Abstract
Ectopically expressed hTERT enables p16INK4A(−) human mammary epithelial cells to proliferate in the absence of growth factors, a finding that has led to the hypothesis that hTERT has growth regulatory properties independent of its role in telomere maintenance. We now show that telomerase can alter the growth properties of cells indirectly through its role in telomere maintenance, without altering growth stimulatory pathways. We find that telomere dysfunction, indicated by 53BP1/phosphorylated histone H2AX foci at chromosome ends, is present in robustly proliferating human mammary epithelial cells long before senescence. These foci correlate with increased levels of active p53. Ectopic expression of hTERT reduces the number of foci and the level of active p53, thereby decreasing sensitivity to growth factor depletion, which independently activates p53. The continuous presence of hTERT is not necessary for this effect, indicating that telomere maintenance, rather than the presence of the enzyme itself, is responsible for the increased ability to proliferate in the absence of growth factors. Our findings provide a previously unrecognized mechanistic explanation for the observation that ectopically expressed hTERT conveys growth advantages to cells, without having to postulate nontelomeric functions for the enzyme.
Keywords: EGF, phosphorylated histone H2AX, insulin, senescence, telomerase
In many cases, ectopic expression of hTERT is sufficient to increase telomerase activity in human somatic cells and prevent replicative senescence, without conferring other attributes of tumorigenic transformation (1–3). However, several studies have suggested that hTERT has functions unrelated to telomere maintenance, which might facilitate tumor cell proliferation and survival. For example, ectopic hTERT expression in human mammary epithelial cells (HMEC) has been shown to confer resistance to TGF-β-induced growth arrest (3) or to promote expression of genes involved in proliferation (4). In other studies and cell types, ectopic hTERT expression caused accelerated cell proliferation, reduced growth factor requirements, altered gene expression, enhanced DNA repair, or increased tumorigenesis (5–7). Wound healing was accelerated in transgenic mice that overexpress mTERT in epidermal keratinocytes (8). These mice were more prone to spontaneous tumorigenesis, suggesting that high telomerase activity may cooperate with age-dependent genetic alterations to promote tumorigenesis. Given the speculation that hTERT might have telomere-independent activities that favor oncogenesis, we set out to determine the mechanism responsible for the apparent effects of hTERT on HMEC proliferation.
Results
hTERT Decreases Sensitivity to Growth Factor Deficiency.
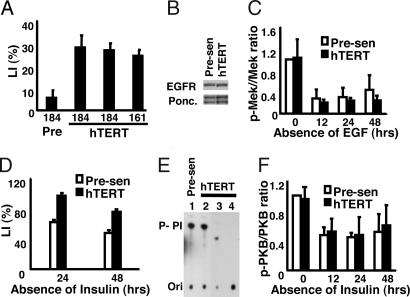
The proliferation of presenescent p16(−) HMEC stringently depends on signaling from the EGF receptor (EGFR). HMEC reversibly arrest growth 24 h after EGF removal and addition of a blocking EGFR monoclonal antibody or a tyrosine kinase inhibitor (EGF-deficient conditions) (9). To determine whether hTERT influences the ability of HMEC to arrest growth under these conditions, we used retroviruses to express hTERT in presenescent HMEC from two individuals (specimens 184 and 161). In complete growth medium, control and hTERT-transduced HMEC had identical doubling times, morphologies, and lack of senescence-associated β-galactosidase staining (10) [see the supporting information (SI)]. Nonetheless, hTERT caused resistance to EGF depletion, as determined by the ability to incorporate tritiated thymidine into DNA during the final 24 h of a 48-h incubation under EGF-deficient conditions. Under these conditions, the relative labeling index (LI) was consistently 3- to 5-fold higher in hTERT-transduced HMEC compared with control HMEC (Fig. 1A).
Fig. 1.
