Abstract
Recent studies have established that a significant fraction of prostate cancers harbor a signature gene fusion between the 5′ region of androgen-regulated TMPRSS2 and an ETS family transcription factor, most commonly ERG. Studies on the molecular mechanisms and functional consequences of this important chromosomal rearrangement are currently limited to the VCaP cell line derived from a vertebral bone metastasis of a hormone-refractory prostate tumor. Here we report on the NCI-H660 cell line, derived from a metastatic site of an extrapulmonary small cell carcinoma arising from the prostate. NCI-H660 harbors TMPRSS2-ERG fusion with a homozygous intronic deletion between TMPRSS2 and ERG. We demonstrate this by real-time quantitative polymerase chain reaction, a two-stage dual-color interphase fluorescence in situ hybridization (FISH) assay testing for TMPRSS2 and ERG break-aparts, and single-nucleotide polymorphism oligonucleotide arrays. The deletion is consistent with the common intronic deletion found on chromosome 21q22.2-3 in human prostate cancer samples. We demonstrate the physical juxtaposition of TMPRSS2 and ERG on the DNA level by fiber FISH. The androgen receptor-negative NCI-H660 cell line expresses ERG in an androgen-independent fashion. This in vitro model system has the potential to provide important pathobiologic insights into TMPRSS2-ERG fusion prostate cancer.
Keywords: TMPRSS2-ERG, prostate cancer, cell line, gene fusion, translocation
Introduction
Recent work demonstrates that nearly half of prostate-specific antigen (PSA)-screened prostate cancers harbor TMPRSS2-ETS fusions [1–3]. ERG (21q22.3), ETV1 (7p21.2), or ETV4 (17q21) is activated by a genetic rearrangement that fuses the 3′ end of either gene to the 5′ end of androgen-regulated TMPRSS2 (21q22.2). This generates an androgen-responsive fusion oncoprotein, leading to overexpression of the respective ETS family member [1,2]. TMPRSS2-ETS fusion is the first mechanistic explanation for dominant oncogene activation in a significant fraction of prostate cancers. TMPRSS2-ERG fusion is the most common rearrangement, which is significantly associated with prostate cancer-specific mortality [4]. Therefore, gene fusion status represents an important subclassification of prostate cancer from a biologic and a clinical standpoint.
TMPRSS2-ERG fusion occurs most frequently through intronic deletion between TMPRSS2 and ERG on 21q22.2- 3 [3]. None of the classic prostate carcinoma cell lines (DU145, PC3, or LNCaP) harbors TMPRSS2-ETS fusions. Understanding the functional biology of underlying gene fusion is currently limited to the androgen-dependent VCaP cell line that was established from the vertebral bone metastasis of a hormone-refractory prostate tumor [5]. The VCaP cell line represents an in vitro model for TMPRSS2-ERG gene fusion but has unusual genomic features in that it shows copy number gain on chromosome 21q, which has not been observed in > 300 prostate cancer samples studied to date [1,3,4]. In addition, VCaP has at least one normal TMPRSS2 and ERG gene, which makes it difficult to study TMPRSS2-ERG rearrangement in vitro in the presence of wild-type copies of both fusion partners. In VCaP, fusion transcripts encode TMPRSS2 fused to exon 4 of ERG, corresponding to the ERGa described by Tomlin et al. [1].
Here we present the androgen-independent NCI-H660 cell line as a novel in vitro model for prostate cancer. NCI-H660 cells harbor TMPRSS2-ERG gene fusion through a common intronic deletion on chromosome 21 in homozygous form. We propose it as a potential cell culture system to study and manipulate TMPRSS2-ERG fusion in vitro.
Materials and Methods
Cell Lines and Xenografts
The prostate cancer cell lines NCI-H660, PC3, DU145, LNCaP, 22Rv1, and CA-HPV-10 were purchased from the American Type Culture Collection (Manassas, VA) and maintained according to the provider's instructions. The VCaP cell line is derived from a vertebral metastatic lesion as part of the Rapid Autopsy Program at the University of Michigan [5]. Normal prostate stromal cells (PrSCs) were obtained from Cambrex Bio Science (East Rutherford, NJ).
LuCaP 23.1, 35, 49, 58, 73, 81, 86.2, 92.1, 93, 96, and 115 have been established by R.L.V. and have been previously described [3]. Of note, LuCaP 49 (established from an omental fat metastasis) and LuCaP 93 are hormone-insensitive [androgen receptor (AR)-negative] small cell prostate cancers with a neuroendocrine phenotype.
