig. 3.
Midgestation and germ-line effects of methyl donor supplementation. (A) F1 phenotypes. Shown is the effect of methyl donor supplementation on mid-gestation supplemented offspring. Dams were supplemented with methyl donors from E8.5 to E15.5, and offspring phenotypes were scored as in Fig. 1B. Results are expressed as the percentage of supplemented offspring with the same phenotype as unsupplemented controls, and the percentage of offspring of each coat color is shown beneath each graph. (B) F2 phenotypes. Shown is the heritable/grandparental effect of methyl donor supplementation. Phenotypes of F2 offspring from pseudoagouti female mice shown in A are expressed as a percentage of control offspring (offspring from pseudoagouti dams that had never been supplemented) with the same phenotype. (C) A schematic diagram illustrating the effect of methyl donor supplementation in the germ line. Epigenetic changes to Avy in primordial germ cells exposed to methyl donors during differentiation (Left) are maintained throughout gametogenesis and embryogenesis. Thus, pseudoagouti F1 mice that are genetically and phenotypically identical but were subject to different diets in utero (Left vs. Right), can produce phenotypically different F2 offspring.
We selected (F1) pseudoagouti Avy/a females that had been exposed to methyl donors in utero from E8.5 to E15.5, and mated them to a/a males without any further methyl donor supplementation; this strategy takes advantage of the tendency for the Avy epigenotype to be partially stable in the female germ line (9, 16). The phenotypes of the second generation (F2) offspring were compared with phenotypes of pups born to pseudoagouti females with no history of exposure to methyl donor supplementation. Phenotypes of these F2 mice were significantly shifted toward the pseudoagouti (Fig. 3B). Thus a pseudoagouti dam who was exposed to methyl donor supplementation only when she was in utero gives rise to phenotypically different offspring than does an otherwise (genetically and phenotypically) identical female who had no exposure to methyl donor supplementation (Fig. 3C); this grandparental effect is directly attributable to the epigenetic state of the Avy allele.
It is likely that the inherited modification induced by methyl donor supplementation is placed on the Avy allele at the point, during mid- to late gestation, when epigenetic marks are reset in the differentiating primordial germ cells that later give rise to the F2 generation (11). Because nutritional supplementation ceased when the F1 mice were still in utero, our evidence indicates that the effect on Avy epigenotype in these primordial germ cells is retained throughout gametogenesis as well as during the fertilization and development of the F2 embryo (Fig. 3C).
Discussion
Our results indicate that methyl donors can affect the germ-line epigenetic state of the Avy allele, and that this effect is stable for at least one generation without further exposure to the supplementary methyl donors. Previous work had shown that mice exposed in utero to higher levels of methyl donors are more likely to display the pseudoagouti phenotype that is associated with methylation and transcriptional silence of the IAP retrotransposon that drives the viable yellow phenotype (4, 8, 10). We find that this effect on the somatic epigenotype of Avy occurs only when the allele is derived from the sire, and that it does not require exposure during early embryogenesis. The spectrum of Avy phenotypes is altered in mice in the generation (F2) that succeeds the one exposed to methyl donors during midgestation. The implications of this finding are worth considering.
The germ-line alteration of the Avy epigenotype is definite evidence that an environmental factor can produce a phenotypic effect by inducing an epigenetic modification in the mammalian germ line, and that such a modification can persist through the epigenetic resetting that takes place during gametogenesis and embryogenesis. A number of reports have described heritable phenotypic effects in mammals induced by environmental agents, some well defined and others less so (13–15, 17, 18). There has been wide speculation that such effects are epigenetic in origin, and in one system, heritable changes in methylation were detected in response to endocrine disruptors (13). But because the specific loci that mediate the described phenotypic effects are not known in any of these cases, the evidence that the phenotypic effects result from epigenetic modifications is inconclusive. On the other hand, the effect of dietary methyl donors on Avy is indisputably one in which the inheritance of the environmental effect is based on epigenetic modification of a specific locus.
