Abstract
We determined the crystal structure of the Escherichia coli nucleoid-associated HUαβ protein by x-ray diffraction and observed that the heterodimers form multimers with octameric units in three potential arrangements, which may serve specialized roles in different DNA transaction reactions. It is of special importance that one of the structures forms spiral filaments with left-handed rotations. A negatively superhelical DNA can be modeled to wrap around this left-handed HUαβ multimer. Whereas the wild-type HU generated negative DNA supercoiling in vitro, an engineered heterodimer with an altered amino acid residue critical for the formation of the left-handed spiral protein in the crystal was defective in the process, thus providing the structural explanation for the classical property of HU to restrain negative supercoils in DNA.
Keywords: HU-multimers, nucleoid, x-ray crystallography
The bacterial nucleoid is distinguished from the eukaryotic chromosome structure by the ostensible lack of an ordered nucleosomal structure, which characteristically influences gene expression pattern. In Escherichia coli, the 4.6-Mb circular DNA chromosome with a 1.6-mm circumference is condensed 1,000-fold to a diameter of 1 μm in the nucleoid (1). Although it has been suggested that DNA compaction in the nucleoid is caused by extensive negative supercoiling, the involvement of several proteins, e.g., HU, HNS, FIS, and SeqA, also in the compaction is now widely accepted (1, 2). However, the precise structure of the nucleoid, and the exact role of these proteins in the nucleoid formation is unknown. Despite the deficiency in our knowledge about the nucleoid physical structure and any influence a structure may have on gene expression, it was found that specific amino acid changes in the E. coli HU qualitatively change the morphology and the transcription profile of cells profoundly (3). The latter observation suggests that HU may be involved in nucleoid structure in an influential way not appreciated previously.
HU contains two homologous subunits, α and β, 9.5 kDa each. HU also exists as αα and ββ homodimers, in different stages of cell growth (4). Although HU binds to DNA nonspecifically, it shows a propensity for binding to distorted, bent, or Holliday-junction DNA, an ability that may aid DNA compaction (5). HU and topoisomerase I acting in concert also regulate DNA supercoiling both in vivo and in vitro (6, 7). In HU-deficient mutants, partially relaxed DNA is compensated by altered levels of topoisomerase I or DNA gyrase (8, 9). Indeed, E. coli nucleoids in the absence of HU are significantly enlarged in volume (10).
That specific mutants of HU protein qualitatively change the cell transcription profile suggests that the HU-mediated DNA structural change results in a defined architecture (topological or otherwise) that guides the transcription pattern in cell. Although HU also plays specific roles in many DNA metabolic reactions (11–14), by well studied mechanisms, a relation between an HU-influenced nucleoid structure and global gene expression profile is elusive. It was also suggested that HU antagonizes, not aids, DNA supercoiling (15).
We report here the structure of the E. coli HUαβ heterodimer crystals determined by x-ray diffraction. The crystal structure of the heterodimer, unlike the structures of HU homodimers from E. coli and other bacteria, show stacked dimers with octameric repeats in three different forms, two of which exhibit distinct spiral multimers. The replacement of the β-E38 residue, critical for interdimeric interface in the left-handed spiral, by alanine affected the negative DNA supercoiling capacity in vitro. We discuss the significance of the findings in nucleoid biology.
Results
X-Ray Structure.
The diffraction data were collected from a crystal with a tetragonal space group I41 with unit cell dimensions of a = b = 82.915 Å and c = 61.048 Å with one heterodimer in an asymmetric unit. The parameters of data collection and refinement are presented in Table 1. The local environments of the residues 12 and 13 support the assignment of the αβ heterodimers. In the α-subunit, E12 and K13 form two salt bridges with β-K18 of an adjacent dimer and β-E34 of the same dimer, respectively (Fig. 1a). In the β-subunit, A12 and G13 have no crystal contact (Fig. 1b). The 2Fo − Fc omit map of α-E12 and α-K13 shows apparent electron density at the site of the deleted residues at cutoff of 1.0 and the corresponding Fo − Fc omit map shows positive electron density at cutoff of 3.0, especially on the location of the side chain of α-K13 (Fig. 1c). The Ramachandran plot shows that 92.6% of residues fall in the most favored region, 5.8% fall in the additional allowed region, and 1.7% fall in the generously allowed region [supporting information (SI) Fig. 8].
Table 1.