Tissue Samples
Lymph node specimens were collected from the radical prostatectomy series at the University of Ulm (Ulm, Germany) [6]. Hormone-refractory metastatic samples were part of the Rapid Autopsy Program for prostate cancer (University of Michigan Specialized Program of Research Excellence) and consist of histologically confirmed prostatic tumors involving solid organs (e.g., liver and lung) or distant lymph nodes [7,8]. All samples were collected as part of institutional review board protocols at each respective institution.
Polymerase Chain Reaction (PCR) and Real-Time Quantitative PCR (qPCR) for TMPRSS2-ERG Fusion Transcripts
We used the following primers for the detection of TMPRSS2-ERG, as described by Tomlins et al. [1]: TMPRSS2:ERG_f and TMPRSS2:ERG_r as reverse primers in ERG exon 4 for real-time qPCR, and exon 5–6_r as reverse primer in ERG exon 6 for reverse transcription (RT) PCR. Exon 5–6_f and exon 5–6_r primers were used for the detection of ERG. Total RNA was isolated with Trizol (Invitrogen, Carlsbad, CA) and reverse-transcribed using TaqMan reverse transcription reagent in the presence of random hexamers and oligo dT primers (Applied Biosystems, Foster City, CA).
RT-PCR amplifications were performed using 30 ng of cDNA as template in a final volume of 50 µl using the Platinum Taq DNA Polymerase kit (Invitrogen) at an annealing temperature of 63°C. Amplified PCR fragments were cloned and sequenced as described [9].
For qPCR, the amount of each target gene relative to GAPDH was determined for each sample using the comparative threshold cycle method (Applied Biosystems), as described [1]. For androgen stimulation experiments, 2x Power SYBR Green Master Mix (Applied Biosystems) and 25 ng of both forward and reverse primers were used for ERG, PSA, and HMBS [10]. A 2x TaqMan Universal PCR Master Mix, a final concentration of 900 nM forward and reverse primers, and a 250-nM probe were used for TaqMan TMPRSS2-ERGa detection using the following primers and probe: TM_ERGa forward, CTGGAGCGCGGCAGGAA; TM_ERGa reverse, CCGTAGGCACACTCAAACAACGA; TM_ERGa probe, TTATCAGTTGTGAGTGAGGAC.
Determining TMPRSS2-ERG Fusion and Deletion Status Using a Two-Stage Dual-Color Interphase Fluorescence In Situ Hybridization (FISH) Assay
To assess the TMPRSS2-ETS fusion status, we applied a two-stage dual-color break-apart FISH assay. First, we identified the rearrangement of TMPRSS2. Second, we tested for ERG break-apart to determine if ERG was the fusion partner for TMPRSS2. The ERG break-apart assay has been described [3]. The TMPRSS2 break-apart assay consisted of the biotin-16-dUTP-labeled BAC clone RP11-662D5 and the digoxigenin-11-dUTP-labeled BAC clone RP11-260O11. These probes span the neighboring centromeric and telomeric regions of the TMPRSS2 locus, respectively. Samples were mounted on Prolong Gold Antifade Reagent with DAPI (Invitrogen) and analyzed under a 60x oil immersion objective using an Olympus BX-51 (Olympus America Inc., Center Valley, PA) fluorescence microscope equipped with appropriate filters, a charge-coupled device camera, and the CytoVision FISH imaging and capturing software (Applied Imaging, San Jose, CA).
Fiber FISH
DNA was isolated from ∼ 5 x 106 cells followed by 24 hours of incubation with 300 µl of Puregene Cell Lysis Solution (Gentra Systems, Minneapolis, MN). About 10 µl of lysed DNA solution was stretched on a poly-l-lysine-coated slide with a coverslip, followed by fixation in ethanol [11]. The slides were hybridized for 16 hours at 37°C with probes specific to TMPRSS2 (RP11-120C17 labeled with biotin- 16-dUTP; red) and ERG (RP11-476D17 labeled with digoxigenin-11-dUTP; green).
