Abstract
Di-myo-inositol 1,1′-phosphate (DIP) is a major osmoprotecting metabolite in a number of hyperthermophilic species of archaea and bacteria. Although the DIP biosynthesis pathway was previously proposed, genes encoding only two of the four required enzymes, inositol-1-phosphate synthase and inositol monophosphatase, were identified. In this study we used a comparative genomic analysis to predict two additional genes of this pathway (termed dipA and dipB) that remained missing. In Thermotoga maritima both candidate genes (in an originally misannotated locus TM1418) form an operon with the inositol-1-phosphate synthase encoding gene (TM1419). A predicted inositol-mono-phosphate cytidylyltransferase activity was directly confirmed for the purified product of T. maritima gene dipA cloned and expressed in Escherichia coli. The entire DIP pathway was reconstituted in E. coli by cloning of the TM1418–TM1419 operon in pBAD expression vector and confirmed to function in the crude lysate. 31P NMR and MS analysis revealed that DIP synthesis proceeds via a phosphorylated DIP intermediate, P-DIP, which is generated by the dipB-encoded enzyme, now termed P-DIP synthase. This previously unknown intermediate is apparently converted to the final product, DIP, by an inositol monophosphatase-like phosphatase. These findings allowed us to revise the previously proposed DIP pathway. The genomic survey confirmed its presence in the species known to use DIP for osmoprotection. Among several newly identified species with a postulated DIP pathway, Aeropyrum pernix was directly proven to produce this osmolyte.
Keywords: comparative genomics; di-myo-inositol-1,3-phosphate biosynthesis; Thermotoga maritima
The most common mechanism of osmoadaptation in microorganisms involves the accumulation of specific organic osmolytes, so-called compatible solutes, amino acids, sugars, and polyols, that can be taken up from the environment or synthesized de novo (1–3). In thermophiles and hyperthermophiles, compatible solutes are generally different from those found in mesophiles (4), and many of them additionally contribute to thermoprotection.
Di-myo-inositol 1,1′-phosphate (DIP) is one of the major compatible solutes in a number of thermophilic archaea of the genera Pyrococcus, Methanococcus, Thermococcus, and Archaeoglobus and bacteria belonging to the genera Aquifex, Rubrobacter, and Thermotoga, including Thermotoga maritima and Thermotoga neapolitana (4–7). DIP levels are highly increased at supraoptimal growth temperatures in both Methanococcus igneus and Thermotogales, suggesting that this solute also plays a thermoprotective role (6, 8).
Based on the study in M. igneus (9) and the reported stereochemistry of DIP (10), the pathway of DIP biosynthesis was originally proposed to occur in four steps: (i) synthesis of l-myo-inositol-1-phosphate (L-I-1-P) from glucose-6-phosphate by NAD+-dependent L-I-1-P synthase [inositol-1-phosphate synthase (IPS)]; (ii) hydrolysis of some of the L-I-1-P by inositol monophosphatase (IMP); (iii) coupling of the L-I-1-P with CTP to form CDP-inositol by CTP:inositol monophosphate cytidylyltransferase (IMPCT); and (iv) generation of DIP by condensation of CDP-inositol with inositol via a DIP synthase (DIPS).
Two of these enzymes, IPS and IMP, are not solely committed to DIP biosynthesis as inositol derivatives play an important role in cell wall biogenesis, signaling, and other pathways in a broad spectrum of species including animals, plants, fungi, and mesophilic bacteria (11–13). Both enzymes are broadly conserved, and examples of each have been well characterized biochemically and structurally, including IMP and IPS from the hyperthermophiles Methanocaldococcus jannaschii (14), Archaeoglobus fulgidus (15–19), T. maritima (20), and Thermococcus kodakaraensis (21).
However, two additional enzymes required for DIP biosynthesis have not yet been identified in any organism (3). We used a subsystems-based comparative genomic analysis [as implemented in the SEED platform (22)] to explore the DIP biosynthesis pathway (further termed DIP pathway) in thermophilic species with completely sequenced genomes. Analysis of chromosomal clustering and co-occurrence profiles implicated two previously uncharacterized genes (termed dipA and dipB; see Fig. 1A) as candidates for the two missing genes of the DIP pathway. Both representative genes from T. maritima were cloned and expressed in Escherichia coli. The in vitro reconstitution of the entire pathway in the crude lysate of E. coli and the analysis of the individual enzymatic steps revealed the role of each gene in the DIP pathway. Whereas the dipA gene was confirmed to encode a predicted IMPCT enzyme, the product of the dipB gene involved in the next step of the pathway appeared to generate a previously unknown phosphorylated DIP intermediate (P-DIP) precursor via the condensation of CDP-inositol with L-I-1-P. The existence of this novel enzymatic activity (termed here PDIPS) instead of the previously postulated DIPS suggested a revision to the previously proposed version of the pathway, where the last step is a dephosphorylation of P-DIP by a phosphatase of the IMP family (Fig. 1B).
Fig. 1.
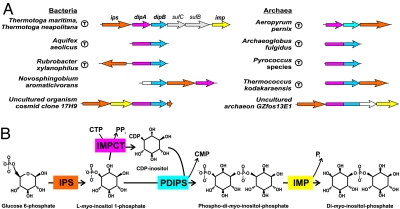
Reconstruction of the DIP biosynthesis pathway in prokaryotic genomes. (A) Genome context of DIP pathway genes in bacterial and archaeal genomes. (Hyper)thermophilic species are marked by a circle with “T.” Orthologs are shown by matching colors of individual genes or respective segments of fused genes. Predicted novel genes (dipA and dipB) are fused (dipAB) or occur in chromosomal clusters with each other and, often, with the two known genes of the DIP pathway (ips and imp). (B) A diagram of the revised DIP synthesis pathway. IPS, EC 5.5.1.4; IMPCT, 2.7.7.–; PDIPS, 2.7.8.–; IMP, 3.1.3.25.