Statistics of data collection, molecular replacement, and refinement
| Diffraction datasets | |
| Resolution, Å | 2.5 |
| Unique reflection | 7,730 |
| Rmerge* (outer bin) | 0.039 (0.399) |
| Compl., % (outer bin) | 92.8 (72.9) |
| Redundancy | 6.2 |
| Refinement | |
| No. of residues | 167 |
| No. of nonhydrogen atoms | 1,068 |
| No. of solvents | 24 |
| No. of Ni | 1 |
| No. of Cl | 1 |
| Resolution range, Å | 20–2.5 |
| R† (outer) | 0.226 (0.357) |
| Rfree† (outer) | 0.258 (0.405) |
| rmsd bond, Å | 0.017 |
| rmsd bond angle, ° | 1.514 |
| rmsd chiral, ° | 0.088 |
*Rmerge = ∑|I − 〈I〉|/∑I, where I and 〈I〉 are the observed intensity and the mean intensity of the same reflection from multiple measurements. Summation is over all reflections.
†R = ∑(|Fo| − |Fc|)/∑|Fo|, where Fo is the measured structure factor and Fc is the calculated structure factor from the model.
Fig. 1.

Snapshots of electron density maps of residues 12 and 13 as finger prints of the α and β chains. (a) E12 and K13 of the α-subunit. (b) A12 and G13 of β. (c) The same region of a with E12 and K13 omitted. The 2Fo − Fc map cutoff at 1 is in green. The positive Fo − Fc map cutoff at 3 is in red. The red and blue text indicates separate HU dimer molecules.
The assignment of the Ni- and a Cl-ions was as follows. A very massive electron density was found that does not fit with any amino acid residues and other molecules in the mother liquor except Ni- and Cl-ions. Detectable anomalous signals for heavier atoms were observed in this spot. Residues 55–74 of α-chain and 56–73 of β that are known as DNA binding arms, as well as the residue I16 located in the turn between helix 1 and helix 2 in β, and the side chains of α-K18, α-K73, β-K9, β-D15, β-K18, β-K75, and β-K83 are invisible. Although required for crystallization, the DNA fragment was missing in the electron density map. The crystals when dissolved in buffer showed a spectrum with significant OD at 260 nm, whereas pure HUαβ solution had no distinct absorbance at that wavelength. It is possible that a trace amount of DNase activity, intrinsic or extraneous, in the HU preparation damaged the DNA so as not to show any ordered electron density. Indeed, we detected a very low DNase activity in the preparation when a 32P-labeled pBR332 plasmid DNA was incubated overnight at 37°C with the HU preparation supplemented with 10 mM MgCl2. Because different DNA duplexes (SI Table 2) used in crystallization enabled HUαβ to form the same type of crystals, perhaps DNA molecules acted as a “general salt” in crystallization.
General Structure.
The secondary structural elements of the HUαβ heterodimer are comparable to the previously reported structures of HU homodimers of Thermotoga maritima, Bacillus stearothermophilus, Anabaena, and HUαα of E. coli (16–20). The superimposition of the structures of the α- and the β-subunits gives an rms deviation (rmsd) of 0.895 Å per 69 Cα atoms. The structures of the E. coli HUαβ heterodimer reported here and that of the αα homodimer (16) are superimposable with an rmsd of 0.807 Å per 140 Cα atoms. The superimposition of two αβ heterodimers, when the α- and β-subunits of one dimer are reversed, gave an rmsd of 0.905 Å per 139 Cα atoms. Both α and β chains in the heterodimer consist of three helices and three antiparallel strands: helix 1 (residues 2–13), helix 2 (17–38), helix 3 (82–90), strand 1 (40–45), strand 2 (47–54), and strand 3 (75–81).
The B-factor profiles indicate that the β-subunit residues, particularly the first 20 and the last 10, are slightly less ordered than the ones in the α-subunit in the crystal packing environment. The folding of a monomer is maintained by core hydrophobic residues; residues L6, I7, I10, A11, A21, and L25 interact with each other to generate a V-shape between helix 1 and helix 2, whereas residues α-I32 (β-V32), L36, V42, F50, V52, and P77 hold helix 2 and the three strands in a given conformation in both subunits. Most of these residues also participate in dimerization (see below), making the formation of monomers and dimers interdependent.
Dimerization.
The two monomers interwind into a dimer as a unit. As reported in the HU homodimers (16–20), the heterodimers also hold extensive intersubunit hydrophobic interactions. The residues that contribute to the hydrophobic core environment are listed in Fig. 2. However, in contrasts to the symmetric intersubunit configuration of the E. coli αα homodimer (16), the intersubunit interactions of the αβ heterodimer reported here are asymmetric (SI Fig. 9). The details of the dimerization interface are provided in SI Materials and Methods.
Fig. 2.
