Abstract
Tetraploid embryo complementation assay has shown that mouse ES cells alone are capable of supporting embryonic development and adult life of mice. Newly established F1 hybrid ES cells allow the production of ES cell-derived animals at a high enough efficiency to directly make ES cell-based genetics feasible. Here we report the establishment and characterization of 12 new F1 hybrid ES cell lines and the use of one of the best (G4) in a gain- and loss-of-function genetic study, where the in vivo phenotypes were assessed directly from ES cell-derived embryos. We found the generation of G4 ES cell-derived animals to be very efficient. Furthermore, even after two consecutive rounds of genetic modifications, the majority of transgenic lines retained the original potential of the parental lines; with 10–40% of chimeras producing ES cell-derived animals/embryos. Using these genetically altered ES cells, this success rate, in most cases, permitted the derivation of a sufficient number of mutants for initial phenotypic analyses only a few weeks after the establishment of the cell lines. Although the experimental design has to take into account a moderate level of uncontrolled damage on ES cell lines, our proof-of-principle experiment provides useful data to assist future designs harnessing the power of this technology to accelerate our understanding of gene function.
Keywords: hybrid, tetraploid complementation assay, vasculogenesis, ES cells
The advent of mouse ES cells (1, 2) has revolutionized the genetic approaches addressing gene function. It helped transform the mouse into the ultimate mammalian model system with significant relevance to human biology. Mutating all of the genes in the mouse has become not only feasible but also a substantial international project supported by the European Union, the National Institutes of Health, and Genome Canada (3). Before this effort, thousands of ES cell lines had already been created, each representing a specific mutation, and many more are currently under way. Introduction of these cells back into the mouse through germ-line transmission is a time-, labor-, and cost-intensive endeavor, followed by the tedious task of phenotypic analyses. Therefore, an assay system, which could accelerate the production of mutant embryos or animals and at the same time provide information about in vivo phenotypes for elucidating gene function in normal developmental and pathological processes, would be of major value.
Two specific properties of mouse ES cells render them exceptional tools for genetic research: (i) virtually unlimited proliferation capacity and (ii) pluripotent developmental potential. Their proliferation capability leads to the production of large number of cells and therefore the occurrence and the identification of very rare events, such as homologous recombination or gene trap insertion. The pluripotent developmental potential of ES cells allows them to contribute to the germ line when reintroduced into an embryonic environment through chimera formation. However, an additional property, currently not broadly recognized, will enable more powerful functional assays to be used for revealing mutant phenotypes. It is well known that ES cells are capable of supporting the development of the entire embryo (4) and the life of the adult mouse (5), when the trophoblast and the primitive endoderm lineages are provided by tetraploid carrier embryos (tetraploid embryo complementation assay). In the early years of this technology only inbred-derived ES cell lines were available and the rate of development of these ES cell-derived embryos/animals was poor. For a long time, only the R1 ES cell line (5) allowed the production of ES cell-derived animals, and even the construction of ES cell-derived embryos showed moderate to low efficiency (6). This situation changed dramatically when ES cell lines derived from F1 hybrid embryos were analyzed. Their superior developmental potential became evident both in cloning by nuclear transfer and in tetraploid embryo complementation assays (7).
The use of hybrid ES cell lines has increased the efficiency for producing completely ES cell-derived embryos and animals to a level, which renders the technology easy to integrate into mouse genetics laboratories. In this report we present an experimental design for the production of mutant mice ready for phenotyping, using new lines of hybrid ES cells in the tetraploid embryo complementation assay. Our proof-of-principle study describes the development of a gain- or loss-of-function screen for testing of biological activity of a large number of gene products. We also point out possible pitfalls associated with the tetraploid embryo complementation assay, which can be avoided with careful experimental design. This system demonstrates prudent use of research time, space and expenses compared with currently available methods. The increased efficiency could further impact our approach to mouse genetics.
Results
Establishment of 129×C57BL/6 F1 Hybrid ES Cell Lines.
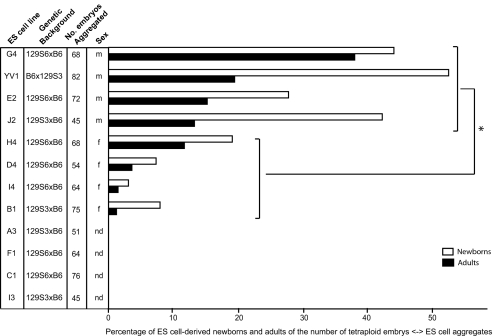
We generated 12 F1 hybrid ES cell lines derived from embryos obtained by crossing 129 and C57BL/6 animals. In all cases except one (YV1), the maternal origin was 129 (either 129S3/SvImJ or 129S6/SvEvTac), whereas the paternal origin was C57BL/6Ncr (B6) (Fig. 1). Of all blastocysts used, 25% became established ES cell lines. An early passage number of all 12 lines was tested by the tetraploid embryo complementation assay (passage number 5–7) to determine the efficiency of generating live animals (Fig. 1). Efficiency was calculated as the percentage of newborns and adults obtained of the number of embryos (chimera aggregates) transferred in recipient females. Interestingly, there was a significant difference between the male and female lines in generating animals, on average 21.6% and 4.6%, respectively. Despite the relatively poor developmental potential of the female ES cell lines, one of them, designated H4, still supported the development to term of 19% of embryos, and 12% survived to adulthood. The ability for each line to support development to term did not correlate with the ability of corresponding ES cell-derived newborns to survive to adulthood (data not shown). Both male and female lines displayed similar average survival efficiency after birth: 48% and 52.7%, respectively. One ES cell line, designated G4, showed superior developmental potential over the other F1 hybrid lines, by generating 30 newborns of the 68 ES cell ⇔ tetraploid embryo aggregates transferred (44% of survival to term). Of those 30, 26 animals (87%) survived to adulthood and were fertile (five of five tested). On this basis this line was used for further studies.