The comparative genomic analysis of the entire collection of species with completely sequenced genomes integrated in the SEED database (http://theseed.uchicago.edu/FIG/index.cgi) confirmed the presence of the DIP pathway in all archaeal and bacterial species where it was previously detected by biochemical methods and allowed us to infer this pathway in several additional microorganisms. Among them, an archaeon Aeropyrum pernix was experimentally confirmed to produce this osmolyte.
Results
Prediction of Novel DIP Pathway Genes by Comparative Genomics.
Genome context analysis including chromosomal gene clustering, protein fusions, and occurrence profiles were applied to the available genomes of the DIP-producing microorganisms to identify candidate genes for the last two steps of the DIP biosynthesis pathway. The detailed results of this analysis are captured in the SEED subsystem available online (http://theseed.uchicago.edu/FIG/subsys.cgi; see “Di-Inositol-Phosphate biosynthesis”) and illustrated in Fig. 1 and Table 1.
Table 1.
Phylogenetic distribution of the DIP biosynthesis pathway in bacteria and archaea
| Organisms | Four-step DIP biosynthesis pathway |
DIP production | |||
|---|---|---|---|---|---|
| IPS | IMPCT | PDIPS | IMP | ||
| T. maritima | TM1419 | TM1418a | TM1418b | TM1415 | Known |
| T. neapolitana | 020_1986 | 020_1988 | 020_1989 | 020_1993 | Known |
| Aquifex aeolicus | aq_1763 | aq_1367N | aq_1367C | aq_1983 | Known* |
| Rubrobacter xylanophilus | Rxyl021258 | Rxyl021259N | Rxyl021259C | Rxyl021693 | Known |
| Novosphingobium aromaticivorans | Saro3074 | Saro3073 | Saro3075 | Saro2521 | Predicted |
| Uncultured bacterium 17H9 | 17H9_20 | 17H9_22N | 17H9_22C | 17H9_21 | Predicted |
| A. pernix | APE1517 | APE1514 | APE1516 | APE1798 | Confirmed |
| Ar. fulgidus | AF1794 | AF0263N | AF0263C | AF2372 | Known |
| Pyrococcus horikoshii | PH1605 | PH1219N | PH1219C | PH1897 | Predicted |
| Pyrococcus abyssi | PAB1989 | PAB2433N | PAB2433C | PAB0189 | Predicted |
| Pyrococcus furiosus | PF1616 | PF1058N | PF1058C | PF2014 | Known |
| Thermococcus kodakaraensis | TK2278 | TK2279N | TK2279C | TK0787 | Known* |
| Uncultured archaeon GZfos13E1 | GZ13E1_33 | GZ13E1_32N | GZ13E1_32C | GZ13E1_31 | Predicted |
Genes encoding four enzymes of DIP pathway are shown by standard GenBank identificators. N and C superscripts denote N-terminal and C-terminal domains in the fusion IMPCT–PDIPS proteins. Organisms experimentally shown to produce DIP are indicated in the last column (4, 7). DIP production in A. pernix was predicted and confirmed in this study.
*Cases when DIP production was confirmed in subspecies related to but distinct from those with available genomic sequences.
An observed chromosomal clustering of two previously uncharacterized genes (e.g., APE1514 and APE1516 in A. pernix) with an ips gene (e.g., APE1517) in a number of thermophilic archaea and bacteria (Fig. 1A) provided us with the first strong evidence of their possible role in the DIP biosynthesis pathway. In some of these species, the aforementioned genes are colocalized with the imp gene, revealing an additional link to this pathway. Moreover, orthologs of these two hypothetical genes (termed here dipA and dipB) are always either located next to each other on the chromosome or, more often, fused together to form a single gene (dipAB), further supporting their strong functional coupling.
An additional scanning of the metagenomic libraries of environmental samples revealed clusters of contiguous ips, imp, and dipAB genes in two uncultured organisms: an archaeon from Eel River sediment from northwestern California and a bacterium from a deep-sea sediment of east Pacific nodule province, China (Fig. 1A).
Additional evidence supporting the possible involvement of these genes in DIP synthesis came from their occurrence only in the genomes containing ips and imp genes (Table 1). This co-occurrence profile is not in contradiction with the existence of many microbial genomes that contain ips and imp genes but do not contain dipA-dipB genes. For example, in Mycobacterium tuberculosis and other actinobacteria IPS plays an essential role in the production of the major inositol-containing thiol and cell wall lipoglycans (23).
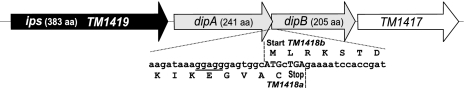
Notably, dipA and dipB genes identified in this study are absent from the current GenBank annotation of the T. maritima genome as the respective locus TM1418 was deemed to contain an authentic frame shift. An alternative interpretation was available in the SEED database (corresponding protein IDs: fig|243274.1.peg.1893 and fig|243274.1.peg.1892 at http://theseed.uchicago.edu/FIG/index.cgi) (Fig. 2) suggesting the presence of the two protein-encoding genes (here referred to as TM1418a and TM1418b) overlapping by 7 bp. Although most species appear to contain a fusion of dipA and dipB genes within a single reading frame dipAB, the existence of two separate genes in T. maritima was additionally supported by the presence of single-gene orthologs in A. pernix and Novosphingobium aromaticivorans.