Single-Nucleotide Polymorphism (SNP) Array Analysis
Genomic DNA was isolated from a total of 25 samples (cell lines, punch biopsies of xenografts, lymph nodes, and visceral organ metastases) according to standard procedures. DNA was hybridized according to the manufacturer's protocol (Affymetrix, Santa Clara, CA) [12], either to Affymetrix 10K or 50K Xba SNP arrays. Arrays were scanned with a GeneChip Scanner 3000 (Affymetrix). Data were analyzed using the informatics platform dChipSNP [13]. For each array type, preprocessing included array data normalization to a baseline array using a set of invariant probes and subsequent processing to obtain single intensity values for each SNP on each sample using a model-based (PM/MM) method [14]. We then merged the datasets on common SNPs (both represented on the 10K and the 50K arrays), ending with a set of ∼ 8K SNPs.
Androgen Stimulation
Treatments with the synthetic androgen R1881 and the AR antagonist flutamide were performed as described [1].
Results
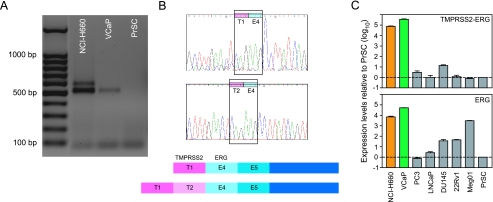
The NCI-H660 cell line was originally described as a small cell lung carcinoma [15]. Subsequently, it was correctly identified as an extrapulmonary small cell carcinoma originating from the prostate gland [16,17]. The line is derived from a lymph node metastasis taken from a patient before therapy. The patient was a 63-year-old white male who was diagnosed with extrapulmonary small cell carcinoma in 1983. He died from this cancer 18 days after the initial diagnosis and was autopsied with multiple metastatic sites (Bruce E. Johnson, personal communication, Dana Farber Cancer Institute, Boston, MA). NCI-H660 has been reported to be AR-negative by Western blot analysis, and its growth has been reported to be androgen-independent [18]. When we tested the NCI-H660 cell line for TMPRSS2-ERG fusion, we detected two RT-PCR products (Figure 1A). Cloning and sequencing identified the transcripts as ERGa, a previously reported fusion type [1], plus an additional isoform containing the first two exons of TMPRSS2, juxtaposed to exon 4 of ERG (fusion type VI according to Wang et al. [9]) (Figure 1B). The two-sequence verified TMPRSS2-ERG fusion transcripts in NCI-H660 are identical to the ones found in human tumor samples [9]. Real-time qPCR demonstrated expression of TMPRSS2-ERG gene fusion in the NCI-H660 cell line, leading to about a 6000-fold elevated ERG expression compared to normal PrSCs (Figure 1C). For comparison, the VCaP cell line shows about 50,000 times higher ERG expression, and Meg01, a leukemia cell line with known overexpression of the ERG oncogene, expresses about 3000 times more ERG than PrSCs (Figure 1C). The prostate carcinoma cell lines PC3, LNCaP, and DU145, and the xenograft cell line 22Rv1 neither harbor TMPRSS2-ERG fusion nor show high ERG expression levels (Figure 1C).
Figure 1.
Characterization of TMPRSS2-ERG fusion and ERG expression in the NCI-H660 prostate cancer cell line. (A) RT-PCR using a forward primer in exon 1 of TMPRSS2 and a reverse primer in exon 6 of ERG revealed two transcripts of TMPRSS2-ERG fusion in NCI-H660 (first lane). For comparison, VCaP expressed only one fusion transcript (second lane), whereas normal PrSCs did not harbor TMPRSS2-ERG fusion (third lane). (B) The identity of the fusion transcripts was verified by sequencing. The shorter transcript consisted of TMPRSS2 exon 1 (T1) fused to ERG exon 4 (E4). This transcript was also found in the VCaP cell line. The longer transcript was identified as TMPRSS2 exons 1 and 2 (T1 and T2), fused to ERG exon 4 (E4). Upper panel: sequencing profiles of T1-E4 and T2-E4 transcripts. Lower panel: corresponding schemes of the two fusion types. (C) Real-time qPCR for TMPRSS2-ERG expression (upper panel) and ERG expression (lower panel) on several cancer cell lines. The standard (PrSCs) was expressed as 1 (dashed line). Orange bars correspond to TMPRSS2-ERG fusion through deletion in NCI-H660, leading to a ∼ 6 x 103-fold overexpression of ERG. Green bars correspond to TMPRSS2-ERG fusion in VCaP, leading to a ∼ 5 x 104-fold overexpression of ERG. Grey bars correspond to the other prostate cancer cell lines tested (PC3, LNCaP, DU145, and 22Rv1) that do not harbor TMPRSS2-ERG fusion and show baseline or moderately elevated ERG expression. Meg01 is a leukemia cell line without TMPRSS2-ERG fusion, but with overexpression of ERG (∼ 3 x 103-fold), which served as a positive control for ERG expression.