Residues that contribute to the hydrophobic core of E. coli HU. Those in red directly contribute to dimerization. The underlined residues are different in α and β chains. The italicized residues are those whose counterparts in the opposite chain are polar or charged.
The sequence difference and the resulting asymmetric dimeric interactions, but not the basic folding, are driving forces for the multimeric arrangement of HUαβ heterodimers, and thus the potential for the formation of higher-ordered nucleoprotein complexes as described below.
HU Multimers and Interdimeric Interfaces.
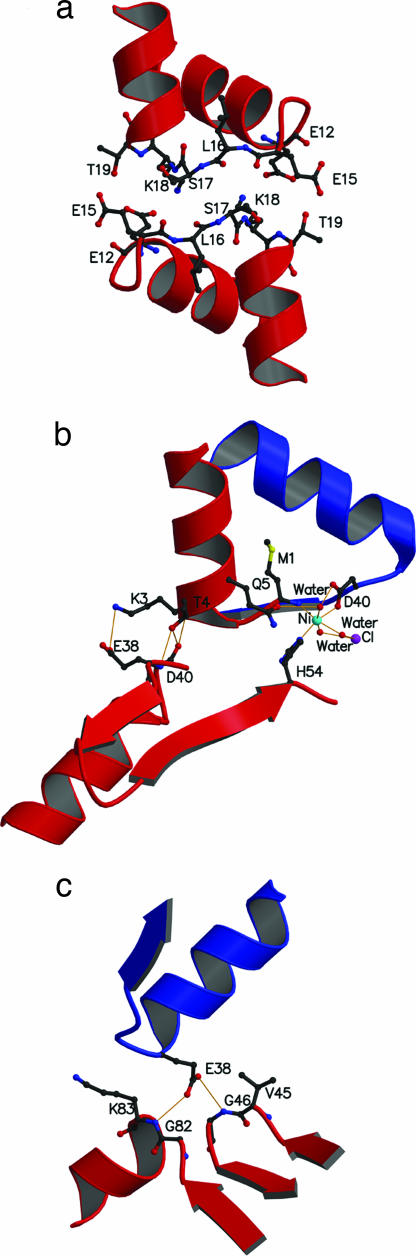
The crystal packing data showed that the heterodimer forms oligomers in three possible arrangements, I, II, and III (Fig. 3). A total of three interdimeric interfaces (a, b, and c) exist in the higher oligomers (Fig. 4). In interface “a,” which has the buried solvent-accessible surface of 566.1 Å2, van der Waals interactions are involved between residues 15–19 (E, L, S, K, and T) in-between helix 1 and helix 2 of an α-subunit with the same in an adjacent but symmetrically inverted α-subunit in an antiparallel mode (Fig. 4a). The side chains of α-E12 and α-E15 interact with the side chains of α-K18 and α-T19, respectively. This interface is present in arrangement I only (Fig. 3 a and b). Interestingly, the residue I16 located in the turn between helices 1 and 2 in β is invisible in our heterodimer structure indicating that the turn may potentially interact with another protein.
Fig. 3.
Top views of crystal packing: an octameric structure with three possible arrangements. (a) The crystal packing lattice. (b) Arrangement I octamer. (c) Arrangement II octameric unit of a right-handed spiral multimer. (d) Arrangement III octameric unit of a left-handed spiral multimer. Red and blue represent the α- and β-subunits, respectively.
Fig. 4.
Diagrams of the interdimeric interfaces of E. coli HUαβ protein. (a) Interface “a” involves interactions between residues (E12 and E15-L16-S17-K18-T19) of two adjacent symmetry-related α-subunits located in the turn between helix 1 and helix 2. (b) Interface “b” shows extensive interactions involving a Ni-ion, a Cl-ion, three H2O molecules, α-M1, α-K3, α-T4, α-Q5, and α-D40 from one dimer, and α-E38, α-H54, and β-D40 from the adjacent dimer. (c) Interface “c” involves the side chain of the β-E38, which rotates to form two H-bonds with the backbone amines of α-G46 and α-K83, respectively, from the interacting adjacent dimer. Red and blue represent segments of the subunits of α and β, respectively.
Interface “b” has the largest buried solvent accessible surface (684.9 Å2) and shows extensive interactions mainly between two adjacent α-subunits (Fig. 4b). First, a Ni-ion, identified by RefMac5 software, participates in the interdimer interactions at this interface by forming coordination bonds with the α-M1 amide group, the β-D40 side chain, the α-H54 side chain of the adjacent α-subunit, and three water molecules. Two of the water molecules are H-bonded with a Cl-ion on the opposite side of the Ni-ion. The α-Q5 side chain also participates in the Ni H-bond network. Thus, the Ni-ion participates both in monomer–monomer and dimer–dimer interactions. In addition, α-K3 forms a salt-bridge with α-E38 of the adjacent dimer. The α-T4 forms three H-bonds with the adjacent dimer; the α-T4 amide and side chain form H-bonds with the α-E38 carbonyl, whereas the α-T4 side chain forms an H-bond with the α-D40 amide. Interface “b” is present in both arrangements I and II (Fig. 3 a and c).