hTERT suppresses the decline in DNA synthesis caused by EGF or insulin withdrawal but does not affect EGFR or downstream signal transduction. (A) Indicated cells were incubated in complete (EGF+) or deficient (EGF−) medium for 48 h, with 3H-thymidine added for the final 24 h. Radiolabeled nuclei were identified by autoradiography. LI values were calculated by dividing the percentage of labeled nuclei in the test media by the percentage in complete medium. Two independent isolates of 184-TERT and one isolate of 161-TERT were assayed in addition to presenescent 184 HMEC infected with LXSN control vector. Additional assays (data not shown) of both uninfected and empty retroviral vector-infected presenescent HMEC showed LI values similar to that displayed for the vector-infected 184 controls. (B) Immunoblot of total cell lysates of presenescent and hTERT-transduced HMEC growing at comparable rates in complete medium, probed with an antibody to EGFR. Ponceau-stained bands served as loading controls. (C) Immunoblots of total cell lysates of cells incubated in EGF-deficient medium for indicated intervals were probed with antibodies against phosphorylated Mek1/2 and total Mek1/2. The relative intensities of the phosphorylated Mek1/2 protein bands were quantified by densitometry, normalized to the levels of total Mek1/2, and expressed as ratios of the level in presenescent HMEC in complete medium (t = 0). (D) Presenescent and hTERT-transduced HMEC were cultured in the presence or absence of insulin for 24 or 48 h with 3H-thymidine added for the final 24 h. LI values were determined as in A. (E) Phosphatidylinositol-kinase activity assays were performed as described by Whitman et al. (34) after immunoprecipitation of the indicated cell extracts by antibodies against phosphotyrosine (lanes 1 and 2). Negative controls were performed with nonspecific IgG (lane 3) or no antibody (lane 4). (F) Immunoblots of total cell lysates of cells incubated in EGF-deficient medium for indicated intervals were probed with antibodies against total and phosphorylated PKB. The relative intensities of the p-PKB protein bands were quantified by densitometry, normalized to the levels of total PKB, and expressed as ratios of the level in presenescent HMEC in complete medium (t = 0). Data are presented ± SD.
To test the possibility that hTERT up-regulated EGFR expression, we compared the levels of total EGFR protein expressed in control and hTERT-transduced HMEC growing at comparable rates in complete medium. The cells expressed equivalent EGFR levels in repeated experiments (Fig. 1B). Moreover, under EGF-deficient conditions, increasing the concentration of blocking antibody 4-fold, from 5 to 20 μg/ml, did not remove the differences in LI (data not shown), indicating that the antibody was present at a saturating concentration.
We next examined signaling events downstream of the EGFR to determine whether hTERT affected growth factor receptor-mediated signal transduction. We compared the levels of active (phosphorylated) Mek1/2 in control and hTERT-transduced HMEC. As expected, levels of phosphorylated Mek1/2 were high in both cell types when actively proliferating (Fig. 1C; 0 h) and significantly decreased 12 h after transfer to EGF-deficient conditions. No significant difference was observed between control and hTERT-transduced cells in the amount of phosphorylated Mek1/2 at any time point examined after blockage of EGFR signaling. Thus, the differences in LI could not be explained by hTERT-mediated changes in EGFR abundance or signaling.
Although presenescent HMEC can proliferate in the absence of insulin, optimal proliferation and the LI are significantly reduced when insulin is omitted from the medium (Fig. 1D). hTERT-transduced HMEC, by contrast, maintained significantly higher levels of DNA synthesis in the absence of insulin. Because phosphatidylinositol-kinase (PI3K) is a downstream effector of insulin signaling, we examined PI3K activity in control and hTERT-transduced HMEC growing at the same rate in complete medium. We assayed anti-phosphotyrosine or anti-PI3K p85 subunit immunoprecipitates for PI3K activity using an in vitro kinase assay (Fig. 1E and data not shown). We observed no significant difference in PI3K activity between control and hTERT-transduced cells, suggesting that the effect of hTERT on LI was independent of PI3K. In addition, we examined PKB/Akt phosphorylation at different times after insulin deprivation (Fig. 1F). As for phosphorylated Mek1/2, no increase in phosphorylated PKB was noted for the hTERT-transduced HMEC that could account for the elevated LI in these cells in the absence of insulin.
hTERT Reduces p53 Phosphorylation.
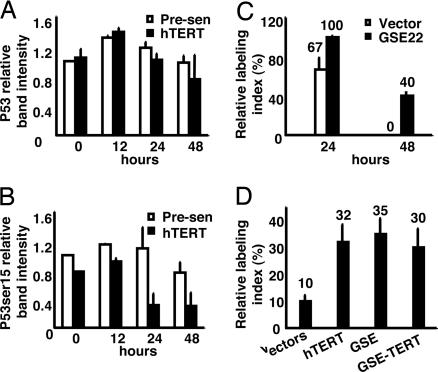
p53 activity contributes to serum deprivation-mediated growth arrest (11). We therefore examined both total and phosphorylated p53 in control and hTERT-transduced HMEC under EGF-deficient conditions. Total p53 levels were similar in proliferating control and hTERT-transduced HMEC (Fig. 2A). After EGF withdrawal, p53 levels increased slightly and transiently as reported for serum deprivation (11). p53 is posttranslationally modified in response to stress signals, most notably by phosphorylation on serine 15 in response to DNA damage, leading to its stabilization and activation as a transcription factor. The levels of p53 phosphorylated on serine 15 (phospho-ser15) were consistently lower in proliferating hTERT-transduced HMEC compared with proliferating control cells (Fig. 2B). Moreover, although phospho-ser15 p53 first increased and then decreased after EGF withdrawal, at all time points tested, the levels were consistently lower in hTERT-transduced compared with control HMEC. Thus, hTERT reduced the level of activated (phospho-ser15) p53 in cells in both complete and EGF-deficient media. Similar results were observed in HMEC deprived of insulin (see the SI).