Fig. 2.
A reconstructed DIP pathway operon in T. maritima. The novel dipA and dipB genes identified by comparative genomic analysis overlap by 7 bp. Start and stop codons are in capital letters. A possible ribosomal-binding site is underlined.
A long-range homology analysis allowed us to tentatively assign both genes specific functional roles in the DIP synthesis pathway. The dipA gene in T. maritima encodes a putative 241-aa cytoplasmic protein that belongs to the large family of sugar phosphate nucleotidyltransferases (COG1213). Among characterized members of this protein family are glucose-1-phosphate cytidylyltransferase from Salmonella typhimurium, 2-C-methyl-d-erythritol 4-phosphate cytidylyltransferase from E. coli, and mannose-1-phosphate guanylyltransferase from yeast [see supporting information (SI) Fig. 8]. Based on these observations, we tentatively assigned the role of missing IMPCT to the dipA gene product.
The T. maritima dipB gene encodes a 205-aa protein that belongs to the CDP-alcohol phosphatidyltransferase class-I protein family (COG0558). A BLAST search revealed similarity of DipB to phosphatidylinositol synthases from eukaryotes, phosphatidylglycerophosphate synthases from bacteria, and phosphatidylserine synthases from yeast and bacteria (see SI Fig. 9). This similarity is particularly striking in the region between amino acids 70 and 100, showing a perfect match to the consensus sequence D(X)2DG(X)2AR(X)2N(X)5G(X)3D(X)3D, characteristic of several alcohol phosphatidyltransferases (24). Based on the overall similarity of chemical reactions catalyzed by the previously characterized proteins from the CDP-alcohol phosphatidyltransferase family, we conjectured that DipB is the most likely candidate for the role of the missing DIPS.
Experimental Characterization of Novel DIP Pathway Enzymes from T. maritima.
To assess the activity of the product of the T. maritima gene dipA predicted to encode a soluble IMPCT enzyme, this gene was cloned and overexpressed in E. coli (see SI Materials and Methods). The recombinant protein with the N-terminal His6 tag was purified by NiNTA chromatography and enzymatically characterized. Because the product of T. maritima gene dipB was predicted to be membrane-bound and, hence, insoluble, its activity was tested in a crude lysate of E. coli where this gene was expressed as a part of the T. maritima operon ips-dipA-dipB cloned in the pBAD expression vector. The results of these enzymatic analyses are described in the following subsections. However, so far we failed to express DipB enzyme alone in the active form, which may reflect the importance of an apparent tight association of DipA and DipB proteins (that are more often fused in one polypeptide in other species) for the proper membrane insertion and activity of DipB enzyme.
Enzymatic Characterization of IMPCT.
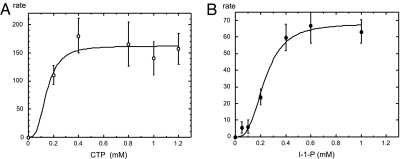
The predicted enzymatic activity of the purified recombinant T. maritima IMPCT enzyme was confirmed by the formation of CDP-inositol as characterized by 31P NMR spectroscopy and mass spectrometry (MS). The IMPCT reaction converts the CTP and L-I-1-P into CDP-inositol and inorganic pyrophosphate. We used the detection of the latter product for quantitative enzymatic characterization of IMPCT using the coupled colorimetric assay for pyrophosphate. Because of significant substrate inhibition observed at higher concentrations of L-I-1-P (>1 mM), only an estimate of the apparent Km ≈ 0.2 mM was obtained for one of its specific substrates, L-I-1-P, at fixed saturating concentration of the second substrate (1.5 mM CTP). With L-I-1-P fixed at 1.0 mM, the Vmax at 80°C was 160 ± 10 mmol min−1 mg−1, and the apparent Km for CTP was 0.16 ± 0.04 mM (Fig. 3). The IMPCT appears to be specific for l-inositol-1-phosphate because no CDP-inositol was produced when d-inositol-1-phosphate (Cayman Chemical) was incubated with the recombinant enzyme. This enzyme is also highly selective for CTP because no activity could be detected by the pyrophosphatase coupled assay in the presence of ATP, GTP, or UTP. Moreover, no detectable adduct formation could be detected by 31P-NMR spectra even after extensive incubation with any of these alternative NTPs. Deoxy-CTP could serve as a substrate, although the rate was ≈20-fold lower (with 1.5 mM L-I-1-P and 3 mM deoxy-CTP) than with CTP. IMPCT activity was reduced >25-fold in the absence of added MgCl2, and it was fully suppressed by 5 mM EDTA. These observations confirm the participation of Mg2+ in its catalytic mechanism, characteristic of this nucleotidyltransferase family.
Fig. 3.
IMPCT kinetics. (A) Dependence of rate (μmol·min−1·mg−1) at 80°C for CDP-inositol formation by recombinant IMPCT on CTP concentration with L-I-1-P fixed at 1.0 mM. (B) Dependence of the IMPCT rate on L-I-1-P concentration with the CTP fixed at 1.5 mM. The Mg2+ cofactor concentration was 5 mM, and the assay pH was 8.0. Data are shown only up to 1.2 mM substrate because at higher concentrations rates are significantly decreased, suggestive of substrate inhibition. Data were fit with a cooperative model, and n = 3 (the average of best fit data for each substrate). The lower specific activities observed for the IMPCT dependence on L-I-1-P is because these rates were acquired with an older preparation of IMPCT.
Identification of PDIPS Enzyme and Reconstitution of the Entire DIP Pathway.