Fig. 1.
The efficiency of generating completely ES cell-derived newborns and adult mice from different F1 hybrid ES cell lines with tetraploid embryo complementation assay. The genetic background first shows the maternal component. P < 0.05. The YV1 line contains an overall expressed yellow fluorescent protein transgene; Tg(ACTB-EYFP) 2 Nagy. m, male; f, female.
Characterization of the G4 ES Cell Line.
Next we investigated whether the G4 ES cell line could retain its developmental potential after extensive culture. The cells were reevaluated by tetraploid embryo complementation assays after increasing the passage numbers. At passages 5, 8, 9, 11, and 14, we obtained 44%, 38%, 26%, 33%, and 32% of viable newborns, respectively. With an increase in passage number, no significant drop in efficiency was observed. Hence, we concluded that the G4 line was capable of maintaining its developmental potential at least for nine additional passages.
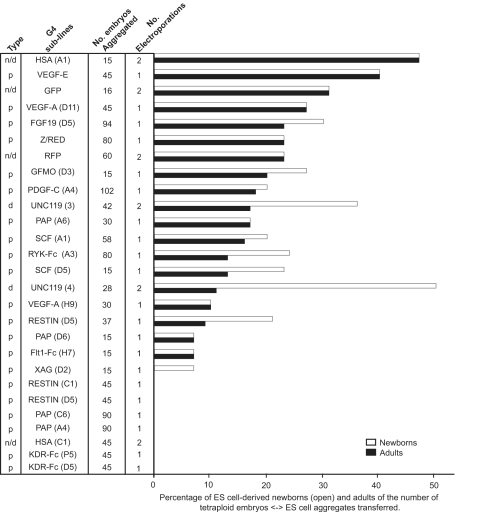
We also addressed the critical question of whether the G4 line would maintain its properties after genetic manipulation (detailed later), which required electroporation and subcloning. Data were collected from 28 transgenic sublines where no specific phenotype was expected. These sublines contained either neutral or Cre recombinase conditionally expressed transgenes. Twenty-two of these lines were subjected to one round of electroporation, and six lines were subjected to two consecutive electroporations. In total, 20 of 28 of these sublines (15 of the single and five of the double electroporated sublines) resulted in development to term (Fig. 2). The success ratio was found to be similar to the ratio obtained during the testing of the parental F1 hybrid ES cell lines (Fig. 1). Most likely the 8 transgenic lines, which were unsuccessful in generating live mice, were damaged during in vitro culture independent from the presence of the transgene. Based on this series of experiments, the probability of deriving undamaged sublines from the G4 ES cell line is 0.71 (20/28). Therefore, if two independent sublines containing the same neutral transgene insertion were tested with tetraploid embryo complementation assay, the probability of obtaining live offspring from at least one of the two lines is 1–0.292 = 0.92, which is in agreement with the fact that we could derive animals from 16 of the 18 different transgenes (different constructs) (Fig. 2). Importantly, the average efficiency of deriving animals remained the same for the two consecutively electroporated and subcloned ES cell lines or for just a single round of electroporation (Fig. 2). These results strongly support the fact that the developmental potential of the G4 ES cell line is robust, and is unaffected by a series of genetic manipulations, subcloning and extensive in vitro culture.
Fig. 2.
The performance of single electroporated (parental) and double electroporated (neutral transgenic) ES cell lines in the tetraploid embryo complementation assay. p, parental; d, daughter; n, neutral.
Developmental Effect of Transgene Expression Determined by Using ES Cell-Derived Embryos.
Next, we tested the feasibility of in vivo assessments of phenotypes associated with transgene expression directly from the G4 F1 hybrid ES cell line using the tetraploid embryo complementation assay.
Cre recombinase conditional transgenes (Fig. 3) were constructed by combining the following sequence of components: (i) a CMV enhancer combined with chicken β-actin promoter (pCAGG) (8) followed by (ii) a loxP-flanked β-galactosidase neomycin fusion gene (βgeo) and three polyadenylation signals (3xpA) (9), (iii) a gene of interest (GOI), and (iv) an IRES-puromycin-pA cassette. In cells, this construct confers neomycin resistance and lacZ expression, but it does not allow expression of the GOI until Cre recombinase-mediated excision occurs. The recombinase removes the βgeo, which brings the GOI-IRES-puromycin-pA under the transcriptional control of the pCAGG promoter. These cells become puromycin-resistant and express the GOI.
Fig. 3.
Structure and basic properties of conditional and activated transgenes.
The GOIs selected for this experiment fell into three categories: (i) known angiogenic/angiostatic factors: VEGF-A (10), VEGF-E (11), SCF (12), FGF-2 (13), KDR-Fc (14), Flt1-Fc (15), IL4 (16), and Restin (17); (ii) unknown (no specific expectation): PDGF-C (18) FGF-19 (19), PAP (20), Ryk-Fc (21), GFMO (22), and UNC-119 (23); (iii) expected neutral factors: HSA (24), EGFP (25), and DsRed-MST (26). These conditional transgenic vectors were electroporated into G4 ES cells and subjected to G418 selection. For each transgene, sublines were screened for single-copy, single-site integration by Southern blot analysis, and for high levels of transgene expression by lacZ staining (data not shown). Two or three lines for each GOI were selected and used in further studies. They did not express the GOI, therefore they were also considered neutral and referred to as “parental” lines. “Daughter” cell lines with active transgene expression were then derived by a subsequent electroporation of the Cre expression vector. Excision of the βgeo cassette and activation of GOI was determined by puromycin selection. The resulting puromycin-resistant lines per electroporation were pooled and frozen for further study.