Fig. 2.
p53 mediates hTERT effects on growth arrest in EGF-deficient medium. (A and B) Immunoblots of total cell lysates of the indicated cells incubated in EGF-deficient medium for indicated intervals were probed with antibodies against total p53 or Ser-15-phosphorylated p53. The relative intensities of the total p53 or Ser-15-phosphorylated p53 protein bands were quantified by densitometry, normalized to the levels of prominent Ponceau-stained proteins, and expressed as ratios of the level in presenescent HMEC in complete medium (t = 0). (C) Vector control or GSE22-transduced HMEC were incubated in complete or EGF-deficient medium for 24 or 48 h, with 3H-thymidine added for the final 24 h. Labeled nuclei were identified by autoradiography and quantified. (D) Presenescent HMEC transduced with vector control, vector control and GSE22 or hTERT individually, or GSE22 plus hTERT were incubated in complete or EGF-deficient medium for 72 h with 3H-thymidine added for the final 24 h. Labeled nuclei were identified by autoradiography and quantified.
To further confirm whether activation of p53 by mitogen withdrawal influences the ability of HMEC to arrest growth, we used a retrovirus to express the dominant interfering p53 genetic suppressor element, GSE22 (12, 13). GSE22-expressing HMEC maintained a high LI 48 h after blockage of EGFR signaling (Fig. 2C), confirming that the effect of EGF depletion on growth was mediated in part through p53 activity. Similar results were obtained with HMEC grown in medium depleted of insulin or in the presence of TGF-β (see the SI). The effect of GSE22 on growth of the presenescent cells depleted of growth factors was very similar to the effect of hTERT. Therefore we compared the LI of HMEC expressing hTERT, GSE22, or both 72 h after blockage of EGFR signaling. We replenished the EGF-deficient medium after 48 h and determined the LI during the final 24 h of the 72-h incubation. Control cells had a LI of 10%, whereas cells expressing either hTERT or GSE22 alone had LI of 30–40% (Fig. 2D). The effects of hTERT and GSE22 were not additive, suggesting that hTERT and p53 act in the same pathway.
Active DNA Damage Signaling in Proliferating HMEC Before Replicative Senescence.
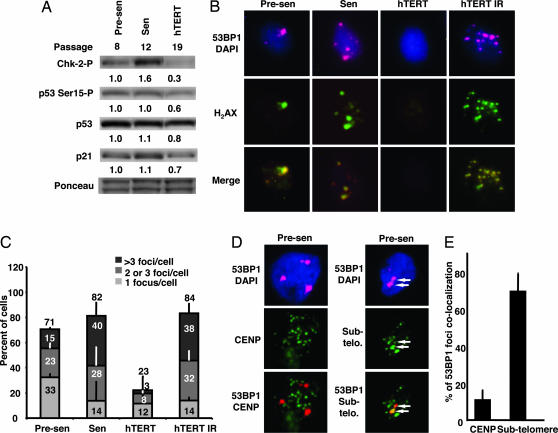
The finding that the level of phospho-ser15 p53 was higher in the control presenescent HMEC than in the hTERT-transduced HMEC, while both cell types proliferated at the same rate in complete growth medium, suggested that an intrinsic source of p53 activation was present in the control cells, and that this stress might be relieved by hTERT transduction. Therefore, we asked whether telomeric DNA damage signaling, which is p53 dependent, might be activated to some extent before replicative senescence and whether such telomeric DNA damage signaling might be suppressed by ectopic hTERT. To test this idea, we first examined mediators of DNA damage signaling in presenescent and hTERT-transduced HMEC, and compared the status of these mediators with that in senescent HMEC known to accumulate high levels of these mediators (Fig. 3A). In addition to phospho-ser15 p53, the actively proliferating presenescent cells contained detectable levels of phospho-thr68 Chk2. Relative to presenescent cells, hTERT-transduced HMEC had consistently lower levels of phospho-thr68 Chk2, phospho-ser15 p53, and total p21, a finding consistent with reduced p53-dependent damage signaling in the hTERT-transduced HMEC.
Fig. 3.
DNA damage signaling occurs at chromosome ends in proliferating presenesent HMEC. (A) Presenescent (Presen), senescing but still proliferative (Sen), and hTERT-transduced (hTERT) HMEC in complete medium were harvested at the indicated passages while subconfluent. Cell lysates were analyzed by immunoblotting for phosphorylated Chk2, Ser-15-phosphorylated p53, total p53, and p21. Ponceau-stained bands were used as loading controls. Normalized staining intensities obtained by densitometry are displayed below each lane. (B) Presenescent, senescent, hTERT-transduced, and X-irradiated (10 Gy) HMEC were immunostained for 53BP1 (red), phospho-H2AX (green), and DNA (blue). (C) The percentage of cells displaying one, two to three, or more than three phosphorylated H2AX foci in presenescent HMEC, senescent HMEC, hTERT-transduced HMEC, and irradiated hTERT-transduced HMEC was tabulated. For each population, 200–400 cells in five to six separate fields were counted. (D) Presenescent cells were costained for 53BP1 and either CENP (a centromeric marker) or subtelomeric DNA (a marker of chromosome ends). Two 53BP1 foci that colocalize with subtelomeric DNA are indicated by white arrows. (E) The percentages of 53BP1 signals that colocalized with CENP or subtelomeric DNA signals were quantitated and compared. For each population, 200–400 cells in five to six separate fields were counted.