Because the product of the dipB gene was expected to be a membrane-protein, its activity was tested in the crude lysate of E. coli carrying the operon ips-dipA-dipB (TM1419, TM1418a, and TM1418b) from T. maritima. This experimental setup allowed us to combine testing of individual enzymatic steps with the pathway reconstitution as none of the respective enzymatic activities or metabolites are present in the E. coli host.
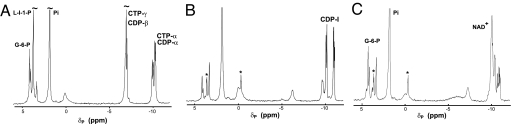
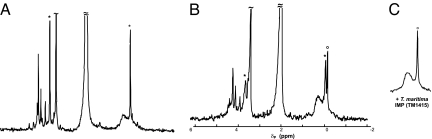
The 2,546-bp PCR-amplified segment of T. maritima chromosome was cloned into the pBAD-TOPO expression vector and transformed to E. coli BL21. The crude cell lysate was obtained after induction with arabinose. Product formation was monitored by 31P NMR after incubation with respective substrates at 80°C and compared with available standards and with the samples prepared using the same host strain of E. coli carrying the same vector with the β-galactosidase gene as a negative control. The 1H-coupled 31P-NMR analysis of the reaction mixture obtained after incubation of the cell lysate with L-I-1-P and CTP confirmed the formation of CDP-inositol and of a novel phosphodiester compound with a chemical shift similar to that of DIP (Fig. 4). The identity of this compound as P-DIP was later confirmed by MS analysis and by its conversion to DIP upon incubation with an exogenous IMP phosphatase (see below).
Fig. 4.
31P NMR (202.2 MHz) 1H-coupled spectra of soluble fractions from incubations (at 80°C) of E. coli BL21-DIP lysed cells with DIP precursors. (A) Control mixture with 3 mM L-I-1-P, 6 mM CTP, 3 mM MgCl2, and 6 mM EDTA incubated for 2 h. (B) Lysed cells incubated for 2 h with 3 mM L-I-1-P, 6 mM CTP, and 3 mM MgCl2. (C) Lysed cells incubated for 2.5 h with 3 mM glucose-6-phosphate, 1.5 mM NAD+, 3 mM CTP, and 3 mM MgCl2. After centrifugation of cell debris, EDTA (6 mM) was added to all samples before analysis by 31P NMR spectroscopy. The asterisks indicate the appearance of the DIP-related peaks; the one at −0.2 ppm is the phosphodiester, and the one at ≈3 ppm is the phosphomonoester.
The results obtained in this study provided an unambiguous assignment of the observed P-DIP synthase (PDIPS) activity to the product of the dipB gene as the properties of IPS and IMPCT encoded by the two other genes of the operon were characterized separately (in ref. 17 and in this study, respectively). Addition of the free inositol to the mixture did not lead to any additional formation of DIP, confirming that formation of the P-DIP intermediate via condensation of CDP-inositol with L-I-1-P (and not with free inositol as for a previously postulated DIPS enzyme) is the main and likely the only possible route of DIP synthesis. These findings allowed us to propose a revised version of the DIP pathway shown in Fig. 1B.
Only lysates of cells that contained all three genes of a T. maritima DIP operon displayed all of the resonances (including a number of novel peaks) consistent with the proposed pathway and not present in control samples (Fig. 4). Under the conditions used, most of the L-I-1-P was converted to CDP-inositol as monitored by two resonances at −10.2 and −11.0 ppm (each appearing as an AB quartet). Likewise, most of the added CTP was consumed (little intensity for the β-phosphorus remained at −22 ppm, not shown in these graphs). A new triplet resonance appeared at −0.2 ppm, consistent with synthesis of a phosphodiester such as DIP; another new resonance ≈4 ppm (a doublet in proton-coupled spectra) consistent with a phosphomonoester had the same integrated intensity as the new phosphodiester peak. The same products in the 31P-NMR spectrum were produced when 3 mM glucose-6-phosphate was incubated with 1.5 mM NAD+, 6 mM CTP, and 3 mM MgCl2 (Fig. 4C), indicating that all three enzymes function under these conditions.
Neither of the two peaks in the 31P-NMR spectra corresponding to CDP-inositol and P-DIP appeared in the absence of Mg2+ or in the presence of 6 mM EDTA. In the presence of Mg2+, both of them increased with the time of incubation and exhibited strong temperature dependence. Under our experimental conditions and at the temperatures examined (2 h of incubation at 65, 75, and 85°C), nearly all of the L-I-1-P was converted to CDP-inositol. Activation energy of its conversion to P-DIP is 58 ± 17 kJ/mol, as estimated from the Arrhenius plot (see SI Fig. 10).