The buried solvent accessible surface of interface “c” is 290.1 Å2. Interface “c” has two H-bonds between the β-E38 side chain of one dimer and the two amide groups between α-G46 and α-K83 of another adjacent dimer (Fig. 4c). β-E38 and β-G39 make a turn between helix 2 and strand 1 that serves as a turning point interwinding the monomers in dimers. The α-G46 residue is located in the hydrophobic β-turn between strands 1 and 2, and belongs to the conserved motif (G-F-G) at region 46–48. The residue α-K83 is a transition point between strand 3 and helix 3. All of the residues that participate in the interdimer interactions in interface “c” are conserved (except E38) in HU family and located on turns. The NMR studies of B. stearothermophilus HU showed that the residues G39, F47, and G82 possess higher flexibility in solution; the higher flexibility of the turn may allow participation in protein-protein interactions (19). The interface “c” is present in arrangements I and III (Fig. 3 a and d).
As mentioned, we observed three potential multimeric arrangements (I, II, and III) in the crystal lattice. In I (Fig. 3b), two of the αβ dimers interact with each other by using interface “a” to form a planar tetramer with the β-hairpin arms of each dimer available for interaction with DNA. By using interfaces “b” and “c,” this tetramer is stacked on another planar tetramer, in which each αβ heterodimer is arranged in an inverted fashion such that the two DNA binding domains are interior.
The oligomerization in arrangement II (Fig. 3c) uses interface “b.” In this, each dimer is stacked on another to make an indefinite spiral staircase. It takes four heterodimers to make a 360° rotation of the spiral, which is right-handed. Interestingly, arrangement III, which uses interface “c” for oligomerization, also generates a continuous spiral with four stacked heterodimers making a full rotation except that the spiral is left-handed (Fig. 3d). In both arrangements II and III the DNA binding domains in each dimer is projected outward from the spiral-body. In all three cases, interdimeric bonds, and so interdimeric surfaces are limited. Thus oligomers may not form in solution. In fact, HU is known to exist as dimers in solution (21, 22).
Model of HU–DNA Complex.
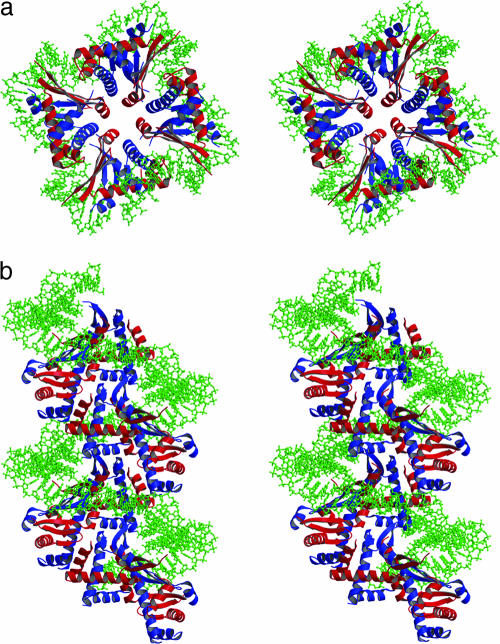
Because crystal packing arrangements II and III can generate right- and left-handed spirals of stacked heterodimers (Fig. 3 c and d), the two configurations were modeled with DNA by using Protein Data Bank entry 1P71 coordinates of the Anabaena HU–DNA complex (20). The HUαβ dimer-DNA model is shown in SI Fig. 10. In the two DNA–HU(αβ) complexes, the right-handed spiral could not accommodate a continuous piece of DNA for interaction with each DNA binding loop in an octamer unit in modeling. However, the left-handed spiral was modeled with DNA in an interpretable manner (Fig. 5 a and b). We assumed that the E. coli heterodimer and the Anabaena homodimer share the same DNA binding site and a similar binding mode. In this model, each dimer covers 16–17 bp with ±1 nt uncertainty caused by one base flipping-out in each strand in the Anabaena complex. It left one DNA bend angle in each heterodimer–DNA complex identical to one of the two bend angles in the Anabaena complex. However, it was not clear which monomer (α or β) is associated with the bend angle. Four HUαβ dimers form one turn in the spiral, which is wrapped by a 64- or 68-bp DNA segment in the spiral groove.