DNA damage and dysfunctional telomeres are marked by the presence of nuclear foci containing phosphorylated histone H2AX (H2AX) and 53BP1 (14–16). To determine whether proliferating cultures of presenescent HMEC contain such foci, we examined them by immunofluorescence (Fig. 3B). Despite the similar proliferation rates and LI in complete growth medium, >70% of proliferating presenescent HMEC, but <25% of proliferating hTERT-transduced HMEC, displayed one or more 53BP1/H2AX foci (Fig. 3C). The lower number of foci in the hTERT-transduced HMEC was not due to a defect in DNA damage detection or signaling, because these cells remained fully capable of forming 53BP1/H2AX foci in response to ionizing radiation. The finding that the majority of presenescent HMEC contained small numbers of foci indicates that the proliferating cells in these cultures must be able to tolerate some degree of DNA damage signaling while continuing to proliferate. The fact that hTERT-transduced HMEC consistently contained fewer foci suggested that hTERT caused a reduction in DNA damage signaling. We obtained similar results in human diploid fibroblasts (see the SI), indicating that this effect was not restricted to p16(−) HMEC.
To determine whether the DNA damage signaling in presenescent HMEC emanates from chromosome ends, we combined immunohistochemical detection of 53BP1 foci with FISH detection of subtelomeric DNA (Fig. 3D). We probed for subtelomeric DNA, reasoning that in some cases telomeric DNA might be too short to detect at chromosome ends exhibiting signs of DNA damage. Nearly 70% of the 53BP1 foci detected in presenescent HMEC colocalized with subtelomeric FISH signals, whereas only 10% colocalized with centromeric CENP proteins (Fig. 3E). Thus, proliferating presenescent HMEC harbor a low but detectable level of telomeric DNA damage before replicative senescence, and this damage is suppressed by hTERT.
Effects of hTERT Require Catalytic Activity and Telomere Maintenance but Not Continuous Expression.
To determine whether the hTERT reverse transcriptase domain must be intact and capable of telomere maintenance in vivo to prevent growth arrest under EGF-deficient conditions, we compared the activities of two mutant constructs to that of wild-type hTERT. One mutant harbored inactivating amino acid substitutions in the reverse transcriptase domain (17), whereas the other contained a carboxyl-terminal HA epitope tag that suppresses telomerase activity in vivo (18). Neither mutant rendered HMEC resistant to growth arrest in response to EGF-deficiency (data not shown). Thus, the ability of hTERT to prevent growth arrest of HMEC depended on its action at telomeres in vivo.
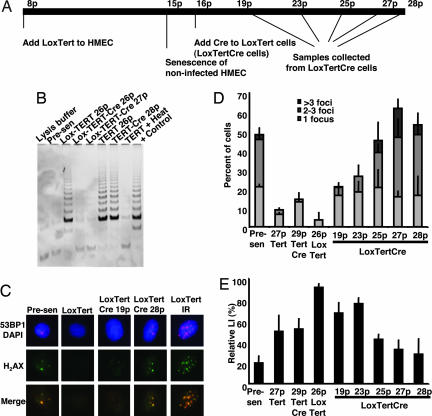
To determine whether continuous telomerase activity is required to prevent growth arrest under EGF-deficient conditions, we used a retrovirus carrying hTERT flanked by loxP excision sites for Cre recombinase (Lox-hTERT) (19). We transduced presenescent HMEC with Lox-hTERT or hTERT viruses. Lox-hTERT HMEC expressed abundant telomerase activity and continued to proliferate after control cells senesced at passage 15 (Fig. 4 A and B). The Lox-hTERT-transduced HMEC were subsequently superinfected at passage 16 with a retrovirus encoding Cre-recombinase, which reduced telomerase activity back to barely detectable levels. Cre-recombinase had no effect on the abundant telomerase activity in HMEC transduced with hTERT lacking Lox sites. Lox-hTERT, Lox-hTERT+Cre, and hTERT+Cre HMEC proliferated at equivalent rates through passage 27. Thereafter, Lox-hTERT-Cre started to show signs of senescence such as a reduction in proliferation rate, changes in morphology, and positive staining for senescence-associated β-galactosidase.
Fig. 4.