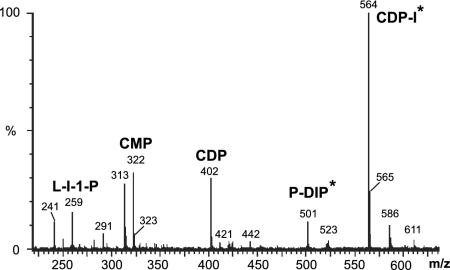
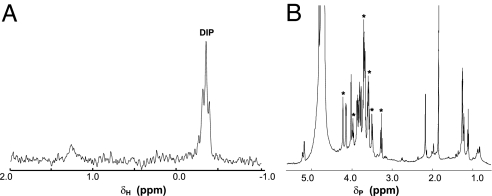
Electrospray MS analysis of the reaction mixture revealed two major peaks not present in control samples at 564.2 m/z, consistent with CDP-I, and at 501.2 m/z, consistent with the hypothesized P-DIP (Fig. 5). The peak corresponding to the genuine DIP (421 m/z) appeared only after longer incubation, likely because of the action of endogenous phosphatases. To test the hypothesis that IMP-catalyzed dephosphorylation of the P-DIP precursor may be the last step of the DIP pathway, we first used the exogenous IMP from M. jannaschii (25). The samples containing P-DIP were incubated at 85°C with 20 μg of pure recombinant IMP in the presence of an additional 5 mM MgCl2 (added to offset the EDTA added to obtain 31P spectra that clearly showed P-DIP) for 20 min. The reaction was stopped by cooling and addition of 7 mM EDTA before obtaining a 31P spectrum. This treatment led to the appearance of a new peak 0.2 ppm upfield of the original one (and identical to the resonance of the authentic DIP), accompanied by a dramatic decrease of the phosphomonoester peak (Fig. 6). Similar results were obtained by using the recombinant IMP-like phosphatase from the T. maritima (TM1415) expressed and purified as previously described (20). In this case, all of the P-DIP was converted to DIP under the same incubation conditions.
Fig. 5.
Mass spectrum of the soluble fraction from incubation (at 80°C) of E. coli BL21-DIP lysed cells that had been incubated for 2 h with 3 mM L-I-1-P, 6 mM CTP, and 3 mM MgCl2. Identities of the major peaks are indicated. The asterisks highlight ions detected only in spectra from incubations with cell lysates containing the three DIP biosynthetic enzymes IPS, IMPCT, and DIPS.
Fig. 6.
1H-decoupled 31P NMR (202.2 MHz) spectra of the phosphomonoester and phosphodiester region of supernatants from incubation of E. coli BL21-DIP lysed cells with L-I-1-P and CTP at 85°C for 2.5 h (A), then treated with M. jannaschii IMP (B) or T. maritima IMP (C). In A the asterisks mark the new peaks associated with activity of the DIP gene products. Note that IMPase activity reduces the original phosphodiester peak and leads to a new one (marked with ○) with the same chemical shift as authentic DIP. The DIP-related phosphomonoester peak is also significantly reduced.
Interestingly, the formation of both CDP-inositol and P-DIP occurred only when incubation was carried out with an “uncleared” lysate, in suspension containing membrane fragments, consistent with the expectation that PDIPS is an integral membrane protein. The apparent absence of IMPCT activity in the cleared lysate (after removal of cell debris by centrifugation) indicates that this enzyme, perfectly soluble when overexpressed as a single gene, may form a tight complex with membrane-bound PDIPS when expressed as a part of an operon.
Verification of the Predicted DIP Production in A. pernix.
The comparative genomic analysis allowed us to infer the presence of the DIP pathway in a number of species with completely sequenced genomes where DIP production has not been previously reported (Table 1). To test this conjecture, we checked for DIP production in the hyperthermophilic archaeon A. pernix. 31P-NMR spectrum of the ethanol extract of these cells showed one major resonance at −0.3 ppm consistent with DIP, without any indication of P-DIP presence (Fig. 7A). The 1H NMR spectrum (Fig. 7B) showed resonances consistent with DIP and mannosylglycerate. The identity of the resonances for these two compounds was confirmed by TOCSY and HMQC experiments (6, 26). 13C chemical shifts were obtained from an HMQC experiment and were consistent with published 13C and 1H shifts for these molecules (27). The MS analysis revealed the presence of 421.2 m/z peak consistent with the DIP anion and the 267.2 m/z peak likely corresponding to mannosylglycerate.
Fig. 7.
NMR spectra of A. pernix cell extracts. (A) 31P NMR (202.2 MHz) spectrum of an ethanol extract of A. pernix JCM 9820. (B) 1H NMR (202.2 MHz) spectrum of the same extract. 1H resonances belonging to DIP (determined from a TOCSY spectrum) are indicated by asterisks; other resonances in this extract in the region of DIP belong to mannosylglycerate.
Discussion
Although our current knowledge of major metabolic pathways and respective genes in well studied model bacteria such as E. coli is nearly comprehensive, it can be only partially projected on the rapidly growing number of sequenced genomes of more distant species. Deep-branched bacteria and archaea, including many extremophiles, represent a particular challenge because of a substantial number of unique pathways supporting their divergent lifestyles. In this study we used comparative genomics to address one such challenge and to predict two previously uncharacterized (missing) genes completing the biosynthetic pathway of DIP, a major osmoprotecting metabolite in a number of thermophilic archaea and bacteria (Fig. 1B).
The key evidence implicating two candidate genes termed dipA and dipB with the DIP pathway was provided by their clustering on the chromosome with the genes encoding IPS and IMP (Fig. 1A), the two previously characterized components of this pathway. Additional support for these conjectures came from the observed co-occurrence profile and a frequent fusion of dipA and dipB into one contiguous gene dipAB. This type of genome context analysis has been successfully applied for identification of missing genes in many metabolic pathways (for review, see ref. 28). General class functions tentatively assigned by homology to the protein products of genes dipA (sugar phosphate nucleotidyltransferase, COG1213) and dipB (CDP-alcohol phosphatidyltransferase, COG0558) were consistent with their expected functional roles in the DIP pathway.
Enzymatic activities of proteins encoded by the genes dipA and dipB were experimentally assessed by cloning and expression of the representative genes from T. maritima. Remarkably, these missing genes in T. maritima were additionally missed at the level of primary gene annotations as the respective chromosomal locus TM1418 was excluded from the original list of predicted proteins based on the postulated “authentic frame shift.” Nevertheless, our results suggest that both protein encoding genes (termed here TM1418a and TM1418b to preserve the original nomenclature) should be added to the public annotations of the T. maritima genome. An overlap of the two adjacent reading frames (Fig. 2) likely reflects a translational coupling often observed for tightly coregulated prokaryotic genes (29).