Fig. 5.
Stereoviews of modeled HUαβ octameric unit of the left-handed multimer with DNA fragment as a repeating unit of a spiral structure. The α-subunit is in red, and the β-subunit is in blue. (a) Top view. (b) Side view. The HUαβ multimeric spiral is wrapped by DNA in a left-handed solenoidal structure.
DNA Supercoiling by HU.
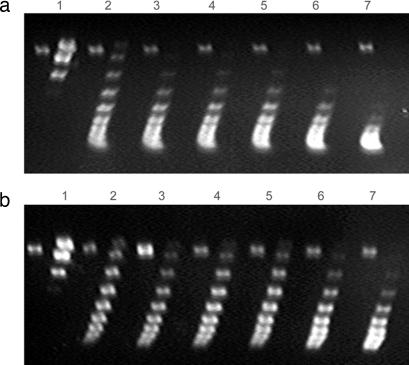
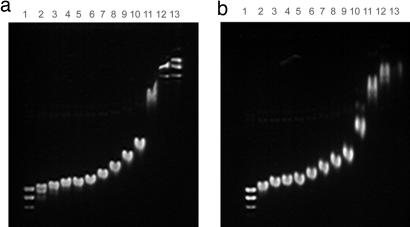
The left-handed αβ spiral-DNA model provides the structural explanation of the previously reported finding that HU restrains negative supercoils in DNA (1). To test the idea that structure III generates negative supercoils, a mutant HUαβ was made in which E38 in β was changed to an alanine disrupting a critical interaction at interface “c.” We tested the DNA supercoiling activity of the wild-type HUαβ and the altered HUα+β-E38A as described in SI Materials and Methods (Fig. 6). Although the results confirmed the ability of wild-type HU to generate and retain negative superhelicity in DNA when the DNA incubation with HU was followed by treatment with topoisomerase I (Fig. 6a), it is clear that the mutant HU protein was substantially defective in DNA supercoiling (Fig. 6b), suggesting an in vivo role of structure III. The wild-type and the mutant HU bind to DNA with comparable affinities by EMSA (Fig. 7).
Fig. 6.
Two-dimensional agarose gel electrophoretic patterns of circular DNA plasmids. Five nanomolar relaxed plasmid samples were treated with wild-type HUαβ (a) and HUα+β-E38A (b) at 0, 1.79, 2.08, 2.50, 3.13, 4.17, and 6.25 μM concentrations (lanes 1–7, respectively) in the presence of topoisomerase I (see text).
Fig. 7.
EMSA of HU-DNA. Five nanomolar relaxed plasmid samples were treated with wild-type α+β+ (a) or mutant α+β-E38A (b) HU at 0, 1.25, 1.39, 1.56, 1.79, 2.08, 2.50, 3.13, 4.17, 6.25, 12.5, 25.0, and 50 μM concentrations (lanes 1–13, respectively).
Discussion
The α- and β-chains of HU heterodimer in E. coli have 30% difference in amino acid sequence. The nonhomology is very likely responsible for causing the asymmetric heterodimeric structure, which is distinct from the more or less symmetric structure of the homodimers in E. coli and other bacteria (16–20). Whether the αα and the ββ homodimers in E. coli have any function different from the heterodimer, other than the instability of the ββ homodimer (23), is not known.
The crystal structure of the HUαβ heterodimer suggests that it may form higher oligomers. The observed asymmetry in the heterodimer allows the subunits to bind DNA differently compared with the way the homodimers bind and provide a structural basis for binding of DNA to multimeric HU. Although HU exists as dimers in solution, it does form oligomers under certain conditions (6, 24). The oligomers may also form when bound to DNA. Consistently, HU binding to DNA is cooperative (24, 25). Cooperativity is usually the result of interactions between adjacent DNA binding protein molecules. We believe that HU uses interface “c” defined above for cooperative binding.