The effects of hTERT on growth arrest do not require its continuous expression. (A) Timeline indicating the passage numbers at which cells were infected with the different retroviruses and when samples were collected for assays. (B) Relative telomerase activity in the indicated samples is shown by the intensity of regularly spaced PCR products in assays performed with a commercial telomerase detection kit (Intergen, Purchase, NY). (C) Presenescent, Lox-TERT, two different passages of Lox-TERT-Cre, and irradiated Lox-TERT-transduced HMEC were immunofluorescently stained for 53BP1 (red), phospho-H2AX (green), and DNA (blue). (D) The percentages of cells displaying one, two to three, or more than three phosphorylated H2AX foci were tabulated for the indicated HMEC. For each population, 200–400 cells in five to six separate fields were counted. (E) Indicated cells were incubated in complete medium or EGF-deficient medium containing the tyrosine kinase inhibitor AG1478 for 48 h with BrdU added for the final 24 h. Labeling indices were then calculated.
We quantitated 53BP1/H2AX foci in Lox-hTERT and Lox-hTERT+Cre cells at different passages (Fig. 4 C and D). The percentage of Lox-hTERT cells bearing one or more foci was significantly lower than that of control presenescent cells. Notably, the percentage of cells bearing foci was not significantly increased immediately after excision of hTERT with Cre but instead increased slowly over time. Importantly, an inverse correlation was observed among the percentages of cells bearing one or more foci and the relative LI of cells incubated under EGF-deficient conditions (Fig. 4E). Whereas resistance to growth arrest signals initially required hTERT activity at telomeres in vivo, the resistance was maintained for several cell generations after hTERT excision. Similar results were obtained for cells exposed to TGF-β (see the SI).
Discussion
This work provides a mechanistic explanation for reports that ectopically expressed hTERT conveys growth advantages to cells. We have demonstrated that these properties can be ascribed to the known activities of telomerase: stabilizing and lengthening telomeres. First, we have shown that robustly proliferating HMEC cultures already express symptoms of DNA damage well before replicative senescence and that these symptoms are ameliorated by transient expression of hTERT. Second, we have shown that growth arrest due to growth factor deprivation proceeds through the same p53-dependent pathway that is activated by DNA damage or telomere dysfunction. Third, we have shown that telomerase reduces p53 signals generated by telomere dysfunction, thereby raising tolerance for activated p53 generated by growth factor deprivation. And finally, we have shown that the continuous presence of telomerase is not necessary for this effect, indicating that telomere maintenance, rather than the presence of the enzyme itself, is responsible for the increased ability to proliferate in the absence of growth factors.
Our results indicating that introduction of hTERT into actively growing presenescent HMEC positively affects their ability to proliferate in the short-term absence of EGFR/insulin signaling agree with a recent study showing that ectopically expressed hTERT confers a growth advantage to HMEC in medium lacking EGF and pituitary extract (4). However, in contrast to that study, we found no evidence that hTERT alters expression of EGFR or downstream components of growth factor signaling pathways. It is likely that the differences in growth regulatory gene expression observed in the previous study were the consequence of using control cells at passages at which short telomeres were already starting to impinge upon the growth rates of the mass cultures. In our study, we were careful to compare control and hTERT-transduced HMEC growing at comparable rates in complete medium. We found that instead of altering growth factor signaling pathways, hTERT altered the susceptibility of HMEC to p53-dependent signals for growth arrest by alleviating the p53-dependent DNA damage signaling emanating from chromosome ends.
The majority of proliferating HMEC contain low but detectable levels of activated DNA damage-responsive proteins (phospho-thr68-Chk2, phospho-ser15-p53, p21) and nuclear foci (containing phosphorylated 53BP1 and H2AX) before replicative senescence. In these cells, most of the 53BP1/H2AX foci are at or near chromosome ends, suggesting that they are caused by dysfunctional telomeres. These symptoms of DNA damage are significantly reduced by hTERT expression. Although a previous study (20) indicated that a single irreparable DNA break may be sufficient to induce p53-dependent growth arrest, dysfunctional telomeres are not synonymous with irreparable DNA breaks. As telomeric DNA on individual chromosome ends decreases, their ability to recruit and retain telomeric proteins may decrease, increasing the interval the ends spend in an “unprotected” state or conformation that can be recognized by DNA damage pathways. One or a few transiently exposed ends may not be sufficient to signal growth arrest or senescence but may be sufficient to lead to a transient ATM/p53-dependent damage response (21), increasing the basal level of activated p53. When combined with other p53-dependent signals, such as that conferred by growth factor depletion, the combined signal may exceed a threshold beyond which growth arrest occurs.