Eighteen daughter lines expressing 14 different GOIs from the first two categories of transgenes were used in tetraploid embryo complementation assays. Only one activated daughter cell line (Unc119) resulted in adult animals (Table 1). To determine the stage of embryonic lethality associated with the expression of each transgene, ES cell-derived embryos were dissected and analyzed at different time-points during gestation. In each case, one or two experiments resulted in enough pregnant females, which were sufficient to establish the developmental stage of lethality (Table 1). In addition, most lines resulted in a sufficient number of embryos to enable us to perform a preliminary phenotypic analysis.
Table 1.
Observed embryonic lethality due to overexpression of factors anticipated to give a phenotype
| Transgenic ES cell lines | Type of factor | No. of embryos aggregated | Status of embryos at time of dissection | Embryonic stage at dissection |
Stage of embryonic lethality observed | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | … | T | ||||||
| VEGF-A (H9) | A | 45 | Dead | 19 | E9.5 | ||||||||||
| Live | 0 | ||||||||||||||
| VEGF-A (D11) | A | 45 | Dead | 27 | E9.5 | ||||||||||
| Live | 0 | ||||||||||||||
| VEGF-E | A | 64 | Dead | 56 | E9.5 | ||||||||||
| Live | 0 | ||||||||||||||
| SCF (A1) | A | 90 | Dead | 3 | 3 | 4 | 13 | E15.5 | |||||||
| Live | 4 | 8 | 6 | 0 | |||||||||||
| SCF (D5) | A | 81 | Dead | 0 | 1 | 7 | 7 | E13.5 | |||||||
| Live | 4 | 4 | 2 | 0 | |||||||||||
| FGF-2 | A | 180 | Dead | 2 | 2 | 2 | 3 | 4 | P1 | ||||||
| Live | 4 | 5 | 7 | 12 | 0 | ||||||||||
| PDGF-C (A4) | U | 44 | Dead | 2 | 13 | E12.5 | |||||||||
| Live | 4 | 0 | |||||||||||||
| KDR-Fc | S | 79 | Dead | 41 | E9.5 | ||||||||||
| Live | 0 | ||||||||||||||
| Flt-1 Fc (H7) | S | 47 | Dead | 20 | E9.5 | ||||||||||
| Live | 0 | ||||||||||||||
| IL-4 (A1) | S | 120 | Dead | 6 | 2 | 5 | E12.5 | ||||||||
| Live | 2 | 3 | 0 | ||||||||||||
| FGF-19 (A5) | U | 60 | Dead | 2 | 4 | 5 | E13.5 | ||||||||
| Live | 3 | 3 | 0 | ||||||||||||
| PAP (A4) | U | 114 | Dead | 1 | 2 | 6 | E12.5 | ||||||||
| Live | 3 | 6 | 0 | ||||||||||||
| PAP (A6) | U | 38 | Dead | 3 | 1 | E13.5 | |||||||||
| Live | 2 | 1 | |||||||||||||
| Ryk-Fc (A3) | U | 40 | Dead | 0 | 0 | 2 | 6 | P1 | |||||||
| Live | 5 | 5 | 4 | 1 | |||||||||||
| GFMO (D3) | U | 14 | Dead | 6 | 0 | 4 | P1 | ||||||||
| Live | 5 | 1 | 0 | ||||||||||||
| XAG (D2) | U | 23 | Dead | 0 | 0 | 1 | P1 | ||||||||
| Live | 4 | 4 | 0 | ||||||||||||
| UNC119 (3) | U | 40 | Dead | 0 | 4 | 2 | Viable at | ||||||||
| Live | 6 | 7 | 7 | birth | |||||||||||
| UNC119 (4) | U | 34 | Dead | 0 | 0 | 2 | Viable at | ||||||||
| Live | 3 | 8 | 3 | birth | |||||||||||
The numbers of live vs. dead embryos dissected at different stages of pregnancies with ES cell-derived embryos expressing factors anticipated to give developmental perturbations. A, angiogenic; S, angiostatic; U, unknown; T, at term; P1, postnatal day 1.
To further validate the assay, parental ES cell-derived animals with conditional transgenes for VEGF-A, Flt1-Fc, VEGF-E, SCF, and dsRed-MST were crossed with Cre recombinase transgenic partners (Cre deletor) (9). Twenty-five percent of the offspring were double transgenic where the Cre recombinase has activated the GOI expression. When examined at different time-points, these embryos recapitulated the phenotypes observed in their ES cell-derived counterparts. For example, VEGF-A transgenic expression resulted in lethality by embryonic day 9.5 (E9.5). The embryos lacked blood in the yolk sac and the embryo proper. In addition the yolk sac vessels were abnormally dilated and failed to develop proper capillary structure (Fig. 4 A–D). The expression of Flt1-Fc a secreted extracellular domain of a VEGF receptor caused a complete lack of blood and endothelial cells in the yolk sac from the ES cell-derived embryos [supporting information (SI) Fig. 5A] as well as in embryos resulted from the cross between conditional Flt1-Fc (germ line of the H7 line; see Fig. 2) and a Cre deletor (Fig. 4 E–H). The absence of dorsal aorta and decreased vascular network (data not shown) in the embryo proper were consistent with the reported phenotype of the VEGF-A deficient embryos (6). Embryos derived from SCF overexpressing cells died between E14.5 and E15.5 (SI Fig. 5 C and D) showing signs of internal bleeding and overall hemorrhaging starting at E12.5. This phenotype was recapitulated when the parental lines were mated with Cre deletor mice (Fig. 4 I–M).