An enzymatic analysis of the overexpressed and purified product of gene dipA (TM1418a) confirmed its predicted IMPCT activity, including previously inferred features, such as preference for CTP over other NTPs and a strict requirement of Mg2+ for activity. On the other hand, the analysis of the enzymatic activity of the product of gene dipB (TM1418b) provided some unexpected results. In keeping with the initial version of the DIP pathway (9), this enzyme was expected to generate DIP by a direct condensation of CDP-inositol (produced from L-I-1-P by IMPCT) with free myo-inositol (produced from L-I-1-P by IMP). However, the reconstitution of the pathway in the crude lysate of E. coli carrying the entire ips-dipA-dipB operon cloned in the expression vector revealed the formation of the previously unknown intermediary metabolite, P-DIP. Therefore, a condensation reaction catalyzed by the product of the dipB gene actually involves CDP-inositol and L-I-1-P rather than free inositol. Based on these results, the respective enzyme was termed PDIPS. The last step in the revised version of the DIP pathway is the dephosphorylation of the newly identified intermediate (Fig. 1B). This reaction is likely catalyzed by an IMP-like phosphatase as confirmed by the formation of DIP upon addition of the pure recombinant IMP enzymes from M. jannaschii and T. maritima to the reaction mixture containing P-DIP.
These findings illustrate a synergy between bioinformatics and experimental aspects of gene discovery. Although the comparative genomics techniques allowed us to accurately predict two novel genes of the DIP pathway, the consequent experimental analysis of the recombinant enzymes was required to confirm one of the inferred activities (IMPCT) and, more importantly, to revise an original prediction of the second activity (PDIPS) and of the entire pathway.
The phylogenetic pattern of occurrence of the DIP pathway genes is in perfect correlation with the previously established distribution of compatible solutes in thermophiles and hyperthermophiles (Table 1). Moreover, projection of the DIP biosynthesis subsystem across the whole collection of integrated complete and almost-complete genomes (http://theseed.uchicago.edu/FIG/subsys.cgi?user=master:&ssa_name=Di-Inositol-Phosphate_biosynthesis&request=show_ssa) allowed us to infer this pathway in seven archaea and six bacteria (including two uncultured species; see Fig. 1A). This inference was further validated in this study by the experimental detection of DIP in the archaeon A. pernix. Also in agreement with the available data, DIP pathway signature genes dipA and dipB are absent from the available genomes of thermophiles where the presence of DIP was not detected in Thermus thermophilus, Pyrobaculum aerophilum, Methanopyrus kandleri, Sulfolobus solfataricus, and Methanobacterium thermoautotrophicum.
Materials and Methods
Genomes and Bioinformatics Tools.
The bulk of comparative genomic analysis, including subsystem encoding and genome context analysis (chromosomal clustering and phylogenetic profiling), was performed by using the SEED genomic database and tools implemented therein (http://theseed.uchicago.edu/FIG/index.cgi) (22). Complete and nearly complete genomes of bacteria and archaea were from the GenBank database (www.ncbi.nlm.nih.gov/GenBank). Preliminary genome sequence data for T. neapolitana was obtained from The Institute for Genomic Research. We used ClustalX to construct multiple protein alignments (30), Psi-BLAST (31) (www.ncbi.nlm.nih.gov/BLAST) to conduct long-range similarity searches, the PFAM (32) (www.sanger.ac.uk/Software/Pfam) and Conserved Domain databases (33) (www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml) to identify conserved functional domain, and TMpred (34) (www.ch.embnet.org/software/TMPRED_form.html) to predict the occurrence of transmembrane segments.
Cloning and Expression of T. maritima ips-dipA-dipB Operon.
The 2,546-bp segment containing ips and two novel predicted genes, dipA and dipB, was PCR-amplified by using T. maritima MSB8 genomic DNA and oligonucleotide primers ATGgtcaaggtcctgatcctcgg and TCAcctgttgagcaccagaagttc. This fragment was cloned in the pBAD-TOPO expression vector (Invitrogen), and the DNA sequence of the entire fragment was verified. The expression of the operon-encoded genes was performed in E. coli strain BL-21 with arabinose induction. Crude cell lysates [0.2 g cells in 1 ml of 50 mM Tris·HCl (pH 7.5)] were heated at 80°C for 5 min to denature most host cell proteins. This suspension was then incubated at 80°C with substrates for DIP synthesis (usually L-I-1-P and CTP but in several instances with glucose-6-phosphate, NAD+ as well as CTP) and 3 mM Mg2+ for 2–3 h. The sample was cooled to room temperature and centrifuged to remove any cell debris. The supernatant was frozen, lyophilized, and redissolved in D2O with 6 mM EDTA for NMR studies.
Enzyme Assays.
IMPCT activity was assayed by measurement of pyrophosphate production using the EnzChek Pyrophosphate Assay Kit from Invitrogen (35). The recombinant IMPCT (typically 0.5–1.0 mg) was incubated at 80°C with I-1-P (0.2–1.5 mM) and CTP (0.2–1.2 mM) in 100 mM Tris·HCl (pH 8.0) with 5 mM MgCl2 added, for 0.5, 1, and 2 min. For each time point, the reaction was stopped by placing the sample on ice. The pyrophosphate assay used 50 ml of 20× reaction buffer, 200 ml of 2-amino-6-mercapto-7-methylpurine ribonucleoside as substrate for 10 ml of purine nucleoside phosphorylase, 10 ml of inorganic pyrophosphatase, 530 ml of distilled water, and 200 ml of IMPCT sample. This mixture was incubated at room temperature for 1 h. The absorbance at 360 nm (from the 2-amino-6-mercapto-7-methylpurine) for samples compared with a pyrophosphate standard curve was used to calculate IMPCT-specific activity.