The indirect effects of telomere maintenance on p53-dependent functions may explain observations in other systems, as well. Although murine telomeres are, on average, much longer than human telomeres, the shortest telomeres in mouse cells are comparable to those in human cells (22). Reduced p53 stress responses due to telomerase maintenance of short telomeres in murine cells may exert subtle changes in p53-mediated growth arrest and/or apoptosis responses, indirectly allowing accelerated wound repair and continued proliferation of cells bearing premalignant defects. Nevertheless, our results do not rule out the possibility that telomerase does have nontelomeric functions. Findings such as those showing that TERT overexpression in TERC−/− mice causes proliferation of hair follicle stem cells (23) remain to be explained.
Although the role of p53 in growth arrest due to genotoxic stress is well characterized, its role in growth arrest by other causes is less understood. Nonetheless, p53 was shown to be important for the growth arrest caused by ribonucleotide depletion (24), TGF-β (25, 26), and serum withdrawal (11). After EGF/insulin withdrawal, phospho-ser15 p53 ultimately declined in both control and hTERT-transduced HMEC. These findings raise the possibility that p53 and its downstream effector p21 may be important for the initiation, but not maintenance, of growth arrest. Alternatively, the ratio of these proteins to their binding partners, rather than their overall abundance, may be critical for growth arrest under the conditions examined. Gene disruption studies in human fibroblasts show that p21 contributes to cell cycle arrest after serum withdrawal, but cells lacking p21 eventually become quiescent, albeit less efficiently (27). Likewise, human fibroblasts lacking p53 function resist growth arrest due to serum withdrawal but eventually become quiescent (11). Similarly, we found that HMEC expressing hTERT or GSE22 eventually cease proliferation after EGF/insulin withdrawal, albeit less efficiently. Other cell cycle regulators, such as pRb and its regulators, likely cooperate with p53 and p21 to establish and maintain quiescence and may do so independently (28). Nonetheless, we documented a direct role for p53 in growth arrest caused by EGF/insulin withdrawal. Thus, p53 may play a more general role in cell cycle regulation, coordinating growth factor-mediated signals with genome and telomere surveillance.
Materials and Methods
Cells and Cell Culture.
HMEC strains were provided by Martha Stampfer (Lawrence Berkeley National Laboratory) and cultured as described (29). Cultured HMEC undergo a self-selection process (30), resulting in postselection HMEC that do not express p16INK4A (31). Our studies used postselection populations designated presenescent and showing robust proliferation through passage 11. Cultures at passages 11–15 showing increasing percentages of nonproliferative cells with altered morphology are designated senescent. hTERT-transduced HMEC were examined in nonselective medium at passages 17–34, after drug selection for transduced cells was complete. Suppression of EGF signaling was achieved by removing EGF from the culture medium and by blocking EGFR signaling by addition of either an anti-EGFR blocking antibody (mAb 225) (9) or a tyrosine kinase inhibitor AG1478 (Calbiochem; catalog no. 658552). DNA synthesis was assessed by 3H-thymidine or BrdU incorporation during the last 24 h before fixation.
Retroviral Transduction.
HMEC were infected with pLXSN or pBABE-PURO retroviruses containing hTERT (17, 32), GRN365 (hTERT with mutations R631A, D712A, and D868A) (3), hTERT with a carboxyl-terminal HA epitope tag (18), or the dominant p53 genetic suppressor element 22 (GSE22) (12), and selected in G418 (GIBCO) or puromycin (Sigma) (33). In some cases, infection with insertless retroviruses and drug selection reduced the growth of presenescent cells in complete medium (not shown). In such cases, to ensure hTERT-transduced HMEC and controls were growing at equal rates, we used uninfected presenescent HMEC as controls. For reversible manipulation of telomerase expression, HMEC were transduced with pBLoxTSH and pLCRESH retroviruses (19), and selected with histidinol or with puromycin and hygromycin as appropriate.
Antibodies, Immunoblot, and Immunofluorescence Analyses.
We used antibodies against total p53 (Oncogene Research Products, San Diego, CA; Ab-6, catalog no. OP43A), serine 15-phosphorylated p53 (Cell Signaling, Beverly, MA; catalog no. 9284), total Mek-1/2 (Cell Signaling; catalog no. 9122), serine 217/221-phosphorylated Mek-1/2 [Cell Signaling; catalog no. 9121(S)], total PKB/Akt (Cell Signaling; catalog no. 9272), threonine 308-phosphoryated PKB/Akt (Cell Signaling; catalog no. 9275), threonine 68-phosphorylated Chk2 (Cell Signaling; catalog no. 2661), total p21 (BD Biosciences, Palo Alto, CA; clone SX118, catalog no. 556430), serine 139-phosphorylated H2AX (Upstate Biotechnology, Lake Placid, NY; clone JBW301), 53BP1 (Bethyl Laboratories, Montgomery, TX; catalog no. A300-273A), and CENP (Antibodies, Davis, CA; catalog no. 15-235-F).
Fluorescence in Situ Hybridization.