Fig. 4.
Phenotype of embryos from crosses between the conditional transgenic and a Cre deletor line recapitulated the phenotype observed in corresponding ES cell-derived embryos. (A and B) Single transgenic E9.5 embryo and yolk sac (wild type). (C and D) Double transgenic littermate expressing VEGF-A from the activated transgene. The yolk sacs (B and D) were stained for PECAM to visualize the organization of the endothelial cells. (E and F) Single transgenic E9.5 embryo and its cross-section at the caudal level. (G and H) Corresponding double transgenic littermate with Flt1-Fc expression activation. (I) Single transgenic “wild-type” embryo at E14.5. (J and K) A corresponding double transgenic littermate displaying sites of hemorrhage (black arrowheads) and edema (white arrowhead). (L and M) H&E section of wild-type and SCF expressor embryonic livers showing severe necrosis in the latter. (Scale bars: 100 μm.)
Discussion
The developmental potential of the new F1 hybrid ES cell lines, including G4, which supports life of ES cell-derived animals, could impact the phenotype analyses of several widely used mouse genetics approaches. Here we provide a validation of an experimental design that could lead to a gain- or loss-of-function in vivo screen for biological activities of gene products during development and adult life at a higher throughput than has ever been possible to date.
The superior developmental potential of some F1 hybrid ES cells was demonstrated by either tetraploid embryo complementation assay or cloning by nuclear transfer (7). Two-thirds of our newly established F1 hybrid ES cell lines supported the development of ES cell-derived animals to term and beyond. A similar frequency (≈70%) of supportive lines was observed when single-cell-derived sublines were generated from one of the hybrid ES cell lines designated G4 by electroporation of neutral transgenes. This similarity argues against the possibility that the developmental potential variation of the cell lines roots from their different cellular origin inside the founder blastocyst. On the other hand, it supports the model that the loss of developmental potential of the cells is acquired during in vitro culture, likely because of genetic or epigenetic changes.
Some new C57BL/6 inbred embryo-derived ES cell lines also show similar superior developmental potential as the good F1 hybrids, tested with tetraploid embryo complementation assay (M.G. and A.N., unpublished data) or with very high levels of ES cell contribution when the cells were injected under the zona pellucida of morula stage diploid embryos (personal communication). Many factors such as the genetic background of the host embryo, the method of chimera generation and the culture conditions of ES cells can influence the level of contribution of ES cells in chimeras and the behavior of their derivatives in adult tissues and organs (27). These findings suggest that heterozygosity (F1 hybrid origin) of ES cells maybe sufficient but not necessary for this phenomenon. Thus, we feel confident that an efficient tetraploid embryo complementation-based phenotyping will not be restricted to the use of F1 hybrid ES cells in the future.
Four of the eight independently established ES cell lines that supported development to term were female. These behaved significantly poorer than their male counterparts in generating ES cell-derived mice (P < 0.05). The difference between male and female ES cells has never been rigorously studied, although the phenomenon has long been suggested. The frequent loss of one of the X chromosomes has been mentioned as a possible underlying mechanism for the differences observed (28). Here we confirmed this observation with statistically significant differences between the developmental potential of male and female ES cell lines.
The present study showed that 71% of the G4-derived sublines were informative with regards to pre-, peri-, and postnatal phenotypes. The remaining 29%, however, had an embryonic lethal phenotype, which was not associated with transgene expression. These embryonic phenotypes, called “false-positive” phenotypes, warrant caution during experimental designs. The use of two or more parallel subclones can distinguish between true embryonic phenotypes and false positive phenotypes. It is also important to use host embryos that express a reporter, such as GFP (in our case) or lacZ as shown in one of our previous studies describing the VEGF-A knockout (6), to identify any occasional tetraploid cell contribution to the embryo proper (29).
The power of the tetraploid embryo complementation assay in the effort to understand gene function depends on the ability of producing gain- or loss-of-function mutant embryonic stem cell lines. The currently available methods of mutant production have different aspects to consider in conjunction with directly phenotyping from ES cells. For example, the expression of “classical” transgenes is integration site dependent, that is, the genomic location affects the level, pattern and stability of the expression. Thus, the validation of a phenotype associated with a transgene expression when they are coming from different genomic sites is very challenging and labor-intensive. However, there are several ways to generate independent transgenic ES cell lines where the same (or different) constructs are expressed from the same genomic location. One way to achieve this is to target a transgene into a well characterized locus, for example the ROSA26 (30) or HPRT (31). Alternatively, recombinase-mediated cassette exchange could be used to target the same genomic site to derive independent transgenic lines for the same construct or to obtain comparable expression for different constructs. For recombinase-mediated cassette exchange, there are several solutions provided by the Cre (32) and FLPe (33) recombinases and PhiC31 integrase (34). Comparison of independently established mutant and control lines could efficiently pinpoint the phenotype associated with the genetic manipulation performed (6). Alternatively, for transgene expression, one might produce a Cre recombinase conditional transgenic line as we did in this study, where the parental line expresses lacZ before Cre excision. This parental line can be tested for single-copy, single-site transgene insertion, the pattern, the strength and stability of lacZ expression in addition to the developmental potential by tetraploid embryo complementation. The normality of parental ES cell-derived embryos/animals ensures that the parental line is not damaged; therefore the majority (our estimate 71%) of sublines generated after Cre activation of the transgene were informative with regards to the phenotype associated with its expression. For practical reasons, these activated sublines can be pooled for phenotypic analysis as shown in this study from the three examples given. Overall, the parental lines were normal and were capable to support ES cell-derived animals to adulthood. Using pooled daughter cells, we were able to recapitulate phenotypes already observed by VEGF-A knockout by overexpression of Flt1-Fc. In another example we showed that overexpression of VEGF-A by 5-fold led to early hematopoietic and endothelial phenotypes. Similarly, the overexpression of SCF led to mid-gestation lethality, which showed signs of hematopoietic and vascular problems.
