Abstract
The MDM2 protein is an ubiquitin ligase that plays a critical role in regulating the levels and activity of the p53 protein, which is a central tumor suppressor. A SNP in the human MDM2 gene (SNP309 T/G) occurs at frequencies dependent on demographic history and has been shown to have important differential effects on the activity of the MDM2 and p53 proteins and to associate with altered risk for the development of several cancers. In this report, the haplotype structure of the MDM2 gene is determined by using 14 different SNPs across the gene from three different population samples: Caucasians, African Americans, and the Ashkenazi Jewish ethnic group. The results presented in this report indicate that there is a substantially reduced variability of the deleterious SNP309 G allele haplotype in all three populations studied, whereas multiple common T allele haplotypes were found in all three populations. This observation, coupled with the relatively high frequency of the G allele haplotype in both and Caucasian and Ashkenazi Jewish population data sets, suggests that this haplotype could have undergone a recent positive selection sweep. An entropy-based selection test is presented that explicitly takes into account the correlations between different SNPs, and the analysis of MDM2 reveals a significant departure from the standard assumptions of selective neutrality.
Keywords: cancer, p53, population genetics, SNP, entropy
In response to a wide variety of stresses, such as DNA damage or oncogene activation, the p53 tumor suppressor protein is activated and initiates a transcriptional program leading to cell cycle arrest, cell senescence or apoptosis (1). This eliminates clones of cells that have acquired mutations, which arise at a high frequency when DNA replication or the cell cycle proceeds under stress. When the p53 gene is mutated in either the germ line or in a somatic cell, many types of cancers can arise (2). The p53 protein is regulated by a ubiquitin ligase, the MDM2 protein, which binds to p53, blocking its function as a transcription factor, and polyubiquitinates the p53 protein sending it to the proteiosome for degradation (3). The MDM2 gene in turn is positively regulated by p53-mediated transcription, setting up an autoregulatory loop that keeps both proteins at moderate levels. Stress responses perturb this feedback loop, which leads to the initiation of p53-dependent apoptosis.
Functional SNPs in the human genome have been identified in both the p53 and the MDM2 genes (4). In the p53 gene, a SNP (codon 72) results in the change of a proline residue to an arginine at codon 72 of the p53 protein (p53-Pro and p53-Arg, respectively). Multiple groups have shown that p53-Pro is weaker than p53-Arg in its ability to both suppress cellular transformation and induce apoptosis in cell culture (5–8), and can associate with an earlier onset of tumor formation and a poorer tumor response to chemotherapy in humans (7, 9, 10). In the MDM2 gene, a SNP (SNP309) results in a nucleotide change from the wild-type thymine (T) to guanine (G) in the intronic promoter/enhancer region (11). The G allele increases the binding of a transcription factor, SP1, which in turn results in higher levels of MDM2 RNA and protein, the attenuation of the p53 pathway and an enhanced early onset of, and increased risk for, tumorigenesis (11–22). More recent studies of MDM2 SNP309 suggest that primarily female specific hormones, like estrogen, either directly or indirectly, allow for the G allele of SNP309 to accelerate tumor formation in women in four different sporadic cancers (diffuse large B cell lymphoma, soft tissue sarcoma, invasive ductal breast carcinoma and colorectal cancer) (14, 23). Together, these data suggest that functional SNPs in the p53 pathway will play a role in regulating the efficiency of the p53 stress response over a lifetime, and as such the efficacy of the p53 pathway in tumor suppression after exposure to stresses.
To date all of the associations of the G allele of MDM2 SNP309 with the early onset of cancers in patients have been linked to this locus alone and little is known of the haplotypes that contain the G or the T alleles at SNP309. It remains possible that one or several G haplotypes will be associated with early onset of cancer, and it is important to determine this at an early stage in the genetic epidemiological studies of this allele. For that reason, the haplotype structure of 14 different SNPs in the MDM-2 gene was determined by employing three different racial and ethnic populations: one African American population, one Caucasian not selected for ethnicity, and one Caucasian of the Ashkenazi Jewish ethnic group. These populations were chosen for further haplotype analysis, as it had been previously observed that African Americans have a low G allele frequency, non-Jewish Caucasians an intermediate G allele frequency, and Ashkenazi Jewish groups a high G allele frequency (14, 24).
The results presented in this report indicate that there are only a few common (≥1%) G allele haplotypes in all three populations studied, one in the African American and Caucasian data sets and two in the Ashkenazi Jewish data set. The SNPs in the G allele haplotype are thus highly correlated. We suggest that the single G allele haplotype in the African American population could have arisen through admixture with other racial groups, like Caucasians, and thus is a relatively recent mutation. In contrast, multiple common T allele haplotypes were found in all three populations, thus exhibiting reduced correlations between the SNPs. This observation coupled with the relatively high frequency of the G allele haplotype in both Caucasian populations suggests that this haplotype could have experienced recent positive selection pressure. To test this hypothesis, an entropy-based selection test is devised that compares both the frequency and long-range correlations of the allele with a simulated model where the allele is selectively neutral. The results confirm that the probability that the G allele haplotype is selectively neutral is quite low.