NMR Spectroscopy.
Cell extracts were prepared by suspending 0.2 g (wet weight) of cell pellets in 1 ml of 50 mM Tris·HCl (pH 7.5); the suspension was heated to 80°C for 5 min to inactivate most E. coli proteins. The suspension was then incubated with 3 mM L-I-1-P that was generated by incubating glucose-6-P in the presence of NAD+ (Sigma–Aldrich) and Mg2+ with the recombinant IPS from A. fulgidus at 85°C (15), 6 mM CTP, and 3 mM MgCl2 at 80°C for 2–2.5 h. The sample was centrifuged at 14,000 rpm for 15 min to pellet all particulate matter. The supernatant was frozen in liquid N2, lyophilized, and then dissolved in D2O containing 6 mM EDTA. This soluble extract was examined by 1H-coupled 31P-NMR to assess CDP-inositol and DIP formation (36). All NMR spectra were acquired on a Varian INOVA 500 spectrometer as described previously (9).
Electrospray Mass Spectrometry.
Electrospray mass spectrometry of the cell lysates soon after preparation was performed by using negative ion mode in acetonitrile/H2O, 1:1 with 0.1% ammonia using an LCT Classic spectrometer.
Growing of A. pernix for DIP Production.
A. pernix strain JCM 9820, obtained from the American Type Culture Collection (Manassas, VA), was grown in Difco Marine Broth 2216 to which Na2S2O3·5H2O (0.1 g per 100 ml of medium) had been added. The medium was inoculated with ice chips of frozen A. pernix, and the flasks containing the cells were placed in a shaking water bath at 92°C and 120 rpm (37). After a 30-h lag time, the cells began to grow exponentially (doubling time ≈5–6 h). Cells were harvested after ≈40 h after the lag period when the cells reached OD660 ≈0.53; at this point they appeared to be in stationary phase. The cell suspension was centrifuged and the pellet resuspended in 2% NaCl. After centrifugation, the pellet was mixed with 70% ethanol, and the mixture was placed in a bath sonicator for 20 min to ensure lysis of the cells. After centrifugation, the pellet was extracted five times with 70% ethanol and all of the supernatants combined, frozen, and lyophilized. The dried sample was solubilized in D2O, and both 1H and 31P-NMR spectra were obtained.
Supplementary Material
Acknowledgments
We thank Scott Lesley at the Joint Center for Structural Genomics for providing T. maritima chromosomal DNA and Ross Overbeek and the whole team of developers of the SEED genomic platform at the Fellowship for Interpretation of Genomes and Argonne National Laboratory for providing guidance and access to integrated genomes and tools for their comparative analysis. This study was partially supported by the National Institutes of Health Roadmap National Technology Centers on Networks and Pathways Grant 5 U54 RR020843-03, by Department of Energy Biosciences DE-FG02–91ER20025 (to M.F.R.), and by National Institutes of Health Grant 1R01 GM64481 (to B.S.).
Abbreviations
- DIP
di-myo-inositol 1,1′-phosphate
- L-I-1-P
l-myo-inositol-1-phosphate
- DIPS
DIP synthase
- IMP
inositol monophosphatase
- IMPCT
CTP:inositol monophosphate cytidylyltransferase
- IPS
inositol-1-phosphate synthase
- PDIPS
P-DIP synthase.
Note.
When this manuscript was in preparation, Borges et al. (38) proposed a similar revision of the DIP pathway based on the detection of the respective enzymatic activities and intermediary metabolites in crude extracts of A. fulgidus by NMR. The results of their study are in agreement with the results presented here and confirm that the DIP synthesis occurs via a P-DIP intermediate.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0609279104/DC1.