One might think that pooling of daughter cell lines is not a good idea, because in chimera production an uncontrolled mixture of informative and damaged cells are introduced into the tetraploid embryo ⇔ ES cell chimeras. This would be definitely true for blastocyst injection chimeras where 10–15 single cells are injected into the host embryo. For our aggregation chimeras, however, loosely connected clumps of ES cells are used. Therefore, if ES cells are plated sparse, as single cells, 2 days before the aggregation, they clonally grow into small clumps during 48 h. They remain loosely connected, therefore, in most of the cases, single cell-derived groups of cells constitute to the aggregates with tetraploid embryos.
In future designs, the Cre recombinase system could offer an alternative strategy for separating true and false phenotypes of ES cell-derived embryos or animals. For example, entirely flanking a single-copy transgene by loxP sites allows reverting the transgene in daughter cell line to wild type by Cre recombinase-mediated excision. This strategy might be the most economical to learn and validate the effect of a gain of function mutations by ES cell-derived embryos/animals.
Creating loss-of-function mutations in ES cells pose both similar and different challenges. The design for RNAi knockdown can be achieved by stable integrant transgenes (35) similarly to the procedure described above for the gain-of-function strategies. An efficient approach could be to control the integration and copy number of the shRNA expressing transgene either by gene targeting or by recombinase-mediated cassette exchange with Flp or PhiC31, where loxP sites flank the entire transgene itself. When the phenotype of the presumed knockdown is established, Cre excision of the transgene should revert the cells to wild type and therefore the validation of phenotypes should be straightforward. RNAi, however, cannot produce null mutations. If this is required, gene targeting is necessary, creating homozygous knockout ES cell lines. There are two ways to do so: applying high concentration G418 selection on heterozygous cells and identifying sublines homozygous for the targeted allele or by double targeting of the ES cells (36). The first option is technically less demanding but often creates uniparental disomy of the chromosome containing the targeted allele (37).
Uniparental disomy also establishes homozygosity along the entire chromosome, which could create altered function mutation in a nontargeted locus. Caution would be needed to ensure that these possible secondary effects do not give false results. One might question the targeting efficiency of genes in an F1 hybrid ES cell line. Successful targeting of several genes has been performed with the G4 F1 hybrid ES cells in our institution as well as others. No sign of any drop of targeting frequency has been detected by using target vectors with C57BL/6 homology arms, which is isogenic to one of the haploid genomes of the hybrid ES cells. Creating loss of function in gene trapped ES cell lines are seemingly less challenging because the vector insertion creates a mutation in one of the alleles. The remaining allele can be targeted; a step that could create parallel mutant and control lines needed for phenotypic analysis of pools or individual subclones. Alternatively, homozygous gene trap lines can be generated by high concentration G418 selection if the wild-type neo gene was not used for trapping (38).
To fully interpret the phenotypes of mutant ES cell-derived embryos, one has to be aware of the “non-mutant” origin of the trophoblast and the primitive endoderm-derived extra-embryonic membranes (4). Thus, the tetraploid embryo complementation assay allows the examination of the gene function in the embryo proper or postnatal animal. In addition, genes that affect key properties of ES cells, such as pluripotency, cannot be easily manipulated without the use of generating conditional alleles. In such cases, it would be necessary to generate breeding colonies.
Genetic studies performed in the mouse have already provided enormous amounts of information on gene function in normal development, physiology and in disease. During the past two decades, thousands of transgenic mouse lines have been produced, ≈5,000 genes have been knocked out by gene targeting (3) and >10,000 have been mutated by gene trapping (39). These efforts are predicted to continue with at least the same speed. The technology of altering the mouse genome has evolved to the point where the ambitious international effort to mutate all of the mouse genes by the use of ES cells has become realistic (3). The European Community, the National Institutes of Health, and Genome Canada have already launched their programs of systematic targeting and mutating genes in ES cells and banking these lines for future gradual phenotypic analyses. The magnitude of this project could grow equal to that of the human genome sequencing, which was initiated ten years ago and accomplished 5 years later. It is obvious that phenotypic analysis of all these mutations will pose several orders of magnitude higher challenges, calling for new, fast and economical ways of analyzing gene function. Our present study could help to move forward in this direction.
Materials and Methods
Derivation of F1 Hybrid ES Cell Lines.