Results
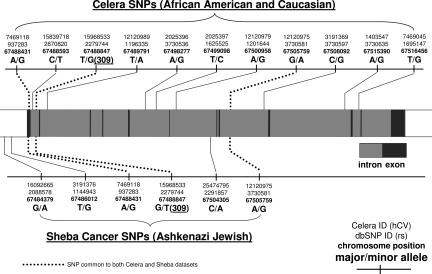
The MDM2 SNPs genotyped from various populations in this study are depicted in Fig. 1, and their frequencies are detailed in Tables 1 and 2. The observed genotype frequencies of the MDM2 SNPs were found not to deviate significantly from Hardy–Weinberg equilibrium within each race and ethnic group, with P values ranging from 0.08 to 1.00. As expected, the assumption of Hardy–Weinberg equilibrium was found to be notably violated when the populations were pooled together. The frequencies of the SNPs common to both studies are, on average, most similar between the Caucasian and Ashkenazi Jewish data sets. However, because the absolute frequencies of the common SNPs across the Caucasian and Ashkenazi Jewish samples do not match exactly, there is no way of combining the haplotypes without incurring severe biases.
Fig. 1.
Schematic diagram of the MDM2 gene and the SNPs genotyped in the present study. The three SNPs common to both the Celera and Sheba data sets are indicated by the dotted lines.
Table 1.
Major allele frequencies of the 11 SNPs genotyped in the Celera study
| SNP ID | dbSNP ID | Major/minor allele | Frequency |
|
|---|---|---|---|---|
| African American | Caucasian | |||
| Cel_1 | rs937283 | A/G | 0.74 | 0.66 |
| Cel_2 | rs2870820 | C/T | 0.94 | 0.66 |
| Cel_3 | rs2279744 | T/G | 0.90 | 0.57 |
| Cel_4 | rs1196335 | T/A | 0.77 | 0.66 |
| Cel_5 | rs3730536 | A/G | 0.90 | 0.57 |
| Cel_6 | rs1625525 | T/C | 0.78 | 0.66 |
| Cel_7 | rs1201644 | A/G | 0.73 | 0.63 |
| Cel_8 | rs3730581 | G/A | 0.90 | 0.57 |
| Cel_9 | rs3730597 | C/G | 1.00 | 0.99 |
| Cel_10 | rs3730635 | A/G | 0.82 | 0.97 |
| Cel_11 | rs1695147 | T/G | 0.41 | 0.80 |
The labeling of major and minor allele was determined by pooling both the African American and Caucasian population samples.
Table 2.
Major allele frequencies of the six SNPs genotyped in the Sheba study
| SNP ID | dbSNP ID | Major/minor allele | Ashkenazi Jewish frequency |
|---|---|---|---|
| She_1 | rs2088578 | G/A | 0.66 |
| She_2 | rs1144943 | T/G | 0.73 |
| She_3 | rs937283 | A/G | 0.74 |
| She_4 | rs2279744 | G/T | 0.54 |
| She_5 | rs2291857 | C/A | 0.63 |
| She_6 | rs3730581 | A/G | 0.57 |
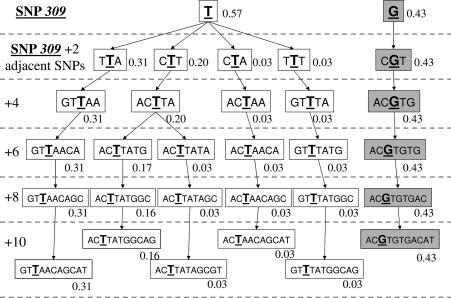
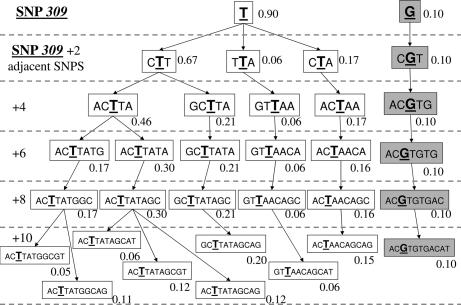
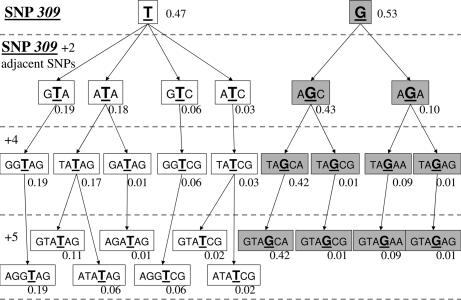
All haplotypes with an expected frequency of at least 1% are presented in Figs. 2, 3, and 4 for Caucasians, African Americans, and Ashkenazi Jewish data sets, respectively. The marginal haplotype frequencies inferred from this calculation are shown in the figures in a SNP309-centric fashion, adding additional SNPs about SNP309 two at a time so as to permit comparisons of subhaplotypes.
Fig. 2.
Inferred haplotype frequencies in the Caucasian population.
Fig. 3.
Inferred haplotype frequencies in the African American population.
Fig. 4.
Inferred haplotype frequencies in the Ashkenazi Jewish population.
