Abstract
The forkhead family protein FOXP3 acts as a repressor of transcription and is both an essential and sufficient regulator of the development and function of regulatory T cells. The molecular mechanism by which FOXP3-mediated transcriptional repression occurs remains unclear. Here, we report that transcriptional repression by FOXP3 involves a histone acetyltransferase–deacetylase complex that includes histone acetyltransferase TIP60 (Tat-interactive protein, 60 kDa) and class II histone deacetylases HDAC7 and HDAC9. The N-terminal 106–190 aa of FOXP3 are required for TIP60–FOXP3, HDAC7–FOXP3 association, as well as for the transcriptional repression of FOXP3 via its forkhead domain. FOXP3 can be acetylated in primary human regulatory T cells, and TIP60 promotes FOXP3 acetylation in vivo. Overexpression of TIP60 but not its histone acetyltransferase-deficient mutant promotes, whereas knockdown of endogenous TIP60 relieved, FOXP3-mediated transcriptional repression. A minimum FOXP3 ensemble containing native TIP60 and HDAC7 is necessary for IL-2 production regulation in T cells. Moreover, FOXP3 association with HDAC9 is antagonized by T cell stimulation and can be restored by the protein deacetylation inhibitor trichostatin A, indicating a complex dynamic aspect of T suppressor cell regulation. These findings identify a previously uncharacterized complex-based mechanism by which FOXP3 actively mediates transcriptional repression.
Keywords: acetylation, deacetylation, regulatory T cell, TIP60, HDAC7
A central theme that has emerged over the last 25 years is that a process of self-regulation of the immune response occurs to limit self-reactivity. Biochemical details of how the immune system distinguishes and regulates self and non-self remain to be fully documented (1). A recently characterized CD4+CD25+ regulatory T cell subset expresses the Foxp3 transcription factor. As a transcriptional repressor of cytokine gene expression (2), Foxp3 was subsequently identified as an essential and sufficient regulator of natural regulatory T cell development and function (3–5).
Mammalian transcriptional repressors can execute their function by either passive or active mechanisms (6, 7). FOXP3 may, for example, function as a passive transcriptional repressor in the case of its association with NFAT and NF-κB (8, 9). In this study, we explore the role of FOXP3 as an active transcriptional repressor by revealing the dynamic FOXP3 ensemble formation with a specific histone acetyltransferase (HAT) and certain class II histone deacetylases (HDACs) in expanded human CD4+CD25+ regulatory T cells (10, 11).
Histone acetylation and histone deacetylation affect chromatin remodeling during T cell development and differentiation (12, 13). HAT and HDAC abnormalities have been associated with leukemia (14, 15), diabetes (16) and other diseases of the immune system (17–19). The linkage of HAT and HDAC as components of a single complex permits dynamic responsiveness to extracellular stimulation (18, 20). The HAT TIP60 (Tat-interactive protein, 60 kDa), originally isolated as an HIV-1 TAT-interactive protein (21), functions as either a transcriptional coactivator or transcriptional corepressor (22, 23). Activated TIP60 can exert its acetyltransferase activity on a variety of proteins, including histone H2A, H3, and H4 (21), protein kinase ATM (24), and transcription factors such as c-myc (25) and p53 (26, 27). TIP60 also functions as a transcriptional corepressor of STAT3 in part through the recruitment of HDAC7 (28, 29).
Class II HDAC subfamily members include HDAC4, HDAC5, HDAC7, HDAC9, and HDAC10, all of which contain an N-terminal 17-aa MEF2D-binding motif, but only HDAC7 and HDAC9 are highly expressed in CD4+ T cells (30, 31). HDAC7 is highly expressed in CD4+CD8+ double positive T cells and regulates negative selection in the thymus by means of inhibition of Nur77 transcription through the specific recruitment of MEF2D to its binding site on the Nur77 promoter (30). HDAC9 expression is notably higher in mouse Foxp3gfp+CD4+ T cells than Foxp3gfp−CD4+ T cells (31).