The hybrid ES cell lines were established from blastocysts obtained from the natural mating between 129 and C57BL/6 parents (for further details see Results). The blastocysts were collected at E3.5 and plated in four-well dishes (Nunc, Rochester, NY; catalog no. 176740) on mitomycin C-treated mouse embryonic fibroblasts in low glucose DMEM (Invitrogen/GIBCO, Carlsbad, CA; catalog no. 11885-084) supplemented with 25% FBS (HyClone, Logan, UT), 2,000 units/ml leukemia inhibitory factor, 0.1 mM 2-mercaptophenol, 2 mM glutamax (Invitrogen), 1 mM sodium pyruvate, and 0.1 mM nonessential amino acids. After the first disaggregation of the outgrowth, the medium was gradually changed to standard ES cell medium (high-glucose DMEM and 15% FBS including supplements mentioned above).
Plasmid Construction.
The cloning vector, designated as CLIP, was made and used for conditional transgenic expression of cDNAs in ES cells. It contains the following: CMV enhancer-chicken β-actin promoter, loxP, coding sequence for β-galactosidase neomycin fusion protein, 3x pA, loxP, cloning site for the GOI, IRES, puromycin, pA. To make the CLIP vector, a 1.1-kb PstI-EcoRI fragment (IRES-puromycin) from pCAGGS-ires puro plasmid was first subcloned into BglII site of pCALL plasmid (40). A linker (BglII, XhoI, NheI, PmeI, and NotI) was then subcloned into an XbaI site of pCALL-IRES-puro. Individual cDNAs were subcloned from p2TOP-Incyte GOI vector into NheI-PmeI of CLIP. An H5V5 tag was introduced 3′ of some of the cDNAs to assist the detection of transgene expression (Fig. 3).
Generation of Transgenic ES Cell Lines.
Briefly, the G4 ES cells were grown at 37°C in 5% CO2 on mitomycin C-treated mouse embryonic fibroblasts [derived from TgN (DR4)1 Jae embryos] in high-glucose DMEM (Invitrogen), supplemented with 15% ES cell-grade FBS (HyClone and Wisent), 0.1 mM 2-mercaptophenol, 2 mM l-glutamine, 1 mM sodium pyruvate, 0.1 mM nonessential amino acids, and 2,000 units/ml lymphocyte inhibitory factor (41). Approximately 5 × 106 cells were mixed with 20–30 μg of linearized (with ScaI or SfiI) DNA in electroporation buffer (Specialty Media) and electroporated at 250 V and 500 μF using a Bio-Rad Gene Pulser. The cells were plated at a density of 2 × 106 cells per 100-mm plate. Twenty-four hours after electroporation, G418 (166 μg/ml Geneticin; Invitrogen) was added to the medium to select for neomycin-resistant cells. The resulting resistant colonies were picked onto mouse embryonic fibroblast-coated 96-well plates and replica-plated. One replica was stained for lacZ expression, and another was used in Southern blot analyses to detect single-copy/site integration of the transgene (a 760-bp neomycin probe or 3,000-bp βgeo probe was used). The third replica plate was frozen for future expansion of the selected ES cell sublines. The strength and uniformity of transgene expression were analyzed by lacZ expression of the neomycin-resistant colonies (parental ES cell lines) as described by Lobe et al. (9). The strong lacZ-expressing single-site/copy transgenic clones were expanded and frozen down as the parental lines.
Two or three of the selected clones were used to establish the daughter (expression) cell lines as follows: the parental ES cells were transiently transfected with a Cre recombinase expression plasmid, where the pCAGG promoter (8) was driving a nuclear localization signal equipped Cre. Twenty-four hours after electroporation of the Cre expression vector, the cells were subjected to puromycin (1.25 μg/ml; Sigma, St. Louis, MO) selection for 3–4 days. For each ES cell line, the puromycin-resistant colonies were pooled, expanded, and frozen. These daughter ES cell lines were later thawed and prepared for the tetraploid embryo complementation assay.
In Vivo Tetraploid Embryo Complementation Assay.
For each GOI, at least two independent expression lines and the parental conditional cell lines noted in Fig. 2 were subjected to the in vivo complementation assay as described (4, 5). Briefly, the blastomeres of two-cell stage embryos (E1.5) from superovulated ICR (Harlan) females mated to males homozygous for pCAGG-EGFP [Tg(ACTB-EGFP)B5 Nagy] (42) were electrofused by using a CF-150B Pulse Generator (BLS). The fused embryos (EGFP transgenics) were cultured overnight at 37°C in 5% CO2 in KSOM medium (Specialty Media). Two tetraploid embryos at the four-cell stage were aggregated with a clump of 8–15 ES cells. The following day, 12–15 embryos were transferred into each pseudopregnant recipient ICR female (43). Embryos were dissected in ice-cold PBS, and contribution of EGFP-positive cells was visualized by using either a Leica MS5 stereomicroscope with attached MAA-02 Universal light source from BLS or Leica MZ16 FA. Embryos were photographed by using a Retiga 1300C digital camera. The EGFP expression of tetraploid embryos was used to confirm the ES cell origin of the dissected embryos. Wild-type ICR embryos were used for ES cell lines left to term to produce live animals.
Preparation of Embryos and Immunohistochemistry.
Dissection, fixation, embedding, and sectioning of embryos were carried out as previously described by Haigh et al. (44). Immunohistochemistry was performed with PECAM (Pharmingen, San Jose, CA). Whole-mount lacZ analysis was performed between E9.5 and E12.5 according to published protocols (43).
Genotyping.