Previously reported SNP309 G allele frequencies in Northern European Caucasians were ≈33%, but in African Americans they have been noted to be significantly lower, ≈11% (14, 24, 25). A salient feature of the African Americans and Caucasians figures is that, in both populations, the G allele of SNP309 is highly correlated with all of the other SNPs across the entire region of MDM2 covered by the Celera Diagnostic SNP set, resulting in only one G haplotype in both populations. The G allele haplotype is identical in both races, just at a much lower frequency in African Americans (43% vs. 10%), possibly suggesting that its presence in African Americans could be due to an admixture (25). If true, this would lead to the prediction that Africans probably do not carry the G allele of SNP309, which then posits the idea that the G allele could have arisen more recently in evolution.
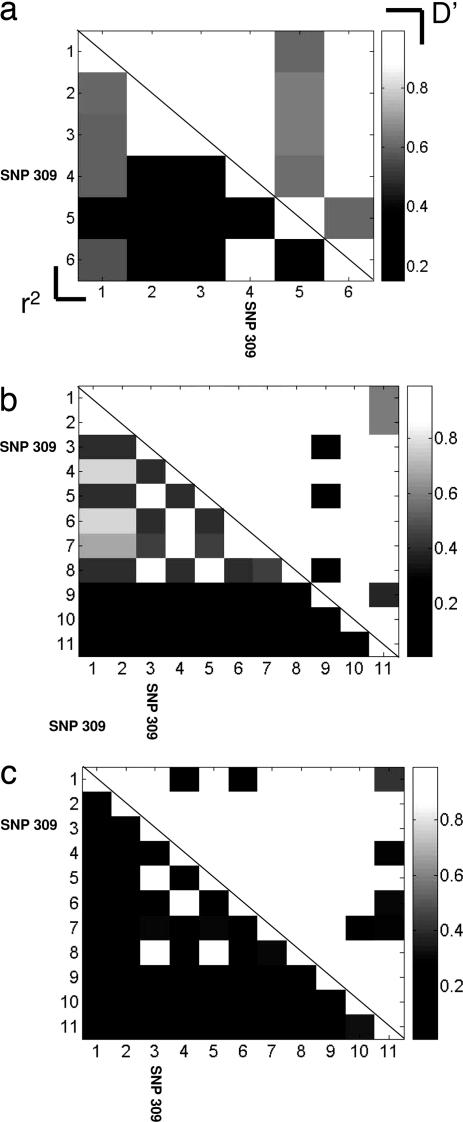
Evidence of recombination was determined by the four-gamete test, serving as a guide to the minimum number of recombination events consistent with the data. Of the possible 55 pairs, there were seven positive tests in both the African American and Caucasians populations, and five out of 15 in the Jewish ethnic population. To directly ascertain the correlations among SNPs of the MDM2 gene in the differing population data sets, the linkage disequilibrium of the MDM2 SNPs was estimated by calculating the standard pairwise values of D′ and r2 (Fig. 5). The linkage disequilibrium estimation analysis of the MDM2 SNPs revealed that the SNP309 locus is absolutely correlated with two other MDM2 SNPs, Cel_5 and Cel_8. These three SNPs seem to always travel together in a haplotype, and thus collectively they represent only one degree of freedom in genetic variation. Hence, there is no detectable simple single mutation precursor of the G allele haplotype of SNP309 amongst the T allele haplotypes in both populations.
Fig. 5.
Pairwise linkage disequilibrium for Ashkenazi Jewish (a), Caucasian (b), and African American (c) populations. The upper right triangle reports the |D′| measure, and the lower left triangle reports the r2 measure. SNP309 is highlighted in all populations.
Independent evolution of subpopulations results in reduced heterozygosity of the total population as detected by Wright's F statistic. For the Caucasian and African American populations, we find that FST = 0.17, mirroring the observation from the allelic Hardy–Weinberg tests that mating was essentially random within each race but not between the races. The data across the races was permuted 110 times and the calculated FST statistic indicated that P < 10−5 (26). The average number of pairwise distances between the two races was Π = 4.3 with P < 10−5.
Although the G allele occurs at intermediate frequencies, it is a striking observation that the number of different G allele haplotypes across the entire gene is dramatically reduced compared with the number of T allele haplotypes even though the T allele frequency is also intermediate in the non-African-American populations. How significant then is the paucity of the G allele haplotypes and what biological implications, if any, does it suggest?
To quantify the total variation of haplotypes within each population for an arbitrary number of SNPs, we appealed to information theory (27) and estimated the entropy, which serves as a unique measure of variability under a few, but very general, mathematical assumptions. To be more specific, we calculated the multientropy H[{X}] from the observed haplotypes arising from a set, {X}, of b SNPs, H[{X}] = −Σi=12bpi(x1x2… xb)log2 pi(x1x2… xb), where each random variable xi denotes one of the two alleles of SNP i. Concavity of the entropy function results, on average, in a negative sampling bias, which can be corrected by using a bootstrap resampling procedure to extrapolate to the infinite sample size (28). Under the standard simple assumptions of a large panmictic population the variability of a stretch of the genome increases with time or, more accurately, number of generations, because of increased probability of recombination and mutation. The total entropy of the African American and Caucasian haplotypes in the Celera data set was calculated to be 3.0 bits and 2.05 bits, respectively, supporting the idea that the Caucasians are a much more recent interbreeding population than the Africans.