Here, we provide evidence that FOXP3 actively represses transcription through its association with HAT TIP60 and HDAC7 and HDAC9 in vivo. We identified the N-terminal 106- to 190-aa proline-rich region of FOXP3, which has little similarity with other FOXP subfamily members, as a critical region for FOXP3 forkhead domain-mediated transcriptional repression, dependent on its dynamic association with TIP60 and HDAC7. Moreover, we demonstrate that FOXP3 is acetylated in primary human regulatory T cells and show that this process is promoted by TIP60. Whereas overexpression of TIP60, but not its HAT-deficient mutant, promotes FOXP3-mediated transcriptional repression, endogenous knockdown of TIP60 relieves this repression.
Results
FOXP3 Is Acetylated, a Process Promoted by HAT TIP60.
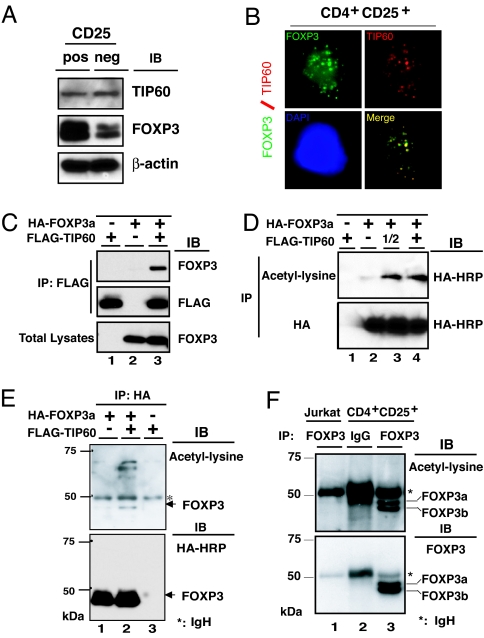
Previously we identified a HAT, the HIV-1 TAT-interactive protein, 60 kDa (TIP60), associated with the C-terminal proline-rich domain of an adaptor protein Cas-Br-M (murine) ecotropic retroviral transforming sequence b (Cbl-b) by yeast two-hybrid screening (B.L. and M.I.G., unpublished data). We found that the N-terminal proline-rich region of FOXP3 could also interact with TIP60. TIP60 expression in human CD4+CD25+ T cells and CD4+CD25− T cell was studied by immunoblotting with rabbit anti-TIP60 polyclonal antibody (Upstate, Temecula, CA), and we found that TIP60 was expressed almost equally in both cell types (Fig. 1A Top). Interestingly, in vitro expanded CD4+CD25− T cells also expressed a small but detectable amount of FOXP3 (Fig. 1A Middle). Next, we observed that endogenous TIP60 colocalizes with FOXP3 in the nucleus of human CD4+CD25+ regulatory T cells, by coimmunostaining with mouse anti-FOXP3, and rabbit anti-TIP60 (Fig. 1B).
Fig. 1.
FOXP3 is acetylated, which is promoted by TIP60. (A) TIP60 expression in both human CD4+CD25+ T cells and CD4+CD25− T cells. FOXP3 and β-actin expression levels were also analyzed by immunoblotting with 221D and anti-β-actin antibodies. (B) Nuclear colocalization of endogenous FOXP3 with TIP60 in CD4+CD25+ T cells. Human CD4+CD25+ T cells were stimulated for 2 h with PMA/ionomycin, fixed, permeabilized, and stained by anti-FOXP3 hFOXY (eBioscience), in conjunction with rabbit anti-TIP60 (Upstate) as indicated. Cell nucleus was demonstrated by DAPI staining (blue panel). (C) FOXP3 associates with TIP60 in vivo. HEK 293T cells were cotransfected with expression plasmids for FLAG-TIP60, or HA-FOXP3a as indicated, immunoprecipitated with anti-FLAG M2, followed by Western blotting with anti-FOXP3 221D, or anti-FLAG M2. (D and E) TIP60 promotes FOXP3 acetylation. (D) HEK 293T cells were cotransfected with HA-FOXP3a and an increasing amount of FLAG-TIP60 as indicated, then immunoprecipitated either with acetylated-lysine Ac-K-103 (Upper) or with anti-HA F-7 probe (Lower), followed by Western blotting with HRP-HA. (E) HEK 293T cells were cotransfected with HA-FOXP3a and FLAG-TIP60 as indicated, then immunoprecipitated with anti-HA, followed by Western blotting either acetylated-lysine (Cell Signaling no. 9441; Upper) or HA-HRP (Lower). (F) FOXP3 is acetylated in human CD4+CD25+ T cells. Nuclear extracts from Jurkat E6.1 T cells and human CD4+CD25+ T cells were immunoprecipitated with anti-FOXP3 hFOXY, or control IgG, then analyzed with rabbit anti-acetyl-lysine (Upstate) (Upper) and reprobed with anti-FOXP3 221D (Lower).