The parental (conditional) ES cell-derived mice were maintained on ICR background. Transmission of the transgene was determined by PCR or lacZ staining of ear punches as described previously (40). Offspring of crosses between conditional ES cell-derived mice and general Cre deletor (pCAGG-Cre) females were genotyped by the following PCR primers: pCAGG-Cre P1, ggttattgtgctgtctcatca; pCAGG-Cre P2, atatcctggcagcgatcgcta; SCF P1, atgaagaagacacaaacttg; SCF P2, gaccgaggagaggggttaagg; PAP P1, atgctgctgtctcaggttca; PAP P2, gaccgaggagagggttaag; VEGF-A P1, tggatccatgaactttctgct; VEGF-A P2, gaattcaccgcctcggcttgtc; Flt1-Fc P1, tggttgtaagccttgcatagtacagtc; Flt1-Fc P2, ctagctagctttaccaggagagtgggag.
Statistics.
χ2 analysis was used to determine the significant difference between male and female F1 hybrid ES cell lines in their ability to give rise to ES cell-derived mice.
Supplementary Material
Acknowledgments
We thank S. MacMaster, S. Tondat, and L. Beyers of the transgenic core facility at the Samuel Lunenfeld Research Institute for doing most of the tetraploid embryo complementation assays; K. Harpal for histology; J. Hsien for maintenance of the mouse lines; and Dr. H. Ding for useful discussions. This work was supported by National Cancer Institute of Canada Grants 16002 and 016343 and a grant from Bayer Pharmaceuticals. A.N. is a Canadian Institutes of Health Research distinguished scientist.
Abbreviations
- GOI
gene of interest
- En
embryonic day n.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0609277104/DC1.
References
- 1.Evans MJ, Kaufman MH. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 2.Martin GR. Proc Natl Acad Sci USA. 1981;78:7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Austin CP, Battey JF, Bradley A, Bucan M, Capecchi M, Collins FS, Dove WF, Duyk G, Dymecki S, Eppig JT, et al. Nat Genet. 2004;36:921–924. doi: 10.1038/ng0904-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nagy A, Gocza E, Diaz EM, Prideaux VR, Ivanyi E, Markkula M, Rossant J. Development (Cambridge, UK) 1990;110:815–821. doi: 10.1242/dev.110.3.815. [DOI] [PubMed] [Google Scholar]
- 5.Nagy A, Rossant J, Nagy R, Abramow-Newerly W, Roder JC. Proc Natl Acad Sci USA. 1993;90:8424–8428. doi: 10.1073/pnas.90.18.8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carmeliet P, Ferreira V, Breier G, Pollefeyt S, Kieckens L, Gertsenstein M, Fahrig M, Vandenhoeck A, Harpal K, Eberhardt C, et al. Nature. 1996;380:435–439. doi: 10.1038/380435a0. [DOI] [PubMed] [Google Scholar]
- 7.Eggan K, Akutsu H, Loring J, Jackson-Grusby L, Klemm M, Rideout WM, III, Yanagimachi R, Jaenisch R. Proc Natl Acad Sci USA. 2001;98:6209–6214. doi: 10.1073/pnas.101118898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Niwa H, Yamamura K, Miyazaki J. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 9.Lobe CG, Koop KE, Kreppner W, Lomeli H, Gertsenstein M, Nagy A. Dev Biol. 1999;208:281–292. doi: 10.1006/dbio.1999.9209. [DOI] [PubMed] [Google Scholar]
- 10.Neufeld G, Cohen T, Gengrinovitch S, Poltorak Z. FASEB J. 1999;13:9–22. [PubMed] [Google Scholar]
- 11.Meyer M, Clauss M, Lepple-Wienhues A, Waltenberger J, Augustin HG, Ziche M, Lanz C, Buttner M, Rziha HJ, Dehio C. EMBO J. 1999;18:363–374. doi: 10.1093/emboj/18.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galli SJ, Zsebo KM, Geissler EN. Adv Immunol. 1994;55:1–96. doi: 10.1016/s0065-2776(08)60508-8. [DOI] [PubMed] [Google Scholar]
- 13.Liekens S, De Clercq E, Neyts J. Biochem Pharmacol. 2001;61:253–270. doi: 10.1016/s0006-2952(00)00529-3. [DOI] [PubMed] [Google Scholar]
- 14.Robb L, Elefanty AG. BioEssays. 1998;20:611–614. doi: 10.1002/(SICI)1521-1878(199808)20:8<611::AID-BIES3>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 15.Fong GH, Rossant J, Gertsenstein M, Breitman ML. Nature. 1995;376:66–70. doi: 10.1038/376066a0. [DOI] [PubMed] [Google Scholar]
- 16.Paul WE. Blood. 1991;77:1859–1870. [PubMed] [Google Scholar]
- 17.Sasaki T, Larsson H, Tisi D, Claesson-Welsh L, Hohenester E, Timpl R. J Mol Biol. 2000;301:1179–1190. doi: 10.1006/jmbi.2000.3996. [DOI] [PubMed] [Google Scholar]
- 18.Ding H, Wu X, Kim I, Tam PP, Koh GY, Nagy A. Mech Dev. 2000;96:209–213. doi: 10.1016/s0925-4773(00)00425-1. [DOI] [PubMed] [Google Scholar]