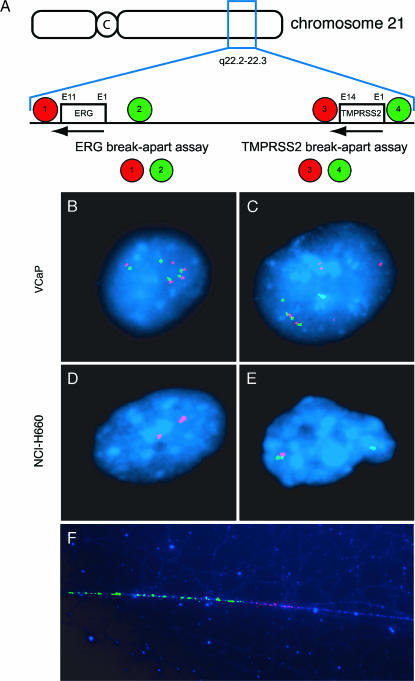
To confirm TMPRSS2-ERG rearrangement in the NCI-H660 cell line, we used a two-stage dual-color break-apart FISH assay testing first for TMPRSS2 and then for ERG break-apart as indirect evidence for TMPRSS2-ERG fusion. The indirect two-stage break-apart assessment is necessary because both genes are located so close to each other (a distance of only ∼ 3 megabases) that a fusion cannot be observed directly by a conventional FISH fusion assay. Fusion through deletion of the region between TMPRSS2 and ERG is indicated by loss of the probes located between the two genes [3]. FISH assays for TMPRSS2 and ERG break-apart are presented schematically in Figure 2A. These assays demonstrated that NCI-H660 cells have biallelic TMPRSS2-ERG fusion through genomic deletion between TMPRSS2 and ERG on chromosome 21q22.2-3 (Figure 2, D and E). The ERG break-apart assay showed that on the centromeric side of the deletion, both telomeric signals (probe 2, green; Figure 2A) are lost. The TMPRSS2 break-apart assay showed one centromeric signal (probe 3, red; Figure 2A), consistent with one deletion reaching this area and one deletion occurring at a more centromeric region. This observation of a homozygous genomic loss between TMPRSS2 and ERG on chromosome 21q22.2-3 has important implications for using this cell line as a prostate cancer model system, as no wild-type ERG allele is expressed. To date, we have observed one homozygous deletion in over 300 fusion-positive prostate cancers (unpublished observation). The fact that genomic deletions between ERG and TMPRSS2 show different borders is in concordance with our earlier observation that the deleted region is variable in length [3]. For the first time, we were able to visualize the fusion of TMPRSS2 and ERG on chromatin fibers of NCI-H660 cells by a high-resolution fiber FISH approach [11] using probes spanning specifically both loci (Figures 2F and W1).
Figure 2.
Dual-color TMPRSS2 and ERG break-apart FISH assays for the detection of TMPRSS2-ERG fusion in NCI-H660 and VCaP. (A) The scheme of the dual-color TMPRSS2 and ERG break-apart FISH assays explains the detection of TMPRSS2-ERG fusion. The location of the genes is indicated relative to the chromosome (boxes); the orientation of the genes is indicated by arrows. C = centromere; E = exon. For both assays, the relative location of differentially labeled telomeric and centromeric BAC probes is indicated by colored circles, with the color indicating the probe color in (B), (C), (D), and (E), and with the number identifying the BACs as follows: 1 = RP11-24A11; 2 = RP11-372O17; 3 = RP11-662D5; 4 = RP11-260O11. (B) FISH image of a VCaP interphase nucleus assessed by dual-color ERG break-apart assay (as illustrated in A). The nucleus contains several juxtaposed red and green signals for amplified wild-type alleles, and separated red and green signals indicating ERG insertion. (C) FISH image of a VCaP interphase nucleus assessed by dual-color TMPRSS2 break-apart assay (as illustrated in A). The nucleus contains several juxtaposed red and green signals for amplified wild-type alleles, and separated red and green signals indicating TMPRSS2 insertion. (D) FISH image of an NCI-H660 interphase nucleus assessed by dual-color ERG break-apart assay (as illustrated in A). The nucleus shows only two single red centromeric signals but no telomeric green signals. This is indicative of a fusion of TMPRSS2 with ERG through homozygous deletion of the intergenic region. (E) FISH image of an NCI-H660 interphase nucleus assessed by dual-color TMPRSS2 break-apart assay (as illustrated in A). The nucleus contains a juxtaposed red and green signal for the wild-type allele, and a single green signal indicating the deletion of one centromeric red probe. This is indicative of the fusion of TMPRSS2 with ERG through deletion of the intergenic region. (F) High-resolution FISH on chromatin fibers of NCI-H660 assessed by a TMPRSS2-specific probe (RP11-121A5; red) and an ERG-specific probe (RP11-476D17; green) (as illustrated in Figure W1).