To determine whether FOXP3 associates with TIP60 in vivo, we cotransfected a HA-FOXP3 (FOXP3a) together with a FLAG-TIP60 expression constructs into 293T cells. We observed that FOXP3 coimmunoprecipitated with TIP60 (Fig. 1C). Because TIP60, as a HAT, acetylates and regulates the function of non-histone transcription factors such as c-Myc (25) and p53 (26, 27), we tested whether FOXP3 is acetylated in vivo. The HA-FOXP3 (FOXP3a)-expressing construct was cotransfected with increasing amounts of FLAG-TIP60 construct into 293T cells. We found overexpressing TIP60 permitted anti-acetyl-lysine mAb Ac-k-103 to immune precipitate increasing amounts of FOXP3 proteins, indicating TIP60 promotes FOXP3 acetylation (Fig. 1D). Reciprocal precipitations were performed as well. As shown in Fig. 1E, overexpression of TIP60 promoted FOXP3 acetylation, as well as acetylation of several unknown proteins, with molecular masses ranging between 50 kDa and 75 kDa, that all exist in the FOXP3 immunoprecipitated complex (Fig. 1E Upper).
Furthermore, to verify whether endogenous FOXP3 is also acetylated under physiological conditions, we immunoprecipitated comparable amounts of nuclear extracts from human FOXP3 expressing CD4+CD25+ regulatory T cells or control Jurkat T cells that lack FOXP3 expression with either monoclonal anti-FOXP3 hFOXY or control IgG, then immunoblotted with rabbit anti-acetyl-lysine polyclonal antibody (Fig. 1F Upper). After stripping, we reprobed with anti-FOXP3 mAb 221D (Fig. 1F Lower). These studies confirmed that endogenous FOXP3 is acetylated in primary human CD4+CD25+ regulatory T cells expanded in vitro.
FOXP3 Associates with Class II HDAC7 in Human CD4+CD25+ T Cells.
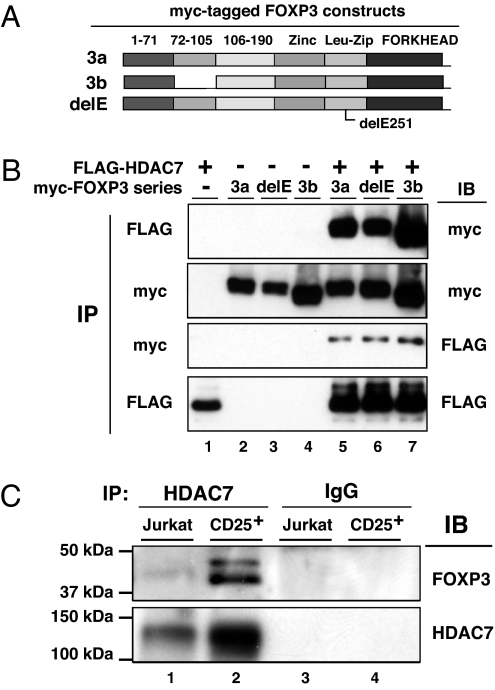
Because the HAT TIP60 associates with and recruits HDAC7 for transcriptional repression in other repressor complexes (28, 32), we tested whether FOXP3 also existed in association with HDAC7. We examined interactions of FOXP3 with HDAC7 using full-length FOXP3a and the exon 2 lacking isoform FOXP3b, as well as the human IPEX patient delE251 mutant FOXP3 (Fig. 2A). We found that HDAC7 associated with both the large and small isoform of FOXP3 (Fig. 2B, lanes 5 and 7). This association is clearly independent of the ability of FOXP3 to undergo dimerization or tetramerization (B.L. and M.I.G., unpublished data) because it is not affected by the IPEX patient mutant delE251 that we have shown exists as a monomer (Fig. 2B, lane 6).