References
- 1.Roberts MF. Front Biosci. 2004;9:1999–2019. doi: 10.2741/1366. [DOI] [PubMed] [Google Scholar]
- 2.Muller V, Spanheimer R, Santos H. Curr Opin Microbiol. 2005;8:729–736. doi: 10.1016/j.mib.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 3.Roberts MF. Saline Syst. 2005;4:5. doi: 10.1186/1746-1448-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Santos H, da Costa MS. Methods Enzymol. 2001;334:302–315. doi: 10.1016/s0076-6879(01)34478-6. [DOI] [PubMed] [Google Scholar]
- 5.Scholz S, Sonnenbichler J, Schafer W, Hensel R. FEBS Lett. 1992;306:239–242. doi: 10.1016/0014-5793(92)81008-a. [DOI] [PubMed] [Google Scholar]
- 6.Ciulla RA, Burggraf S, Stetter KO, Roberts MF. Appl Environ Microbiol. 1994;60:3660–3664. doi: 10.1128/aem.60.10.3660-3664.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Neves C, da Costa MS, Santos H. Appl Environ Microbiol. 2005;71:8091–8098. doi: 10.1128/AEM.71.12.8091-8098.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martins LO, Carreto LS, Da Costa MS, Santos H. J Bacteriol. 1996;178:5644–5651. doi: 10.1128/jb.178.19.5644-5651.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen L, Spiliotis ET, Roberts MF. J Bacteriol. 1998;180:3785–3792. doi: 10.1128/jb.180.15.3785-3792.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van Leeuwen SH, van der Marel GA, Hensel R, van Boom JH. Recl Trav Chim Pays Bas. 1994;113:335–336. [Google Scholar]
- 11.Majerus PW. Annu Rev Biochem. 1992;61:225–250. doi: 10.1146/annurev.bi.61.070192.001301. [DOI] [PubMed] [Google Scholar]
- 12.Drobak BK. Biochem J. 1992;288:697–712. doi: 10.1042/bj2880697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Movahedzadeh F, Smith DA, Norman RA, Dinadayala P, Murray-Rust J, Russell DG, Kendall SL, Rison SC, McAlister MS, Bancroft GJ, et al. Mol Microbiol. 2004;51:1003–1014. doi: 10.1046/j.1365-2958.2003.03900.x. [DOI] [PubMed] [Google Scholar]
- 14.Stec B, Yang H, Johnson KA, Chen L, Roberts MF. Nat Struct Biol. 2000;7:1046–1050. doi: 10.1038/80968. [DOI] [PubMed] [Google Scholar]
- 15.Chen L, Zhou C, Yang H, Roberts MF. Biochemistry. 2000;39:12415–12423. doi: 10.1021/bi001517q. [DOI] [PubMed] [Google Scholar]
- 16.Stieglitz KA, Johnson KA, Yang H, Roberts MF, Seaton BA, Head JF, Stec B. J Biol Chem. 2002;277:22863–22874. doi: 10.1074/jbc.M201042200. [DOI] [PubMed] [Google Scholar]
- 17.Stieglitz KA, Yang H, Roberts MF, Stec B. Biochemistry. 2005;44:213–224. doi: 10.1021/bi048267o. [DOI] [PubMed] [Google Scholar]
- 18.Neelon K, Wang Y, Stec B, Roberts MF. J Biol Chem. 2005;280:11475–11482. doi: 10.1074/jbc.M500469200. [DOI] [PubMed] [Google Scholar]
- 19.Wang YK, Morgan A, Stieglitz K, Stec B, Thompson B, Miller SJ, Roberts MF. Biochemistry. 2006;45:3307–3314. doi: 10.1021/bi052467y. [DOI] [PubMed] [Google Scholar]
- 20.Chen L, Roberts MF. Appl Environ Microbiol. 1999;65:4559–4567. doi: 10.1128/aem.65.10.4559-4567.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sato T, Imanaka H, Rashid N, Fukui T, Atomi H, Imanaka T. J Bacteriol. 2004;186:5799–5807. doi: 10.1128/JB.186.17.5799-5807.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Overbeek R, Begley T, Butler RM, Choudhuri JV, Chuang HY, Cohoon M, de Crecy-Lagard V, Diaz N, Disz T, Edwards R, et al. Nucleic Acids Res. 2005;33:5691–5702. doi: 10.1093/nar/gki866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jackson M, Crick DC, Brennan PJ. J Biol Chem. 2000;275:30092–30099. doi: 10.1074/jbc.M004658200. [DOI] [PubMed] [Google Scholar]
- 24.Yamashita S, Nikawa J. Biochim Biophys Acta. 1997;1348:228–235. doi: 10.1016/s0005-2760(97)00102-1. [DOI] [PubMed] [Google Scholar]
- 25.Chen L, Roberts MF. Appl Environ Microbiol. 1998;64:2609–2615. doi: 10.1128/aem.64.7.2609-2615.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lamosa P, Martins LO, da Costa MS, Santos H. Appl Environ Microbiol. 1998;64:3591–3598. doi: 10.1128/aem.64.10.3591-3598.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Silva Z, Borges N, Martins LO, Wait R, da Costa MS, Santos H. Extremophiles. 1999;3:163–172. doi: 10.1007/s007920050112. [DOI] [PubMed] [Google Scholar]
- 28.Osterman A, Overbeek R. Curr Opin Chem Biol. 2003;7:238–251. doi: 10.1016/s1367-5931(03)00027-9. [DOI] [PubMed] [Google Scholar]
- 29.Eyre-Walker A. J Bacteriol. 1995;177:5368–5369. doi: 10.1128/jb.177.18.5368-5369.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Finn RD, Mistry J, Schuster-Bockler B, Griffiths-Jones S, Hollich V, Lassmann T, Moxon S, Marshall M, Khanna A, Durbin R, et al. Nucleic Acids Res. 2006;34:D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marchler-Bauer A, Anderson JB, Cherukuri PF, DeWeese-Scott C, Geer LY, Gwadz M, He S, Hurwitz DI, Jackson JD, Ke Z, et al. Nucleic Acids Res. 2005;33:D192–D196. doi: 10.1093/nar/gki069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hofmann K, Stoffel W. Biol Chem Hoppe-Seyler. 1993;374:166. doi: 10.1515/bchm3.1993.374.7-12.507. [DOI] [PubMed] [Google Scholar]
- 35.Upson RH, Haugland RP, Malekzadeh MN, Haugland RP. Anal Biochem. 1996;243:41–45. doi: 10.1006/abio.1996.0479. [DOI] [PubMed] [Google Scholar]
- 36.Roberts MF. Methods Microbiol. 2006;35:615–647. [Google Scholar]
- 37.Milek I, Cigic B, Skrt M, Kaletunc G, Ulrih NP. Can J Microbiol. 2005;51:805–809. doi: 10.1139/w05-060. [DOI] [PubMed] [Google Scholar]
- 38.Borges N, Goncalves LG, Rodrigues MV, Siopa F, Ventura R, Maycock C, Lamosa P, Santos H. J Bacteriol. 2006;188:8128–8135. doi: 10.1128/JB.01129-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.