The parent-of-origin-specific effects of methyl donor supplementation on Avy (Fig. 2) may provide an insight into the inheritance of Avy epigenetic state. In isogenic Avy mice, the epigenotype is weakly heritable through the female: the spectrum of coat colors in offspring of Avy/a females differs with maternal phenotype, whereas offspring of Avy/a males exhibit the same spectrum of coat colors regardless of the sire's phenotype (9, 16). That is, the epigenotype of the paternal Avy allele undergoes complete epigenetic resetting during embryogenesis, whereas the maternal Avy allele partly retains its epigenotype. This relative stability of the Avy epigenetic state in the female germ line may relate to its resistance to the influence of increased methyl donors. The paternally inherited Avy allele, being more epigenetically labile in the germ line, may be more vulnerable to environmental influence. Because we examined the effects of methyl donors on the Avy allele only in offspring of pseudoagouti mothers (carrying a silent Avy), further investigation will be required to determine whether all epigenetic states of the maternal Avy allele are resistant to methyl donor supplementation.
We find that the somatic state of the Avy allele is susceptible to methyl donor supplementation even when exposure takes place after the period of epigenetic resetting in early embryogenesis. This result was surprising, because the pattern of mosaicism in Avy mice indicates stability of the epigenetic state in later embryogenesis and thereafter. Taken together with a recent finding that an imprinted locus is sensitive to nutritional intervention even post partum (19), this finding implies that epigenetic states may be sensitive to environmental effects throughout the life cycle. It is notable that the evidence for epigenetic resetting in preimplantation embryogenesis, and the stability of epigenetic states thereafter, is principally based on analysis of CpG methylation (20–23). If, as discussed below, the effect of methyl donor supplementation is mediated by another pathway, this finding might indicate an unsuspected plasticity of somatic epigenetic states.
Epigenetic silencing at Avy is linked to increased CpG methylation of the IAP (4, 8, 9, 24). It is tempting to suppose that methyl donors act directly to increase cytosine methylation on the IAP. CpG methylation is, however, merely one part of a complex of epigenetic modifications that are typical of silent chromatin; available evidence indicates that it is placed on DNA subsequent to epigenetic silencing, and it serves to consolidate and maintain the silent state through interaction with chromatin proteins (24). Studies of the somatic effects of dietary methyl donor supplementation have shown that although supplementation increases the propensity of the allele to silence, the level of CpG methylation on the IAP in supplemented mice is equivalent to that in unsupplemented mice of the same phenotype; i.e., when adjusted for phenotype, the methylation states are equivalent (4). Methyl donors are expected to increase the pool of S-adenosylmethionine, which can donate a methyl group to a variety of proteins as well as cytosine. Thus, the epigenetic effects of dietary methyl donors may be mediated by effects on, for example, histones, which would have the effect of increasing CpG methylation due to silencing of the Avy allele in a higher proportion of cells (24).
The IAP in the Avy allele is a “controlling element”: a transposable element that exerts transcriptional control over a gene near its insertion site, as first described by McClintock (2, 25). Although it is not known how many retroelements act as controlling elements in the mammalian genome, several published examples are available (2, 12, 26), and our work indicates that there may be many more (D.I.K.M. and G. Thomson, unpublished work). Thousands of retroelements have the potential, if active, to behave as controlling elements, with unpredictable effects on phenotype (2). The susceptibility of these elements to perturbation by environmental agents provides another way in which epigenetics can mediate environmental influence on phenotype.
The increasing evidence for the heritability of environmental exposure introduces an added degree of complexity into attempts to disentangle so-called “gene–environment interactions,” often interpreted to signify the interaction of fixed (Mendelian) inheritance with a dynamic environment. It is apparent that the heritability of “environment” through epigenetic settings will make such an effort more difficult. Moreover, in light of the roughly 20-year generation time of humans, our results suggest that current dietary habits may have an influence on grandchildren who will be born decades from now, independent of the diets that their parents consume.
Materials and Methods
Mice and Diets.