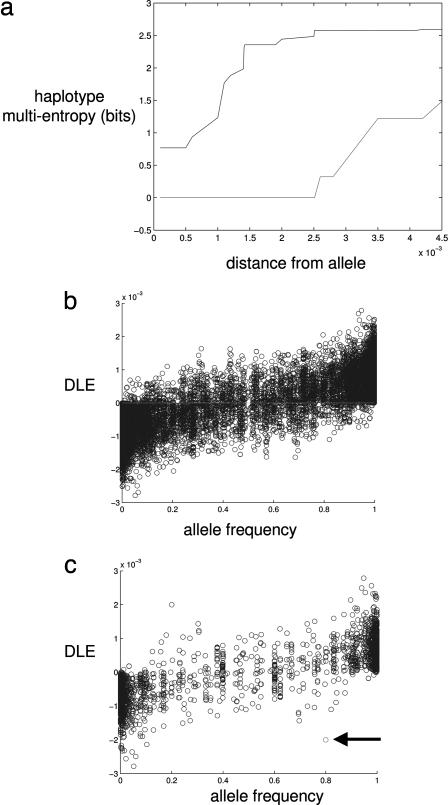
A large level of linkage disequilibrium across the gene entails a low level of multientropy. It is expected from simple considerations of a neutral model of a population of mutating and recombining chromosomes that low frequency (younger) alleles ought to occur on fewer haplotypes than higher frequency (older) alleles, and thus will be in strong linkage disequilibrium across relatively large stretches of the genome. To capture this intuition, we suggest that, under the assumptions of a neutral model, there ought to be a monotonically increasing relationship between the frequency of the allele and the mean entropy of the associated haplotypes. To show this, we generated samples of polymorphic data from a Monte Carlo simulation within a coalescent framework of neutral mutation and homogeneous recombination (29). In Fig. 6a, we show a plot of the entropy of haplotypes around a particular SNP with distance (base pairs) away from the SNP. For low-frequency alleles, the entropy of associated haplotypes rises slowly with distance away from the allele because low-frequency alleles are usually recently occurring mutations that have not had sufficient time to attain linkage equilibrium with surrounding SNPs. Higher frequency alleles, as expected, exhibit a much greater increase in linkage equilibrium and entropy. To make comparisons between the alleles of a particular SNP, we summarize the linkage equilibrium for each allele by calculating the distance-averaged entropy up to some cutoff distance away from the SNP. The cutoff in our study here has a natural upper bound due to the limited number of SNPs genotyped across the genetic region of interest. More generally, a natural cutoff is given by the accuracy by which we can calculate the entropy, and a simple large-sample calculation of the first-order error in entropy estimation shows that the criteria for accurate entropy estimation is a function of N, the number of samples, and b, the number of SNPs, (i.e., 2bN−1 <1). Decreasing the cutoff arbitrarily results in greater variability of the distribution of linkage equilibrium for a given allele and, conversely, increasing the cutoff compromises the accuracy of entropy, and thus there ought to be an optimal intermediate cutoff where both effects are mitigated.
Fig. 6.
Monte Carlo simulation of haplotypes. (a) The conditional multientropy of the adjacent SNPs is plotted for a particular locus where the minor allele (lower curve) had a frequency of 0.19 and the major allele (upper curve) had a frequency of 0.81. The unit of distance is measured in rescaled base pairs. (b) Plot of the DLE values versus frequency of each allele. One thousand Monte Carlo genome samples of size 250,000 base pairs were generated. The mutation rate was set to produce 8,000 SNPs, and the recombination rate was set to 1 cM Mb−1. (c) Selected allele (indicated by arrow) underwent a selective advantage per copy per generation of 0.1
By comparing the differences of average linkage equilibrium for different alleles at a particular SNP, we are able to obviate the systematic error that occurs due to variable recombination rates across the genome. Thus, the summary statistic of the differential mean linkage equilibrium (DLE) for an allele A is given by
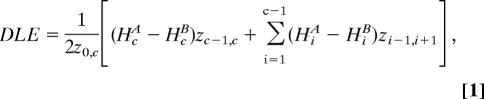 |
where the index B refers to the other allele at the locus, c is the number of SNPs before the cutoff, zi,j is the base pair distance between SNPs situated at loci i and j, and HiA,B is the multientropy for i SNPs extending away from a particular locus, conditional on either allele A or B at locus i = 0. In Fig. 6b, we summarize the data of allele frequency and DLE showing the expected relationship under the neutral model, whereby high-frequency alleles have positive DLE and low-frequency alleles have negative DLE. Any deviation from this would be due to a relaxation of the standard assumptions of a neutral model. An allele with a large frequency but low DLE is suggestive of a selection pressure that has rapidly pushed up the frequency of the allele giving it insufficient time to approach equilibration with surrounding SNPs. As proof of principle, we then generated a Monte Carlo sample of polymorphic data where one allele was preassigned to have a weak positive selection pressure (selective advantage per copy per generation = 0.1) (30). The summary plot of the DLE values for all SNPs (Fig. 6c) shows that the selected allele does indeed significantly deviate from the mean of the null distribution. Indeed, recent work (31, 32) has provided supportive evidence that such haplotype-based methods are far more sensitive to recent selection detection. The P value was estimated by Monte Carlo simulation of a neutral model (31) with 106 trials to provide a fine-scale distribution of neutral DLE for a given allele frequency where the frequencies were quantized into steps of 0.1.