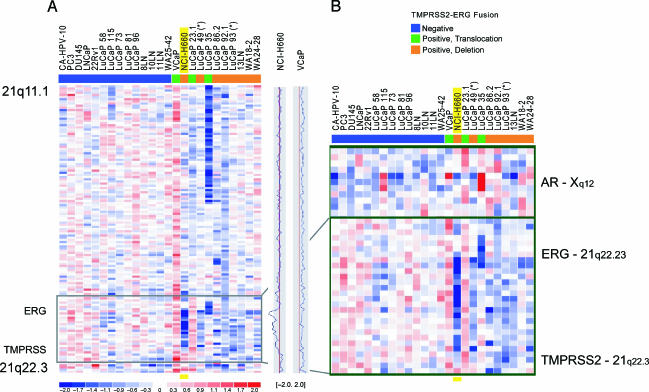
We then used oligonucleotide SNP arrays to further characterize the NCI-H660 cell line and to compare it with other cell lines, xenografts, and prostate cancer samples, including hormone-naïve and hormone-refractorymetastases (Figure 3, A and B). We identified the loss of about three megabases of genomic material on SNPs located between TMPRSS2 and ERG in the NCI-H660 cell line with a strong loss of signal intensity, corresponding to the homozygous deletion in these cells that was detected by FISH (Figure 2, D and E). Toward the telomeric side of the deletion in NCI-H660, the copy number ratio increases, suggesting that one allele is present in that area. This confirms the different lengths of the deletions that were detected by FISH. For comparison, the VCaP cell line showed significant copy number gain on chromosome 21q (Figure 3A). In agreement with SNP data, we found multiple signals for both wild-type TMPRSS2 and wild-type ERG in VCaP nuclei using independent TMPRSS2 and ERG breakapart FISH assays (Figure 2, B and C). Therefore, we conclude that the copy number gain on chromosome 21 leads to copy number gains of wild-type TMPRSS2 and ERG. Interestingly, the two small cell prostate cancer xenografts included in this study (LuCaP 49 and LuCaP 93) both showed TMPRSS2-ERG rearrangement through genomic deletion (denoted by * in Figure 3, A and B).
Figure 3.
Genomic loss between TMPRSS2 and ERG on chromosome 21q22.2-3 in prostate cancer cell lines, xenografts, and metastatic prostate cancer samples. 8K SNP data on a panel of 25 prostate cancer samples revealed genomic deletion between TMPRSS2 and ERG (21q22.2-3) in a subset of samples. (A) Twenty-five prostate cancer samples, including 7 cell lines, 11 xenografts, and 7 prostate cancer metastases, were analyzed for their TMPRSS2-ERG fusion status by qPCR and/or FISH and color-coded as described before [3] (blue, fusion-negative; green, fusion-positive through translocation; orange, fusion-positive through deletion). The plots on the right side of the panel represent the copy number ratio of the NCI-H660 and VCaP cell lines (vertical red lines represent baseline; no copy number variation). It is evident that VCaP shows copy number gain throughout the whole q arm of chromosome 21. (B) Magnification of the black framed box in (A), and status of the AR on chromosome X in these 25 samples. The blue signal in NCI-H660 corresponds to genomic copy number loss between TMPRSS2 and ERG. The strong intensity of this signal is consistent with homozygous loss, as demonstrated by FISH. The boundaries of the intronic deletion of NCI-H660 are very similar to the deletions seen in xenografts and tissue samples. Toward the telomeric side of the deletion in NCI-H660, the signal intensity is weaker, confirming different lengths of the deletions seen by FISH. The androgen-independent NCI-H660 cell line shows a loss in the region of the AR, whereas VCaP, which is known to be androgen-responsive, shows genomic gain in this area.