- 19.Xie MH, Holcomb I, Deuel B, Dowd P, Huang A, Vagts A, Foster J, Liang J, Brush J, Gu Q, et al. Cytokine. 1999;11:729–735. doi: 10.1006/cyto.1999.0485. [DOI] [PubMed] [Google Scholar]
- 20.Iovanna JL, Keim V, Bosshard A, Orelle B, Frigerio JM, Dusetti N, Dagorn JC. Am J Physiol. 1993;265:G611–G618. doi: 10.1152/ajpgi.1993.265.4.G611. [DOI] [PubMed] [Google Scholar]
- 21.Halford MM, Armes J, Buchert M, Meskenaite V, Grail D, Hibbs ML, Wilks AF, Farlie PG, Newgreen DF, Hovens CM, et al. Nat Genet. 2000;25:414–418. doi: 10.1038/78099. [DOI] [PubMed] [Google Scholar]
- 22.Ogawa K, Tanaka K, Ishii A, Nakamura Y, Kondo S, Sugamura K, Takano S, Nakamura M, Nagata K. J Immunol. 2001;166:6404–6412. doi: 10.4049/jimmunol.166.10.6404. [DOI] [PubMed] [Google Scholar]
- 23.Higashide T, McLaren MJ, Inana G. Invest Ophthalmol Vis Sci. 1998;39:690–698. [PubMed] [Google Scholar]
- 24.Mendez CM, McClain CJ, Marsano LS. Nutr Clin Pract. 2005;20:314–320. doi: 10.1177/0115426505020003314. [DOI] [PubMed] [Google Scholar]
- 25.Cubitt AB, Heim R, Adams SR, Boyd AE, Gross LA, Tsien RY. Trends Biochem Sci. 1995;20:448–455. doi: 10.1016/s0968-0004(00)89099-4. [DOI] [PubMed] [Google Scholar]
- 26.Vintersten K, Monetti C, Gertsenstein M, Zhang P, Laszlo L, Biechele S, Nagy A. Genesis. 2004;40:241–246. doi: 10.1002/gene.20095. [DOI] [PubMed] [Google Scholar]
- 27.Schoonjans L, Kreemers V, Danloy S, Moreadith RW, Laroche Y, Collen D. Stem Cells. 2003;21:90–97. doi: 10.1634/stemcells.21-1-90. [DOI] [PubMed] [Google Scholar]
- 28.Zvetkova I, Apedaile A, Ramsahoye B, Mermoud JE, Crompton LA, John R, Feil R, Brockdorff N. Nat Genet. 2005;37:1274–1279. doi: 10.1038/ng1663. [DOI] [PubMed] [Google Scholar]
- 29.Eakin GS, Hadjantonakis AK, Papaioannou VE, Behringer RR. Dev Biol. 2005;288:150–159. doi: 10.1016/j.ydbio.2005.09.028. [DOI] [PubMed] [Google Scholar]
- 30.Friedrich G, Soriano P. Genes Dev. 1991;5:1513–1523. doi: 10.1101/gad.5.9.1513. [DOI] [PubMed] [Google Scholar]
- 31.Szybalski W. BioEssays. 1992;14:495–500. doi: 10.1002/bies.950140712. [DOI] [PubMed] [Google Scholar]
- 32.Araki K, Araki M, Yamamura K. Nucleic Acids Res. 2002;30:e103. doi: 10.1093/nar/gnf102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seibler J, Schubeler D, Fiering S, Groudine M, Bode J. Biochemistry. 1998;37:6229–6234. doi: 10.1021/bi980288t. [DOI] [PubMed] [Google Scholar]
- 34.Belteki G, Gertsenstein M, Ow DW, Nagy A. Nat Biotechnol. 2003;21:321–324. doi: 10.1038/nbt787. [DOI] [PubMed] [Google Scholar]
- 35.Kunath T, Gish G, Lickert H, Jones N, Pawson T, Rossant J. Nat Biotechnol. 2003;21:559–561. doi: 10.1038/nbt813. [DOI] [PubMed] [Google Scholar]
- 36.te Riele H, Maandag ER, Clarke A, Hooper M, Berns A. Nature. 1990;348:649–651. doi: 10.1038/348649a0. [DOI] [PubMed] [Google Scholar]
- 37.Lefebvre L, Dionne N, Karaskova J, Squire JA, Nagy A. Nat Genet. 2001;27:257–258. doi: 10.1038/85808. [DOI] [PubMed] [Google Scholar]
- 38.Lin Q, Donahue SL, Moore-Jarrett T, Cao S, Osipovich AB, Ruley HE. Nucleic Acids Res. 2006;34:e139. doi: 10.1093/nar/gkl728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nord AS, Chang PJ, Conklin BR, Cox AV, Harper CA, Hicks GG, Huang CC, Johns SJ, Kawamoto M, Liu S, et al. Nucleic Acids Res. 2006;34:D642–D648. doi: 10.1093/nar/gkj097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lobe CG, Koop KE, Kreppner W, Lomeli H, Gertsenstein M, Nagy A. Dev Biol. 1999;208:281–292. doi: 10.1006/dbio.1999.9209. [DOI] [PubMed] [Google Scholar]
- 41.Mereau A, Grey L, Piquet-Pellorce C, Heath JK. J Cell Biol. 1993;122:713–719. doi: 10.1083/jcb.122.3.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hadjantonakis AK, Gertsenstein M, Ikawa M, Okabe M, Nagy A. Mech Dev. 1998;76:79–90. doi: 10.1016/s0925-4773(98)00093-8. [DOI] [PubMed] [Google Scholar]
- 43.Nagy A, Gersenstein M, Vintersten K, Behringer R. Manipulating the Mouse Embryo: A Laboratory Manual. 3rd Ed. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 2003. [Google Scholar]
- 44.Haigh JJ, Gerber HP, Ferrara N, Wagner EF. Development (Cambridge, UK) 2000;127:1445–1453. doi: 10.1242/dev.127.7.1445. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.