The physiological relevance of the multimers needs further consideration. The multimer with arrangement I may be involved in specialized functions like playing an architectural role in DNA–multiprotein complexes involving DNA looping, Holliday-junction formation, etc. (26, 27). We also speculate that arrangement II may participate in retaining positive DNA supercoils locally in the chromosome under some conditions. Our modeling shows that a DNA can wrap around the multimer with arrangement III with negative superhelicity. In this model, the asymmetry in the basic heterodimeric structure ensures negative DNA superhelicity when the oligomers of arrangement III bind to a continuous DNA. An HU octamer (a tetramer of the dimers) is the basic repeating unit around which a 64- or 68-bp-long DNA helix makes contacts with the protein creating one toroidal DNA with negative superhelicity. This potential of HU is consistent with the following observations: (i) HU is a major nucleoid protein (28); (ii) HU forms nucleosome-like structures (29, 30); (iii) HU binds to DNA cooperatively (24, 25), presumably by using the interfaces of arrangement III; (iv) HU could form cylindrical structures around which DNA is wound in a toroidal supercoils (6); (v) HU increases the thickness of DNA from 20 to 60 Å (7); (vi) plasmid DNA extracted from HU-deficient cells are partially relaxed (31); (vii) HU makes negative supercoils in DNA in the presence of topoisomerase I in vitro (6); and (viii) a mutant HU defective in generating the potential left-handed spiral observed in the crystal is inefficient in creating negative DNA supercoils as reported here. In the left-handed HU spiral, the residue E38 of a β-subunit forms H-bonds with two backbone amide groups of the α-subunit of a neighboring dimer through interface “c” (Fig. 4c). This interaction is weak, which may allow a dynamic balance between HU dimers and multimers depending on environments. Consistently, higher salt concentrations abolish dimer–dimer interactions (6). Our experiments showing that the HUα+β-E38A mutant showed significantly decreased ability in generating negative DNA supercoils compared with the wild type strongly supports the idea that the left-handed spiral (structure III) is real and wraps DNA around it in generating negative superhelicity.
From the results of DNase I digestion of DNA-bound HU, it was shown previously that the nuclease cut sites are made with a periodicity of 8.5 bp (6). Our modeling shows that a complete superhelical DNA turn around the left-handed HU spiral contains ≈64 or 68 base pairs, i.e., ≈8 or 8.5 base pairs per HUαβ monomer. This is in complete agreement with the results of DNase I digestion (6).
The left-handed superhelical DNA around a tetragonal HU structure III giving rise to a 64- or 68-bp solenoidal structure is reminiscent of eukaryotic nucleosome structures. Indeed, the existence of nucleosome-like structures in bacterial chromosome has been predicted before (29, 30). However, the bacterial “nucleosome” structure deduced here from the multimeric arrangement III may differ from the eukaryotic counterpart. In the eukaryotic nucleosome, each core unit restraining negative superhelical DNA is separated from a neighboring unit by spacer DNA (32), whereas in the bacterial case, octameric HU units wrapped by negatively supercoiled DNA are stacked one on another as in a spiral staircase without spacer DNA. Our results demonstrate that by providing a foundation for negative superhelicity in DNA, HUαβ can form a “nucleosome-like” structure, presumably regionally. This structure would help conversion of a plectonomic negatively supercoiled DNA circles into a more open ring with nucleosomal “bead” distributed as was shown by atomic force microscopy (29).
Materials and Methods
Wild-type and mutant HU proteins were purified from an expression plasmid (pRLM118) donated by R. McMacken (33). The HUα+βE38A mutant was made as described in the SI Materials and Methods. The purified proteins were concentrated by Amicon Centrifugal Filters (Millipore, Billerica, MA) to 16.4 mg/ml in a solution containing Tris·HCl (20 mM, pH 8.0), KCl (0.5 M), and glycerol (5%). Protein concentration was measured by using the BCA Protein Assay Kit (Pierce, Rockford, IL) with BSA as standard. HPLC-purified DNA oligomers (SI Table 2) from Sigma-Genosys (The Woodlands, TX) were dissolved in pure water to a concentration of 10 mM. Crystallization reagents were from Hampton Research (Aliso Viejo, CA). Details of crystallization are provided in SI Materials and Methods. The frozen crystals were used for x-ray diffraction by an in-house x-ray diffractometer with a Mar345 imaging plate detector and a Rigaku RU-3H x-ray generator. Data were collected at 1° rotation for each frame and were reduced by the Denzo program and scaled by the Scalepack program of the HKL package (34). The CCP4 Interface was used in molecular replacement and model refinement (35). A 5% FreeR-flag was inserted in data reduction. The molecular replacement was conducted with the Molrep program by using the dimeric Thermotoga maritima HU coordinates (Protein Data Bank entry 1B8Z) (17) as a search model. The electron density map generated from the experimental absolute structure factor and the phase from the molecular replacement output coordinates was improved to be more traceable by the Density Modification (DM) program with a solvent content of 42%. RefMac 5.0 was used for the rigid body, restraints, and translation–libration–screw refinements. The electron density maps were calculated with the FFFbig program. The omit maps were generated by running the “refine.inp” and “model_map.inp” programs of crystallography and NMR system (36). The starting temperature was set at 2,000 K in the annealing process. The solvent-accessible areas were calculated with the AREAIMOL program. The molecular model was visualized and manipulated with the O program (37). The graphs were generated with MolScript (38), Raster3D (39), and the PyMOL Molecular Graphics System (www.pymol.org). The superposition of the HU protein structure of the Anabaena HU homodimer–DNA complex (20) to the E. coli HU(αβ) heterodimer brings the DNA structural coordinates of the former to the corresponding region of the latter giving rise to an E. coli HU(αβ) heterodimer–DNA complex (SI Materials and Methods).