Because NCI-H660 is derived from an androgen-independent small cell prostate tumor and was shown to be negative for AR on both transcript and protein levels [18], we also analyzed the AR locus on chromosome X for all samples (Figure 3B). NCI-H660 had a loss in that region, as most of the other xenografts and tissue samples in that panel. SNP data confirmed the negative AR status of this cell line.
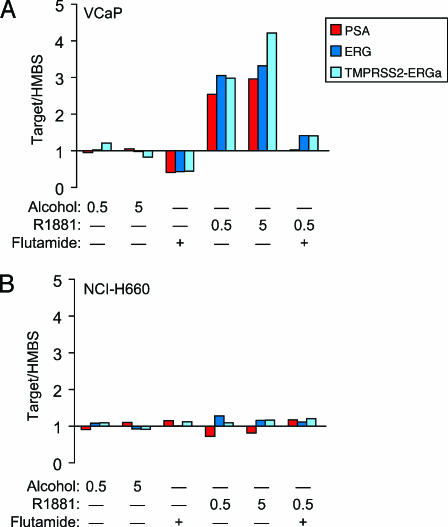
To investigate whether TMPRSS2-ERG fusion results in the androgen regulation of ERG in AR-negative NCI-H660 cells, we assessed the expression of ERG by qPCR in androgen-treated NCI-H660 and VCaP cells. The androgen-sensitive VCaP cell line has been shown to respond to androgen stimulation with increased ERG expression sensitive to bicalutamide and flutamide [1] (Figure 4A). As expected, NCI-H660 does not respond to androgen stimulation with increased expression of PSA or ERG, nor is it sensitive to the AR antagonist flutamide (Figure 4B).
Figure 4.
The androgen stimulation of the VCaP and NCI-H660 prostate cancer cell lines carrying TMPRSS2-ERG fusion. PSA (red bars), ERG (exons 5 and 6; dark blue bars), and TMPRSS-ERG (TMPRSS2 exon 1 to ERG exon 4; light blue bars) expression relative to HMBS in androgen-sensitive VCaP cells (A) and androgen-insensitive NCI-H660 cells (B) was assessed by qPCR. Cell lines were incubated with vehicle or 10 µM of the AR antagonist flutamide for 2 hours before treatment for 24 hours with 0.5 or 5 nM of the synthetic androgen R1881 or vehicle, as indicated. Relative amounts of PSA, ERG, or TMPRSS2-ERG per HMBS were compared for VCaP and NCI-H660.
Discussion
A significant percentage of prostate cancers express a signature gene fusion of the 5′ region of androgen-regulated TMPRSS2 gene to an ETS family transcription factor, most commonly the ERG gene. This is the first demonstration of constitutive oncogene activation in prostate cancer. However, the functional consequences of this chromosomal rearrangement are difficult to predict and study. Reliable preclinical in vitro models, which use homogenous human cellular material that can be studied in large quantities over time, are needed. The NCI-H660 cell line is a novel in vitro model for prostate cancer that closely relates to the molecular signature of clinical cases. We demonstrate that NCI-H660 has an intronic deletion on chromosome 21 that was found to be the most common mechanism underlying TMPRSS2-ERG fusion [3]. This deletion is observed on both alleles as a homozygous loss. Therefore, NCI-H660 expresses neither TMPRSS2 nor ERG in the native genomic context.
NCI-H660 does not express AR and is representative of TMPRSS2-ERG-positive androgen-independent prostate cancer. It has been shown that both TMPRSS2-ERG fusion and the associated deletion occur in androgen-independent prostate cancers and metastases that have no functional AR [3]. The membrane-bound serine protease TMPRSS2 can be expressed in both androgen-dependent and androgen-independent tumors (i.e., this usually androgenresponsive gene can be uncoupled from androgen control) [19]. Androgen-independent expression of TMPRSS2-ERG in the NCI-H660 cell line is in line with this observation. NCI-H660 as an androgen-independent model system might facilitate the search for androgen-independent factors and pathways influencing TMPRSS2-ERG fusion. In contrast to a recent report showing that TMPRSS2-ERG gene fusion is not expressed in AR-negative prostate cancer specimens [20], we and others [21] have found TMPRSS2-ERG and ERG expression in the AR-negative NCI-H660 cell line. This implies that TMPRSS2-ERG fusion is constitutively stimulated in this androgen-independent prostate cancer cell line. Further efforts will be directed at investigating which factors can activate TMPRSS2-ERG fusion with subsequent ERG overexpression, using the NCI-H660 cell line as an in vitro model for TMPRSS2-ERG fusion through deletion.