Cells were fixed with 4% paraformaldehyde for immunostaining, and then incubated in denaturing solution (70% formamide in 2× SSC) at 76°C for 7 min, dehydrated in a 70%, 85%, and 100% ethanol series, and hybridized with a mixture of 41 subtelomeric probes. DNA (50–100 ng) from each of four pools of subtelomeric probes was labeled with biotin by using the BioPrime DNA labeling system (Invitrogen, Carlsbad, CA), and the labeled probes were mixed for hybridization. The probe mixture was denatured at 76°C for 10 min and preincubated at 37°C for 40 min before hybridization for 2 days at 37°C. Posthybridization washes were 50% formamide and 2× SSC (two washes at 45°C, one wash at room temperature, 10 min each). Biotinylated probes were detected by FITC-conjugated avidin and visualized by confocal microscopy.
Supplementary Material
Acknowledgments
We thank H. U. Weier (Lawrence Berkeley National Laboratory) for the subtelomeric DNA probes, Tiffany Tuton for technical assistance with FISH, and Jimmie Fata for help with confocal microscopy. This work was supported by U.S. Army Medical Research and Materiel Command Grant DAMD17-00-1-0308 (to P.Y.), the Office of Energy Research (Office of Health and Biological Research, U.S. Department of Energy) under Contract DE-AC03-76SF00098 (to P.Y., M.J.B., and J.C.), National Institutes of Health Grant AG09909 (to J.C.), and fellowships from Le Fonds pour la Formation de Chercheurs et l'Aide à la Recherche and the Canadian Institutes of Health Research (to A.B.).
Abbreviations
- EGFR
EGF receptor
- GSE22
dominant interfering p53 genetic suppressor element
- H2AX
phosphorylated histone H2AX
- HMEC
human mammary epithelial cells
- LI
labeling index
- Lox-hTERT
hTERT flanked by loxP excision sites for Cre-recombinase.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0700260104/DC1.
References
- 1.Jiang W-R, Jimenez G, Chang E, Frolkis M, Kusler B, Sage J, Beeche M, Bodnar AG, Wahl GM, Tlsty TD, Chiu C-P. Nature Gen. 1999;21:111–114. doi: 10.1038/5056. [DOI] [PubMed] [Google Scholar]
- 2.Morales CP, Holt SE, Ouellette M, Kaur J, Yan Y, Wilson KS, White MA, Wright WE, Shay JW. Nature Gen. 1999;21:115–118. doi: 10.1038/5063. [DOI] [PubMed] [Google Scholar]
- 3.Stampfer M, Garbe J, Levine G, Lichsteiner S, Vasserot A, Yaswen P. Proc Natl Acad Sci USA. 2001;98:4498–4503. doi: 10.1073/pnas.071483998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith LL, Coller HA, Roberts JM. Nat Cell Biol. 2003;5:474–479. doi: 10.1038/ncb985. [DOI] [PubMed] [Google Scholar]
- 5.Xiang H, Wang J, Mao Y, Liu M, Reddy VN, Li DW. Oncogene. 2002;21:3784–3791. doi: 10.1038/sj.onc.1205455. [DOI] [PubMed] [Google Scholar]
- 6.Stewart SA, Hahn WC, O'Connor BF, Banner EN, Lundberg AS, Modha P, Mizuno H, Brooks MW, Fleming M, Zimonjic DB, et al. Proc Natl Acad Sci USA. 2002;99:12606–12611. doi: 10.1073/pnas.182407599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sharma GG, Gupta A, Wang H, Scherthan H, Dhar S, Gandhi V, Iliakis G, Shay JW, Young CS, Pandita TK. Oncogene. 2003;22:131–146. doi: 10.1038/sj.onc.1206063. [DOI] [PubMed] [Google Scholar]
- 8.González-Suárez E, Samper E, Ramirez A, Flores JM, Martin-Caballero J, Jorcano JL, Blasco MA. EMBO J. 2001;20:2619–2630. doi: 10.1093/emboj/20.11.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stampfer MR, Pan CH, Hosoda J, Bartholomew J, Mendelsohn J, Yaswen P. Exp Cell Res. 1993;208:175–188. doi: 10.1006/excr.1993.1236. [DOI] [PubMed] [Google Scholar]
- 10.Dimri GP, Lee X, Basile G, Roskelley C, Medrano EE, Rubelji I, Pereira-Smith OM, Peacocke M, Campisi J. Proc Natl Acad Sci USA. 1995;92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Itahana K, Dimri GP, Hara E, Itahana Y, Zou Y, Desprez PY, Campisi J. J Biol Chem. 2002;277:18206–18214. doi: 10.1074/jbc.M201028200. [DOI] [PubMed] [Google Scholar]