Fig. 2.
FOXP3 associates with HDAC7 in primary CD4+CD25+ T cells. (A) Schematic representation of the myc-tagged FOXP3 constructs used for detection of FOXP3-HDAC7 association. (B) FOXP3 associates with HDAC7. HEK 293T cells were cotransfected with FLAG-HDAC7, myc-FOXP3a (3a), or myc-FOXP3b (3b) as indicated. Cell lysates were either immunoprecipitated with anti-FLAG or anti-myc antibodies, then immunoblotted with indicated Abs. (C) Endogenous FOXP3 associates with HDAC7 in human CD4+CD25+ T cells. Nuclear extracts from Jurkat E6.1 T cells and human CD4+CD25+ T cells were immunoprecipitated with anti-HDAC7 C-18, or control IgG, then analyzed by Western blotting with anti-FOXP3 221D (Upper) and reprobed with anti-HDAC7 KG-17 (Lower).
We next determined that FOXP3 associates with HDAC7 under physiologic conditions. Nuclear extracts from FOXP3 expressing primary human CD4+CD25+ regulatory T cells or an equal number of control Jurkat T cells that lack FOXP3 expression were immune precipitated with either goat anti-HDAC7 (C-18) or control IgG, then immunoblotted with anti-FOXP3 mAb 221D. After stripping, we then reprobed with rabbit anti-HDAC7 (KG-17). These studies established the endogenous FOXP3 association with HDAC7 in in vitro expanded primary human CD4+CD25+ regulatory T cells (Fig. 2C).
N-Terminal 106- to 190-aa Region of FOXP3 as a Transcriptional Repression Domain.
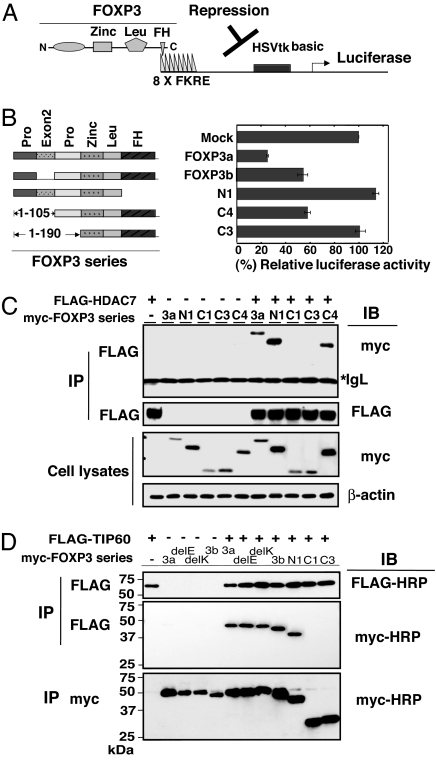
FOXP3 distinguishes itself from other family members including FOXP1, FOXP2, and FOXP4 by its unique N-terminal proline-rich 1- to 190-aa region, supporting a role for this region in regulatory T cell function. To determine which subdomains of FOXP3 are responsible for its transcription repressive activity, we cotransfected a panel of FOXP3 expression constructs together with a firefly luciferase reporter 8xFK1tk-Luc construct (33) driven by a specific upstream promoter/enhancer with eight forkhead-binding sites in 293T cells, which express endogenous TIP60 and HDAC7(Fig. 3A). We found that the large isoform of FOXP3 (FOXP3a) has the maximal transcription repression activity and the repression is apparently mediated by the FOXP3–TIP60–HDAC7 complex, although the forkhead deletion mutation of FOXP3 (N1) has completely lost its repressive activity as expected. The smaller isoform of FOXP3 lacking exon 2 (FOXP3b) still mediated repression, as did the N-terminal 1- to 105-aa deletion mutant C4. However, the N-terminal 1- to 190-aa deletion mutant (C3) with the intact forkhead DNA-binding domain was unable to repress transcription from the reporter (Fig. 3B). These data clearly show that the N-Terminal region between amino acids 106 and 190 is essential and critical to FOXP3-mediated transcriptional repression induced by forkhead domain binding to DNA.
Fig. 3.