In summary, the NCI-H660 cell line is a potent in vitro tool with a genotypic profile that reflects the TMPRSS2-ERG rearrangement found in human prostate cancer. The NCI-H660 cell line should be a valuable model for prostate cancer and has the potential to provide important pathogenic insight and guidance for therapeutic strategies.
Supplementary Material
Acknowledgements
The authors are grateful to Xiao-Wei Sun and Laura Johnson for technical support critical to this study, to Tobias Junt for critical reading of the manuscript, to Levi Garraway for providing us with SNP data, and to Bruce E. Johnson (Department of Medical Oncology, Dana Farber Cancer Institute) for communicating clinical information about the origin of NCI-H660 and for critical reading of the manuscript.
Abbreviations
- AR
androgen receptor; FISH, fluorescence in situ hybridization
- PrSC
prostate stromal cell
- PSA
prostate-specific antigen
- qPCR
quantitative polymerase chain reaction
- RT-PCR
reverse transcription-polymerase chain reaction
- SNP
single-nucleotide polymorphism
Footnotes
This work was supported by the National Institutes of Health Prostate Specialized Program of Research Excellence (SPORE) at the Dana Farber/Harvard Cancer Center (NCI P50 CA090381) and the University of Michigan (NCI P50 CA69568 and R01AG21404 to M.A.R. and A.M.C.), the Swiss Foundation for Medical-Biological Grants SSMBS (SNF 1168 to K.D.M.), the Prostate Cancer Foundation (to F.D.), Deutsche Forschungsgemeinschaft DFG PE1179 (to S.P.), the Department of Defense Fellowship Awards (PC061474 to S.P. and PC040638 to R.B.), and the NW Prostate Cancer SPORE (to R.L.V.).
This article refers to supplementary material, which is designated by “W” (i.e., Figure W1) and is available online at www.bcdecker.com.
References
- 1.Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 2.Tomlins SA, Mehra R, Rhodes DR, Smith LR, Roulston D, Helgeson BE, Cao X, Wei JT, Rubin MA, Shah RB, et al. TMPRSS2:ETV4 gene fusions define a third molecular subtype of prostate cancer. Cancer Res. 2006;66:3396–3400. doi: 10.1158/0008-5472.CAN-06-0168. [DOI] [PubMed] [Google Scholar]
- 3.Perner S, Demichelis F, Beroukhim R, Schmidt FH, Mosquera JM, Setlur S, Tchinda J, Tomlins SA, Hofer MD, Pienta KG, et al. TMPRSS2:ERG fusion-associated deletions provide insight into the heterogeneity of prostate cancer. Cancer Res. 2006;66:8337–8341. doi: 10.1158/0008-5472.CAN-06-1482. [DOI] [PubMed] [Google Scholar]
- 4.Demichelis F, Fall K, Perner S, Andren O, Schmidt F, Setlur SR, Hoshida Y, Mosquera J-M, Pawitan Y, Lee C, et al. TMPRSS2:ERG gene fusion associated with lethal prostate cancer. Oncogene. 2007 doi: 10.1038/sj.onc.1210237. 2007 Jan 22 [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 5.Korenchuk S, Lehr JE, MClean L, Lee YG, Whitney S, Vessella R, Lin DL, Pienta KJ. VCaP, a cell-based model system of human prostate cancer. In Vivo. 2001;15:163–168. [PubMed] [Google Scholar]
- 6.Hofer MD, Kuefer R, Huang W, Li H, Bismar TA, Perner S, Hautmann RE, Sanda MG, Gschwend JE, Rubin MA. Prognostic factors in lymph node-positive prostate cancer. Urology. 2006;67:1016–1021. doi: 10.1016/j.urology.2005.10.055. [DOI] [PubMed] [Google Scholar]
- 7.Rubin MA, Putzi M, Mucci N, Smith DC, Wojno K, Korenchuk S, Pienta KJ. Rapid (“warm”) autopsy study for procurement of metastatic prostate cancer. Clin Cancer Res. 2000;6:1038–1045. [PubMed] [Google Scholar]