Previous tests of selection in the population genetics literature also look for deviations away from the neutrality but do not incorporate information from multiallelic associations even though selection pressure can have a strong effect on levels of linkage disequilibrium, and thus these tests have low power to detect recent selective sweeps (32). Nevertheless, we used a variety of standard selection tests on the population data sets, and the results, as detailed in Table 3, are inconsistent with rejecting the null hypothesis of a neutral model, with each population data set showing significant departure or not from neutrality depending on which selection test is used. Our proposed test of neutrality does not rest on an assumption of independent alleles but is explicitly based on the variability of haplotypes and hence also linkage disequilibrium.
Table 3.
Results of various selection tests
| Population data set | Fu and Li's F | Fu and Li's D | Tajima's D |
|---|---|---|---|
| Caucasian | 1.59 (>0.05) | 0.80 (>0.10) | 2.35 (<0.05) |
| African American | 1.62 (>0.05) | 1.36 (>0.10) | 1.37 (>0.10) |
| Ashkenazi Jewish | 2.24 (<0.02) | 1.04 (>0.10) | 3.44 (<0.01) |
P values are given in parentheses.
When we calculated the frequency versus DLE relationship for the MDM2 SNPs, we found that the G allele of SNP309, along with the minor alleles of SNPs Cel_5 and Cel_8, did indeed show evidence of significant deviation away from the frequency versus DLE relationship determined by the other alleles in the study. To assess significance, we ran neutral simulations with matching number of SNPs, constructed with the inferred average recombination rate parameter of 0.43 cm/Mb−1 (from PHASE v2.1) across the MDM2 gene, and found that the (Bonferroni-corrected) P value was <0.01 for the Caucasians not selected for ethnicity and 0.03 for the Ashkenazi Jewish Caucasians. Another haplotype-based method designed to detect recent selection (31) was used and found that the deviation from neutral simulation results were also significant in both Caucasian populations with P < 0.04. The data suggest that only the G allele haplotype has been pushed up in frequency because of a recent positive selection pressure.
Discussion
It had been shown that MDM2 SNP309 occurs at race- and ethnicity-specific frequencies (14, 24). In this report, these observations were extended to the haplotype structure of this important oncogene. Specifically, in the Caucasian population not selected for ethnicity and in the African American population, only one major haplotype for the G allele of MDM2 SNP309 was observed (Figs. 2 and 3). This observation, plus the low frequency of the G allele haplotype in African Americans (43% in Caucasians vs. 10% in African Americans), supports the idea that the G haplotype could have arisen in this racial group from admixture with Caucasians. This finding allows for the possibility that the T to G change could have occurred more recently in human evolution, namely after the initial migrations of humans out of Africa. It is intriguing to speculate whether the two common G haplotypes in the Ashkenazi Jewish data set are unique to this ethnic Caucasian group, but is impossible to know as the analysis of the two Caucasian populations differed in the MDM2 SNPs genotyped.
Interestingly, one common trait of all three populations in this study was the large number of common (>3%) SNP309 T allele haplotypes compared with the small number of G allele haplotypes. Specifically, anywhere from two common T allele haplotypes were found in the Caucasian population not selected for ethnicity (Fig. 2), to eight in African Americans (Fig. 3), to four in the Ashkenazi Jewish population (Fig. 4). This observation, coupled with the relatively high frequency of the G allele haplotype in the Caucasian populations, suggests that this haplotype could have experienced positive selection pressure. This hypothesis was tested by developing a statistical model of the predicted entropy of haplotypes under the assumption of neutral polymorphisms (Fig. 6). Indeed, the results demonstrate that the G allele haplotype deviates significantly from the neutral model in both the Caucasian population data set and the Ashkenazi Jewish population data set (P = 0.01 and P = 0.03, respectively). Similar results were obtained by using previously published haplotype-based method of detecting selection (31). Together, these data support the hypothesis that the G haplotype may have undergone a recent selective sweep.
Another effect that would also result in the loss of haplotype variability while pushing up the G allele frequency is a founder effect in which the SNP309 T to G mutation occurred late in the parent population. However, this would affect the DLE of all of the SNPs in the founding population, not just SNP309, which is not what we observe. On a related matter, we also point out that the selection arguments drawn here, because of the limited number of SNPs in the study, are contingent on rejecting a specified null hypothesis, constructed from simulating the standard neutral model. Future work may provide genotypes of a much larger region of the genome around MDM2, which would allow us to compare the relative selective pressure of SNP309 with the rest of the genome, assumed to be mostly neutral, and not just with a simulated neutral model (32).
Curiously, there seems to be no observable simple one step mutation to derive the G haplotypes from any of the T allele containing haplotypes. Rather, the G allele is in high linkage disequilibrium with two other MDM2 SNPs (Cel-5 and Cel-8, Fig. 5), suggesting that these three SNPs arose in a narrow window of time on a common haplotype background.