DNA Supercoiling Assays.
The supercoiled circular plasmid pFLEX-gap6 was relaxed by E. coli topoisomerase I. One hundred-nanogram relaxed DNA samples were incubated at 37°C for 30 min with 0.036–0.50 μg of wild-type or mutant HU in 10-μl solutions containing 35 mM Tris·HCl (pH 8.0), 20 mM NaCl, and 5 mM DTT. Calf thymus topoisomerase I (0.5 μl, 5–15 units/μl; Invitrogen) was added, and incubation continued for 2 h. Ten micrograms of proteinase K was then added and incubated for another 30 min. The DNA samples were analyzed by two-dimensional agarose gel electrophoresis in a 4.5 × 4.5-square-inch gel. The first-dimension electrophoresis was run at 0.25× TBE for ≈15 h, and the second-dimension electrophoresis was run at 0.4× TBE containing 1 μg/ml chloroquine for 3 h. The DNA binding assays were as before (40). One hundred-nanogram relaxed plasmid samples were incubated at 37°C for 30 min with 0.025–1.0 μg of wild-type or mutant HU in a 10-μl solution containing 35 mM Tris·HCl (pH 8.0), 20 mM NaCl, and 5 mm DTT. The DNA samples were analyzed by agarose electrophoresis in 4.5 × 4.5-square-inch gels in 0.25× TBE for 15 h.
Supplementary Material
Acknowledgments
We thank our colleagues for assistance, especially S. Kar for her work in HU that prompted us to determine the structure, and T. Soares for purification of proteins. We are grateful to D. Xia, F. Dyda, V. Zhurkin, M. Tolstorukov, P. Rice, and D. Davies for critical reading of the manuscript and helpful suggestions. The work was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research. We used the high-performance computational capabilities of the Helix Systems at the National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
Data deposition: The structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2O97).
This article contains supporting information online at www.pnas.org/cgi/content/full/0611686104/DC1.
References
- 1.Johnson RC, Johnson LM, Schmidt J, Gardner JF. In: The Bacterial Chromosome. Higgins NP, editor. Washington, DC: Am Soc Microbiol; 2005. pp. 65–132. [Google Scholar]
- 2.Azam TA, Hiraga S, Ishihama A. Genes Cells. 2000;5:613–626. doi: 10.1046/j.1365-2443.2000.00350.x. [DOI] [PubMed] [Google Scholar]
- 3.Kar S, Edgar R, Adhya S. Proc Natl Acad Sci USA. 2005;102:16397–16402. doi: 10.1073/pnas.0508032102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Claret L, Rouviere-Yaniv J. J Mol Biol. 1997;273:93–104. doi: 10.1006/jmbi.1997.1310. [DOI] [PubMed] [Google Scholar]
- 5.Castaing B, Zelwer C, Laval J, Boiteux S. J Biol Chem. 1995;270:10291–10296. doi: 10.1074/jbc.270.17.10291. [DOI] [PubMed] [Google Scholar]
- 6.Broyles SS, Pettijohn DE. J Mol Biol. 1986;187:47–60. doi: 10.1016/0022-2836(86)90405-5. [DOI] [PubMed] [Google Scholar]
- 7.Rouviere-Yaniv J, Yaniv M, Germond JE. Cell. 1979;17:265–274. doi: 10.1016/0092-8674(79)90152-1. [DOI] [PubMed] [Google Scholar]
- 8.Bensaid A, Almeida A, Drlica K, Rouviere-Yaniv J. J Mol Biol. 1996;256:292–300. doi: 10.1006/jmbi.1996.0086. [DOI] [PubMed] [Google Scholar]
- 9.Malik M, Bensaid A, Rouviere-Yaniv J, Drlica K. J Mol Biol. 1996;256:66–76. doi: 10.1006/jmbi.1996.0068. [DOI] [PubMed] [Google Scholar]
- 10.Paull TT, Johnson RC. J Biol Chem. 1995;270:8744–8754. doi: 10.1074/jbc.270.15.8744. [DOI] [PubMed] [Google Scholar]
- 11.Ryan VT, Grimwade JE, Nievera CJ, Leonard AC. Mol Microbiol. 2002;46:113–124. doi: 10.1046/j.1365-2958.2002.03129.x. [DOI] [PubMed] [Google Scholar]