The Avy allele arose in the C3H/HeJ strain (27) and was backcrossed into C57BL/6 at The Jackson Laboratory (Bar Harbor, ME). The mice used in this study are descended from the isogenic C57BL/6 Avy colony maintained at Oak Ridge National Laboratories and were rederived at the Victor Chang Cardiac Research Institute in 2001.
Mice were fed ad libitum on NIH-31 diet (control) or methyl-donor supplemented NIH-31 [plus (per kg) 15 g of choline, 15 g of betaine, 7.5 g of l-methionine, 150 mg of ZnSO4, 15 mg of folic acid, and 1.5 mg of vitamin B12] (Specialty Feeds) (8, 10). For mice supplemented throughout pregnancy, methyl donor supplementation of the dam was started 2 weeks before mating and continued throughout pregnancy and lactation. For mice supplemented during midgestation, the date of conception was determined by observing vaginal plugs. Methyl donor supplementation was started on E8.5 and discontinued at E15.5.
Female Avy/a mice selected for breeding were always pseudoagouti, because the epigenotype of the dam influences that of the offspring. Male Avy/a mice selected for breeding were either mottled or pseudoagouti: although the epigenotype of the sire does not influence that of the offspring, yellow mice are inefficient breeders. Control groups consisted of mice bred in exactly the same way but without any methyl donor supplementation.
Phenotype Scoring.
Coat color in Avy mice is tightly linked to the other manifestations of the obese yellow phenotype and to methylation of the Avy allele (4, 6, 9). Coat color in Avy/a offspring was assessed at mouse weaning age (3 weeks) by two trained, independent observers. We used a numerical scale from 1 to 5 such that 1 was completely yellow, 2 was mostly yellow with slight agouti mottling, 3 was ≈50% yellow/agouti mottling, 4 was mostly agouti, and 5 was complete agouti (called pseudoagouti); representative phenotypes are illustrated in Fig. 1B. The scores from the two observers correlated >85% of the time; when they did not agree, the coat color was randomly assigned to one or the other of the scores.
Statistical Analyses.
For statistical analysis, offspring in each treatment group were considered as independent variables. We used the Kruskall–Wallis test to establish that offspring coat color phenotype was independent of the dam by comparing the means of the coat colors of offspring from all dams in a treatment group. This method allowed us to compare all offspring in each treatment group with all offspring in the corresponding control group without reference to the dam. For comparison of treatment groups with controls, we used a Mann–Whitney test (α = 0.05). Parametric analysis (one-way ANOVA and Student's t test) gave very similar results, probably due to our large sample size, allowing parametric testing of ordinal, non-normally distributed data.
Acknowledgments
We thank Amy Cutler for technical assistance and Rob Bryson-Richardson for photography. This work was supported by National Institutes of Health Grants 1 R01 CA115768-01 (to D.I.K.M.) and P60 MD00222 (to K.B.B.), National Health and Medical Research Council Grants 2130408 and 256301 (to D.I.K.M.), and the Victor Chang Cardiac Research Institute.
Abbreviations
- ASP
agouti signaling protein
- En
embryonic day n
- IAP
intracisternal A-particle.
Footnotes
The authors declare no conflict of interest.
See Commentary on page 17071.
This article is a PNAS direct submission.