An interesting model for positive selection of the functional SNP in the p53 gene, codon 72, has been previously proposed to explain the observation that the p53–Pro isoform increases in a monotonic manner in multiple populations as they near the equator (in Africa) (33). As mentioned earlier, the p53–Pro isoform is weaker than p53–Arg in its ability to both suppress cellular transformation and induce apoptosis in cell culture (5–8), and can associate with an earlier onset of tumor formation and a poorer tumor response to chemotherapy in humans (7, 9, 10). The data presented in this and previous reports suggest that Africans will have a very low frequency, if not zero, of the G allele of SNP309, whereas Caucasians and Asians will a much higher G allele frequency (14, 16, 21, 22, 24, 33). Because the G allele of SNP309 has a lower apoptotic frequency than the T allele in some cell types in culture (11, 13), it is tempting to speculate that the presence of the G allele could compensate for higher apoptotic frequencies brought about by the p53–Arg isoform as populations move to northern Asia and Europe. A well regulated p53 pathway has been shown in many organisms to be crucial not only for tumor suppression but also for proper embryonic development (34, 35); therefore, selection pressure on the p53 pathway could be possible. In fact, the p53 pathway also responds to inflammation, raising the possibility that infectious diseases could play a role in the selection of p53 pathway SNPs, like MDM2 SNP309 and codon 72.
Materials and Methods
Sample Populations.
Two separate genotyping assays were carried out for the MDM2 gene, one performed by Celera Diagnostics, and the other performed by the Sheba Cancer Research Center. These two studies assayed a different set of SNPs in the differing populations. A combined total of 14 different SNPs were typed across a region spanning >32,000 bp, including a region 4,000 bp upstream from the first exon (see Fig. 1 and Tables 1 and 2). Both studies included SNP309 (identified as Cel_3 and She_4 in the two data sets).
Celera Diagnostics.
Sample population.
Eleven SNPs were genotyped from a collection of 113 lymphoblastoid cell lines from Caucasian and African American individuals obtained from the Coriell Diversity Set of cell lines that had been previously analyzed for their frequencies of apoptosis and their associated SNPs (24). Subsequent to the publication of those results it was determined, and it was confirmed by the Coriell Institute, that 22 of the cell lines used (each with a different number) in that study were duplicates derived from the same individuals even though this is not indicated in their information. These duplicate cell lines are presented in supporting information (SI) Tables 4 and 5. It was also determined that 32 of the SNP309 MDM2 genotypic assignments made in Harris et al. were incorrect. These genotypes are now corrected and presented in SI Table 6. Because of these changes in the data set, the MDM2 SNP309 is not very significantly associated with a low apoptotic frequency in these cell lines (P = 0.15) as had been reported (24). This is probably due to the observation that an active estrogen-signaling pathway is not present in these cells. High levels of the AKT-1 SNPs (3 and 4) were only weakly associated with a low apoptotic frequency (P = 0.07). The remainder of the observations and figures from these publications remain correct. Because of these duplications in cell lines, the DNA samples were obtained from 91 unrelated individuals, males and females, of which 38 were self-reported Caucasians and 53 were self-reported African Americans. The allele frequencies for this group are presented in Table 1. SNP Cel-9 was monomorphic in the African American cohort and was thus eliminated in the detailed computational analysis of this population. Both races agreed on the major allele at each locus except SNP Cel_11.
MDM2 SNPs and genotyping.
Genotyping of SNPs was performed by allele-specific real-time PCR for individual samples using primers designed and validated in-house (25). Previous analyses showed that the accuracy of our genotyping is better than 99%, as determined by internal comparisons of differentially designed assays for the same marker and comparisons for the same marker across different groups (36).
Sheba Medical Center.
Sample population.
Study participants were all of Jewish Ashkenazi origin previously counseled and tested at the Oncogenic unit, Sheba Medical Center, Tel Hashomer, Israel, because of a family history of breast and/or ovarian cancer. The study was approved by the institutional review boards (Helsinki committees) at Sheba Medical Center and the national IRB for genetics. A control population made up of healthy Ashkenazi women (n = 139) with no family history or personal history of cancer and no BRCA1/2 mutations were accrued through the genetic center at the Sheba Medical Center. These women were referred for genetic testing for nonneoplastic conditions and gave their written informed consent for anonymous testing. To prevent ascertainment bias of the haplotype inferences, we used only the control group in the analyses; thus, it is assumed that the data set in this paper, labeled as the Askenazi Jewish population, is a random sample from the representative (non-cancer risk) population.
DNA preparation.
Genomic DNA was prepared from anticogulated, venous blood samples by using the PUREGene DNA isolation kit (Gentra Systems, Minneapolis, MN) using the protocol recommended by the manufacturer.
MDM2 SNPs and genotyping.
Six SNPs were genotyped in a region in and adjacent to the MDM-2 SNP309 locus spanning 4,000 bp upstream to the eighth intron (Fig. 1).
SNPs for genotyping in the MDM-2 gene were selected from three different databases; www.ensembl.org, www.broad.mit.edu/mpg/haploview, and dbsnp (www.ncbi.nlm.nih.gov/snp) based on their location in the MDM-2 gene. SNP genotyping was performed in 384-well microplates with a high-throughout system of chip-based mass spectrometry (MALDI-TOF) (Sequenomx, San Diego, CA). The allele determination in the sampled DNA was based on MALDI-TOF mass spectrometry of allele-specific primer products. Genotyping assays were designed as multiplex reactions using SpectroDESIGNER software version 2.0.7 (Sequenom).
PCR Primer Design, Amplification, and Primer Extension.