- 12.Ossovskaya VS, Mazo IA, Chernov MV, Chernova OB, Strezoska Z, Kondratov R, Stark GR, Chumakov PM, Gudkov AV. Proc Natl Acad Sci USA. 1996;93:10309–10314. doi: 10.1073/pnas.93.19.10309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stampfer MR, Garbe J, Nijjar T, Wigington D, Swisshelm K, Yaswen P. Oncogene. 2003;22:5238–5251. doi: 10.1038/sj.onc.1206667. [DOI] [PubMed] [Google Scholar]
- 14.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. J Biol Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 15.Schultz LB, Chehab NH, Malikzay A, Halazonetis TD. J Cell Biol. 2000;151:1381–1390. doi: 10.1083/jcb.151.7.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takai H, Smogorzewska A, de Lange T. Curr Biol. 2003;13:1549–1556. doi: 10.1016/s0960-9822(03)00542-6. [DOI] [PubMed] [Google Scholar]
- 17.Nakamura TM, Morin GB, Chapman KB, Weinrich SL, Andrews WH, Lingner J, Harley CB, Cech TR. Science. 1997;277:955–959. doi: 10.1126/science.277.5328.955. [DOI] [PubMed] [Google Scholar]
- 18.Counter CM, Hahn WC, Wei W, Caddle SD, Beijersbergen RL, Lansdorp PM, Sedivy JM, Weinberg RA. Proc Natl Acad Sci USA. 1998;95:14723–14728. doi: 10.1073/pnas.95.25.14723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rubio MA, Kim SH, Campisi J. J Biol Chem. 2002;277:28609–28617. doi: 10.1074/jbc.M203747200. [DOI] [PubMed] [Google Scholar]
- 20.Linke SP, Clarkin KC, Wahl GM. Cancer Res. 1997;57:1171–1179. [PubMed] [Google Scholar]
- 21.Herbig U, Jobling WA, Chen BP, Chen DJ, Sedivy JM. Mol Cell. 2004;14:501–513. doi: 10.1016/s1097-2765(04)00256-4. [DOI] [PubMed] [Google Scholar]
- 22.Zijlmans JM, Martens UM, Poon SS, Raap AK, Tanke HJ, Ward RK, Lansdorp PM. Proc Natl Acad Sci USA. 1997;94:7423–7428. doi: 10.1073/pnas.94.14.7423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sarin KY, Cheung P, Gilison D, Lee E, Tennen RI, Wang E, Artandi MK, Oro AE, Artandi SE. Nature. 2005;436:1048–1052. doi: 10.1038/nature03836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Linke SP, Clarkin KC, Di Leonardo A, Tsou A, Wahl GM. Genes Dev. 1996;10:934–947. doi: 10.1101/gad.10.8.934. [DOI] [PubMed] [Google Scholar]
- 25.Landesman Y, Bringold F, Milne DD, Meek DW. Cell Signal. 1997;9:291–298. doi: 10.1016/s0898-6568(97)89890-7. [DOI] [PubMed] [Google Scholar]
- 26.Cordenonsi M, Dupont S, Maretto S, Insinga A, Imbriano C, Piccolo S. Cell. 2003;113:301–314. doi: 10.1016/s0092-8674(03)00308-8. [DOI] [PubMed] [Google Scholar]
- 27.Brown JP, Wei W, Sedivy JM. Science. 1997;277:831–834. doi: 10.1126/science.277.5327.831. [DOI] [PubMed] [Google Scholar]
- 28.Rivard N, L'Allemain G, Bartek J, Pouysségur J. J Biol Chem. 1996;271:18337–18341. doi: 10.1074/jbc.271.31.18337. [DOI] [PubMed] [Google Scholar]
- 29.Hammond SL, Ham RG, Stampfer MR. Proc Natl Acad Sci USA. 1984;81:5435–5439. doi: 10.1073/pnas.81.17.5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stampfer MR, Yaswen P. Cancer Lett. 2003;194:199–208. doi: 10.1016/s0304-3835(02)00707-3. [DOI] [PubMed] [Google Scholar]
- 31.Brenner AJ, Stampfer MR, Aldaz CM. Oncogene. 1998;17:199–205. doi: 10.1038/sj.onc.1201919. [DOI] [PubMed] [Google Scholar]
- 32.Meyerson M, Counter CM, Eaton EN, Ellisen LW, Steiner P, Caddle SD, Ziaugra L, Beijersbergen RL, Davidoff MJ, Liu Q, et al. Cell. 1997;90:785–795. doi: 10.1016/s0092-8674(00)80538-3. [DOI] [PubMed] [Google Scholar]
- 33.Garbe J, Wong M, Wigington D, Yaswen P, Stampfer MR. Oncogene. 1999;18:2169–2180. doi: 10.1038/sj.onc.1202523. [DOI] [PubMed] [Google Scholar]
- 34.Whitman M, Downes CP, Keeler M, Keller T, Cantley L. Nature. 1988;332:644–646. doi: 10.1038/332644a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.