- 8.Shah RB, Mehra R, Chinnaiyan AM, Shen R, Ghosh D, Zhou M, Macvicar GR, Varambally S, Harwood J, Bismar TA, et al. Androgen-independent prostate cancer is a heterogeneous group of diseases: lessons from a Rapid Autopsy Program. Cancer Res. 2004;64:9209–9216. doi: 10.1158/0008-5472.CAN-04-2442. [DOI] [PubMed] [Google Scholar]
- 9.Wang J, Cai Y, Ren C, Ittmann M. Expression of variant TMPRSS2/ERG fusion messenger RNAs is associated with aggressive prostate cancer. Cancer Res. 2006;66:8347–8351. doi: 10.1158/0008-5472.CAN-06-1966. [DOI] [PubMed] [Google Scholar]
- 10.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-7-research0034. (RESEARCH0034) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, Scherer SW, Lee C. Detection of large-scale variation in the human genome. Nat Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 12.Kennedy GC, Matsuzaki H, Dong S, Liu WM, Huang J, Liu G, Su X, Cao M, Chen W, Zhang J, et al. Large-scale genotyping of complex DNA. Nat Biotechnol. 2003;21:1233–1237. doi: 10.1038/nbt869. [DOI] [PubMed] [Google Scholar]
- 13.Lin M, Wei LJ, Sellers WR, Lieberfarb M, Wong WH, Li C. dChipSNP: significance curve and clustering of SNP-array-based loss-of- heterozygosity data. Bioinformatics. 2004;20:1233–1240. doi: 10.1093/bioinformatics/bth069. [DOI] [PubMed] [Google Scholar]
- 14.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci USA. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carney DN, Gazdar AF, Bepler G, Guccion JG, Marangos PJ, Moody TW, Zweig MH, Minna JD. Establishment and identification of small cell lung cancer cell lines having classic and variant features. Cancer Res. 1985;45:2913–2923. [PubMed] [Google Scholar]
- 16.Johnson BE, Whang-Peng J, Naylor SL, Zbar B, Brauch H, Lee E, Simmons A, Russell E, Nam MH, Gazdar AF. Retention of chromosome 3 in extrapulmonary small cell cancer shown by molecular and cytogenetic studies. J Natl Cancer Inst. 1989;81:1223–1228. doi: 10.1093/jnci/81.16.1223. [DOI] [PubMed] [Google Scholar]
- 17.Lai SL, Brauch H, Knutsen T, Johnson BE, Nau MM, Mitsudomi T, Tsai CM, Whang-Peng J, Zbar B, Kaye FJ, et al. Molecular genetic characterization of neuroendocrine lung cancer cell lines. Anticancer Res. 1995;15:225–232. [PubMed] [Google Scholar]
- 18.van Bokhoven A, Varella-Garcia M, Korch C, Johannes WU, Smith EE, Miller HL, Nordeen SK, Miller GJ, Lucia MS. Molecular characterization of human prostate carcinoma cell lines. Prostate. 2003;57:205–225. doi: 10.1002/pros.10290. [DOI] [PubMed] [Google Scholar]
- 19.Lin B, Ferguson C, White JT, Wang S, Vessella R, True LD, Hood L, Nelson PS. Prostate-localized and androgen-regulated expression of the membrane-bound serine protease TMPRSS2. Cancer Res. 1999;59:4180–4184. [PubMed] [Google Scholar]
- 20.Hermans KG, van Marion R, van Dekken H, Jenster G, van Weerden WM, Trapman J. TMPRSS2:ERG fusion by translocation or interstitial deletion is highly relevant in androgen-dependent prostate cancer, but is bypassed in late-stage androgen receptor-negative prostate cancer. Cancer Res. 2006;66:10658–10663. doi: 10.1158/0008-5472.CAN-06-1871. [DOI] [PubMed] [Google Scholar]
- 21.Clark J, Merson S, Jhavar S, Flohr P, Edwards S, Foster CS, Eeles R, Martin FL, Phillips DH, Crundwell M, et al. Diversity of TMPRSS2-ERG fusion transcripts in the human prostate. Oncogene. 2006 doi: 10.1038/sj.onc.1210070. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.