The detailed PCR and primer extension reactions were performed according to the protocol for high multiplex homogeneous MassEXTEND (hME) procedure (Sequenom application notes), as follows. PCR primers were tagged with 5′-ACGTTGGATG-3′ at the 5′ end to avoid interference with the mass spectra. Amplification of 2.5 ng of cDNA was performed in 2.75 mM MgCl2/200 μM dNTP/0.1 unit of HotStart TaqDNA polymerase (Qiagen) and 1 pmol of each forward and reverse PCR primers in 5-μl total volume. PCR conditions: 95°C 15 min, followed by 45 cycles of 95°C for 30 s, 56°C for 1 min, then 72°C for 1:30 min, with an extension at 72°C for 7 min. Products were then treated with 0.04 unit of shrimp alkaline phosphatase (SAP) (Sequenom) followed by extension cycle, to which 1.2 μM final concentration of extension primer and 0.6 unit of ThermoSequenase (Sequenom) were added to a total reaction of 9 μl with the termination mixture. The extension conditions include a 94°C 2 min with 75 cycles of the following: 94°C for 5 s, 52°C for 5 s and 72°C for 5 s. Quality control and quality assurance were provided by randomly including non-DNA containing well in the chip as well as regenotyping ≈10% of samples for all SNPs on different chips. The error rate of reproducibility due to missed calls was 3.4%.
Haplotying.
The predictions of haplotypes from the genotyping, carried out with the three different populations, were calculated employing the Bayesian algorithm known as PHASE, version 2.1 (37). Note that the limited number of samples in both the Celera and Sheba studies preclude discovery of rare haplotypes. To test Hardy–Weinberg equilibrium of the genotypes, stratified by population, an exact test based on the Markov chain algorithm of Guo and Thompson (38) was used, as implemented by Excoffier et al. (26). This method compares the observed heterozygosity to the expected heterozygosity in each population under study.
Supplementary Material
Abbreviation
- DLE
differential mean linkage equilibrium.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0610998104/DC1.
References
- 1.Jin S, Levine AJ. J Cell Sci. 2001;114:4139–4140. doi: 10.1242/jcs.114.23.4139. [DOI] [PubMed] [Google Scholar]
- 2.Lain S, Lane D. Eur J Cancer. 2003;39:1053–1060. doi: 10.1016/s0959-8049(03)00063-7. [DOI] [PubMed] [Google Scholar]
- 3.Bond GL, Hu W, Levine AJ. Curr Cancer Drug Targets. 2005;5:3–8. doi: 10.2174/1568009053332627. [DOI] [PubMed] [Google Scholar]
- 4.Murphy ME. Cell Death Differ. 2006;13:916–920. doi: 10.1038/sj.cdd.4401907. [DOI] [PubMed] [Google Scholar]
- 5.Bonafe M, Salvioli S, Barbi C, Mishto M, Trapassi C, Gemelli C, Storci G, Olivieri F, Monti D, Franceschi C. Biochem Biophys Res Commun. 2002;299:539–541. doi: 10.1016/s0006-291x(02)02691-8. [DOI] [PubMed] [Google Scholar]
- 6.Dumont P, Leu JI, Della Pietra AC, III, George DL, Murphy M. Nat Genet. 2003;33:357–365. doi: 10.1038/ng1093. [DOI] [PubMed] [Google Scholar]
- 7.Sullivan A, Syed N, Gasco M, Bergamaschi D, Trigiante G, Attard M, Hiller L, Farrell PJ, Smith P, Lu X, Crook T. Oncogene. 2004;23:3328–3337. doi: 10.1038/sj.onc.1207428. [DOI] [PubMed] [Google Scholar]
- 8.Thomas M, Kalita A, Labrecque S, Pim D, Banks L, Matlashewski G. Mol Cell Biol. 1999;19:1092–1100. doi: 10.1128/mcb.19.2.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones JS, Chi X, Gu X, Lynch PM, Amos CI, Frazier ML. Clin Cancer Res. 2004;10:5845–5849. doi: 10.1158/1078-0432.CCR-03-0590. [DOI] [PubMed] [Google Scholar]
- 10.Shen H, Zheng Y, Sturgis EM, Spitz MR, Wei Q. Cancer Lett. 2002;183:123–130. doi: 10.1016/s0304-3835(02)00117-9. [DOI] [PubMed] [Google Scholar]
- 11.Bond GL, Hu W, Bond EE, Robins H, Lutzker SG, Arva NC, Bargonetti J, Bartel F, Taubert H, Wuerl P, et al. Cell. 2004;119:591–602. doi: 10.1016/j.cell.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 12.Alhopuro P, Ylisaukko-Oja SK, Koskinen WJ, Bono P, Arola J, Jarvinen HJ, Mecklin JP, Atula T, Kontio R, Makitie AA, et al. J Med Genet. 2005;42:694–698. doi: 10.1136/jmg.2005.031260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arva NC, Gopen TR, Talbott KE, Campbell LE, Chicas A, White DE, Bond GL, Levine AJ, Bargonetti J. J Biol Chem. 2005;280:26776–26787. doi: 10.1074/jbc.M505203200. [DOI] [PubMed] [Google Scholar]