References
- 1.Wolffe AP, Matzke MA. Science. 1999;286:481–486. doi: 10.1126/science.286.5439.481. [DOI] [PubMed] [Google Scholar]
- 2.Whitelaw E, Martin DI. Nat Genet. 2001;27:361–365. doi: 10.1038/86850. [DOI] [PubMed] [Google Scholar]
- 3.Duhl DM, Vrieling H, Miller KA, Wolff GL, Barsh GS. Nat Genet. 1994;8:59–65. doi: 10.1038/ng0994-59. [DOI] [PubMed] [Google Scholar]
- 4.Waterland RA, Jirtle RL. Mol Cell Biol. 2003;23:5293–5300. doi: 10.1128/MCB.23.15.5293-5300.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jaenisch R, Bird A. Nat Genet. 2003;33(Suppl 1):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 6.Wolff GL, Roberts DW, Mountjoy KG. Physiol Genomics. 1999;1:151–163. doi: 10.1152/physiolgenomics.1999.1.3.151. [DOI] [PubMed] [Google Scholar]
- 7.Wolff GL, Pitot HC. Genetics. 1973;73:109–123. doi: 10.1093/genetics/73.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cooney CA, Dave AA, Wolff GL. J Nutr. 2002;132:2393S–2400S. doi: 10.1093/jn/132.8.2393S. [DOI] [PubMed] [Google Scholar]
- 9.Morgan HD, Sutherland HGE, Martin DIK, Whitelaw E. Nat Genet. 1999;23:314–318. doi: 10.1038/15490. [DOI] [PubMed] [Google Scholar]
- 10.Wolff GL, Kodell RL, Moore SR, Cooney CA. FASEB J. 1998;12:949–957. [PubMed] [Google Scholar]
- 11.Morgan HD, Santos F, Green K, Dean W, Reik W. Hum Mol Genet. 2005;14:R47–R58. doi: 10.1093/hmg/ddi114. [DOI] [PubMed] [Google Scholar]
- 12.Rakyan VK, Chong S, Champ ME, Cuthbert PC, Morgan HD, Luu KV, Whitelaw E. Proc Natl Acad Sci USA. 2003;100:2538–2543. doi: 10.1073/pnas.0436776100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anway MD, Cupp AS, Uzumcu M, Skinner MK. Science. 2005;308:1466–1469. doi: 10.1126/science.1108190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drake AJ, Walker BR, Seckl JR. Am J Physiol. 2005;288:R34–R38. doi: 10.1152/ajpregu.00106.2004. [DOI] [PubMed] [Google Scholar]
- 15.Pembrey ME, Bygren LO, Kaati G, Edvinsson S, Northstone K, Sjostrom M, Golding J. Eur J Hum Genet. 2006;14:159–166. doi: 10.1038/sj.ejhg.5201538. [DOI] [PubMed] [Google Scholar]
- 16.Wolff GL. Genetics. 1978;88:529–539. doi: 10.1093/genetics/88.3.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anway MD, Skinner MK. Endocrinology. 2006;147:S43–S49. doi: 10.1210/en.2005-1058. [DOI] [PubMed] [Google Scholar]
- 18.Dubrova YE. Oncogene. 2003;22:7087–7093. doi: 10.1038/sj.onc.1206993. [DOI] [PubMed] [Google Scholar]
- 19.Waterland RA, Lin JR, Smith CA, Jirtle RL. Hum Mol Genet. 2006;15:705–716. doi: 10.1093/hmg/ddi484. [DOI] [PubMed] [Google Scholar]
- 20.Howlett SK, Reik W. Development (Cambridge, UK) 1991;113:119–127. doi: 10.1242/dev.113.1.119. [DOI] [PubMed] [Google Scholar]
- 21.Lane N, Dean W, Erhardt S, Hajkova P, Surani A, Walter J, Reik W. Genesis. 2003;35:88–93. doi: 10.1002/gene.10168. [DOI] [PubMed] [Google Scholar]
- 22.Monk M. Dev Genet. 1995;17:188–197. doi: 10.1002/dvg.1020170303. [DOI] [PubMed] [Google Scholar]
- 23.Santos F, Hendrich B, Reik W, Dean W. Dev Biol. 2002;241:172–182. doi: 10.1006/dbio.2001.0501. [DOI] [PubMed] [Google Scholar]
- 24.Bird A. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 25.Fincham JR, Sastry GR. Annu Rev Genet. 1974;8:15–50. doi: 10.1146/annurev.ge.08.120174.000311. [DOI] [PubMed] [Google Scholar]
- 26.Speek M. Mol Cell Biol. 2001;21:1973–1985. doi: 10.1128/MCB.21.6.1973-1985.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dickie MM. J Hered. 1962;53:84–86. doi: 10.1093/oxfordjournals.jhered.a107129. [DOI] [PubMed] [Google Scholar]