- 12.Hashimoto M, Imhoff B, Ali MM, Kow YW. J Biol Chem. 2003;278:28501–28507. doi: 10.1074/jbc.M303970200. [DOI] [PubMed] [Google Scholar]
- 13.Kar S, Adhya S. Genes Dev. 2001;15:2273–2281. doi: 10.1101/gad.920301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paull TT, Haykinson MJ, Johnson RC. Biochimie. 1994;76:992–1004. doi: 10.1016/0300-9084(94)90024-8. [DOI] [PubMed] [Google Scholar]
- 15.Dame RT, Goosen N. FEBS Lett. 2002;529:151–156. doi: 10.1016/s0014-5793(02)03363-x. [DOI] [PubMed] [Google Scholar]
- 16.Ramstein J, Hervouet N, Coste F, Zelwer C, Oberto J, Castaing B. J Mol Biol. 2003;331:101–121. doi: 10.1016/s0022-2836(03)00725-3. [DOI] [PubMed] [Google Scholar]
- 17.Christodoulou E, Rypniewski WR, Vorgias CR. Extremophiles. 2003;7:111–122. doi: 10.1007/s00792-002-0302-7. [DOI] [PubMed] [Google Scholar]
- 18.Tanaka I, Appelt K, Dijk J, White SW, Wilson KS. Nature. 1984;310:376–381. doi: 10.1038/310376a0. [DOI] [PubMed] [Google Scholar]
- 19.Vis H, Mariani M, Vorgias CE, Wilson KS, Kaptein R, Boelens R. J Mol Biol. 1995;254:692–703. doi: 10.1006/jmbi.1995.0648. [DOI] [PubMed] [Google Scholar]
- 20.Swinger KK, Lemberg KM, Zhang Y, Rice PA. EMBO J. 2003;22:3749–3760. doi: 10.1093/emboj/cdg351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rouviere-Yaniv J, Kjeldgaard NO. FEBS Lett. 1979;106:297–300. doi: 10.1016/0014-5793(79)80518-9. [DOI] [PubMed] [Google Scholar]
- 22.Roy S, Dimitriadis EK, Kar S, Geanacopoulos M, Lewis MS, Adhya S. Biochemistry. 2005;44:5373–5380. doi: 10.1021/bi047720t. [DOI] [PubMed] [Google Scholar]
- 23.Bonnefoy E, Almeida A, Rouviere-Yaniv J. Proc Natl Acad Sci USA. 1989;86:7691–7695. doi: 10.1073/pnas.86.20.7691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coombs RO, Cann JR. Electrophoresis. 1996;17:12–19. doi: 10.1002/elps.1150170103. [DOI] [PubMed] [Google Scholar]
- 25.Shimizu M, Miyake M, Kanke F, Matsumoto U, Shindo H. Biochim Biophys Acta. 1995;1264:330–336. doi: 10.1016/0167-4781(95)00173-5. [DOI] [PubMed] [Google Scholar]
- 26.Geanacopoulos M, Vasmatzis G, Lewis DE, Roy S, Lee B, Adhya S. Genes Dev. 1999;13:1251–1262. doi: 10.1101/gad.13.10.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kamashev D, Rouviere-Yaniv J. EMBO J. 2000;19:6527–6535. doi: 10.1093/emboj/19.23.6527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rouviere-Yaniv J, Gros F. Proc Natl Acad Sci USA. 1975;72:3428–3432. doi: 10.1073/pnas.72.9.3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Griffith JD. Proc Natl Acad Sci USA. 1976;73:563–567. doi: 10.1073/pnas.73.2.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murphy LD, Zimmerman SB. J Struct Biol. 1997;119:321–335. doi: 10.1006/jsbi.1997.3883. [DOI] [PubMed] [Google Scholar]
- 31.Hillyard DR, Edlund M, Hughes KT, Marsh M, Higgins NP. J Bacteriol. 1990;172:5402–5407. doi: 10.1128/jb.172.9.5402-5407.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 33.Aki T, Adhya S. EMBO J. 1997;16:3666–3674. doi: 10.1093/emboj/16.12.3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 35.Collaborative Computational Project N. Acta Crystallogr D. 1994;50:760–763. [Google Scholar]
- 36.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 37.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 38.Kraulis PJ. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 39.Merritt EA, Bacon DJ. Methods Enzymol. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
- 40.Majumdar A, Adhya S. Proc Natl Acad Sci USA. 1984;81:6100–6104. doi: 10.1073/pnas.81.19.6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.