- 14.Bond GL, Hirshfield KM, Kirchhoff T, Alexe G, Bond EE, Robins H, Bartel F, Taubert H, Wuerl P, Hait W, et al. Cancer Res. 2006;66:5104–5110. doi: 10.1158/0008-5472.CAN-06-0180. [DOI] [PubMed] [Google Scholar]
- 15.Bougeard G, Baert-Desurmont S, Tournier I, Vasseur S, Martin C, Brugieres L, Chompret A, Bressac-de Paillerets B, Stoppa-Lyonnet D, Bonaiti-Pellie C, Frebourg T. J Med Genet. 2005;43:531–533. doi: 10.1136/jmg.2005.037952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hong Y, Miao X, Zhang X, Ding F, Luo A, Guo Y, Tan W, Liu Z, Lin D. Cancer Res. 2005;65:9582–9587. doi: 10.1158/0008-5472.CAN-05-1460. [DOI] [PubMed] [Google Scholar]
- 17.Lind H, Zienolddiny S, Ekstrom PO, Skaug V, Haugen A. Int J Cancer. 2006;119:718–721. doi: 10.1002/ijc.21872. [DOI] [PubMed] [Google Scholar]
- 18.Menin C, Scaini MC, De Salvo GL, Biscuola M, Quaggio M, Esposito G, Belluco C, Montagna M, Agata S, D'Andrea E, et al. J Natl Cancer Inst. 2006;98:285–288. doi: 10.1093/jnci/djj054. [DOI] [PubMed] [Google Scholar]
- 19.Swinney RM, Hsu SC, Hirschman BA, Chen TT, Tomlinson GE. Leukemia. 2005;19:1996–1998. doi: 10.1038/sj.leu.2403941. [DOI] [PubMed] [Google Scholar]
- 20.Zhang X, Miao X, Guo Y, Tan W, Zhou Y, Sun T, Wang Y, Lin D. Hum Mutat. 2005;27:110–117. doi: 10.1002/humu.20277. [DOI] [PubMed] [Google Scholar]
- 21.Dharel N, Kato N, Muroyama R, Moriyama M, Shao RX, Kawabe T, Omata M. Clin Cancer Res. 2006;12:4867–4871. doi: 10.1158/1078-0432.CCR-06-0111. [DOI] [PubMed] [Google Scholar]
- 22.Park SH, Choi JE, Kim EJ, Jang JS, Han HS, Lee WK, Kang YM, Park JY. Lung Cancer. 2006;54:19–24. doi: 10.1016/j.lungcan.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 23.Bond GL, Menin C, Bertorelle R, Alhorpuro P, Aaltonen LA, Levine AJ. J Med Genet. 2006;43:950–952. doi: 10.1136/jmg.2006.043539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harris SL, Gil G, Robins H, Hu W, Hirshfield K, Bond E, Bond G, Levine AJ. Proc Natl Acad Sci USA. 2005;102:16297–16302. doi: 10.1073/pnas.0508390102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Millikan RC, Heard K, Winkel S, Hill EJ, Heard K, Massa B, Mayes L, Williams P, Holston R, Conway K, Edmiston S, de Cotret AR. Cancer Epidemiol Biomarkers Prev. 2006;15:175–177. doi: 10.1158/1055-9965.EPI-05-0692. [DOI] [PubMed] [Google Scholar]
- 26.Excoffier L, Laval G, Schneider S. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- 27.Cover TM, Thomas JA. Elements of Information Theory. New York: Wiley; 1991. [Google Scholar]
- 28.Strong SP, Koberle R, de Ruyter van Steveninck RR, Bialek W. Phys Rev Lett. 1998;80:197. [PubMed] [Google Scholar]
- 29.Hudson RR. Bioinformatics. 2002;18:337–338. doi: 10.1093/bioinformatics/18.2.337. [DOI] [PubMed] [Google Scholar]
- 30.Spencer CCA, Coop G. Bioinformatics. 2004;20:3673–3675. doi: 10.1093/bioinformatics/bth417. [DOI] [PubMed] [Google Scholar]
- 31.Sabeti PC, Reich DE, Higgins JM, Levine HZ, Richter DJ, Schaffner SF, Gabriel SB, Platko JV, Patterson NJ, McDonald GJ, et al. Nature. 2002;419:832–837. doi: 10.1038/nature01140. [DOI] [PubMed] [Google Scholar]
- 32.Voight BF, Kudaravalli S, Wen X, Pritchard JK. PLoS Biol. 2006;4:3. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beckman G, Birgander R, Sjalander A, Saha N, Holmberg PA, Kivela A, Beckman L. Hum Hered. 1994;44:266–270. doi: 10.1159/000154228. [DOI] [PubMed] [Google Scholar]
- 34.Choi J, Donehower LA. Cell Mol Life Sci. 1999;55:38–47. doi: 10.1007/s000180050268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lozano G, Liu G. Semin Cancer Biol. 1998;8:337–344. doi: 10.1006/scbi.1998.0096. [DOI] [PubMed] [Google Scholar]
- 36.Germer S, Holland MJ, Higuchi R. Genome Res. 2000;10:258–266. doi: 10.1101/gr.10.2.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stephens M, Smith NJ, Donnelly P. Am J Hum Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guo SW, Thompson EA. Biometrics. 1992;48:361–372. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.