N-terminal 106–190 aa as the transcriptional repression domain of FOXP3 is essential for TIP60 and HDAC7 association. (A) Schematic representation of FOXP3 binding to 8x Forkhead-binding sites luciferase reporter construct (8x FK1TK-Luc) used in luciferase reporter assay. (B) Luciferase reporter assay using 8x FK1TK-Luc reporter. 293T cells were transfected with the control empty vector (mock), wild-type FOXP3a, FOXP3b, FOXP3 forkhead domain deletion (N1) expression vectors, or FOXP3 deletion mutant del C4 or delC3, plus 8x FK1TK-Luc luciferase reporter and control TK-Renilla luciferase vector as indicated, then analyzed by means of dual luciferase assay normalized with Renilla luciferase activity. Results are means of three separate experiments with SD. (C) HDAC7 associates with 3a, N1, and C4, but not C3 with an additional deletion of 106- to 190-aa region. 293T cells were transfected with a panel of myc-tagged FOXP3 expression vectors, combined with FLAG-HDAC7 as indicated, immunoprecipitated with anti-FLAG M2, then analyzed by Western blotting with indicated Abs. (D) TIP60 associates with 3a, 3b, delE, delK, N1, but not C3 with an additional deletion of 106- to 190-aa region. 293T cells were transfected with a panel of myc-tagged FOXP3 expression vectors, with or without FLAG-TIP60 as indicated, immunoprecipitated with either anti-FLAG or anti-myc mAb, then analyzed by Western blotting with indicated Ab.
Because FOXP3 associates with HDAC7 and TIP60, which also interact with each other and have been suggested to act as transcriptional repressors (28), we tested whether the N-Terminal 106- to 190-aa transcription repression domain was also essential for the association of FOXP3 with HDAC7 and TIP60. A series of myc-tagged FOXP3 expression constructs together with the FLAG-HDAC7 expression construct were transfected into 293T cells (Fig. 3C). Whereas the FOXP3 forkhead deletion mutant (N1) and N-terminal 1- to 105-aa deletion mutant (C4) could still associate with HDAC7, the N-terminal 1- to 190-aa deletion (C3) or N-terminal 1- to 220-aa deletion (C1) limited FOXP3's association with HDAC7 (Fig. 3C).
Similar experiments were performed to map the subdomain of FOXP3 associated with TIP60, which interacts with HDAC7. We found that TIP60 associated with FOXP3a, FOXP3b, delE251 FOXP3a, delK250 FOXP3a and FORKHEAD deleted FOXP3 (N1)(Fig. 3D), and N-terminal 1- to 105-aa deletion mutant (C4) (data not shown), but not the N-terminal 1- to 220-aa deletion (C1) or N-terminal 1- to 190-aa deletion of FOXP3 (C3) (Fig. 3D). Together, the coincidence of the N-terminal 106–190 aa as an essential region for FOXP3 transcription repression activity, as well as the importance of this region for association with transcription corepressors HDAC7 and TIP60, identifies one molecular mechanism by which a tripartite ensemble of TIP60, HDAC7 and FOXP3 functions as a transcriptional repressor in vivo.
Knockdown of Endogenous TIP60 Relieves FOXP3-Mediated Repression in Vivo.
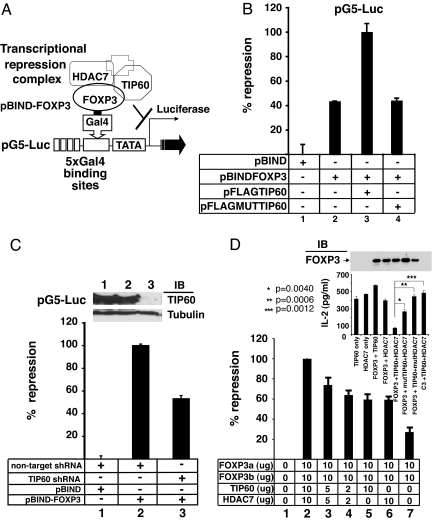
To evaluate whether the molecular mechanism of FOXP3-mediated repression is specifically dependent on TIP60, we established a transcriptional repression assay (illustrated in Fig. 4A). We cotransfected Gal4-FOXP3a fusion protein expressing vector pBIND-FOXP3a, the five Gal4-binding site driven firefly luciferase reporter pG5luc (Promega), pMSVβgal control vector (34), together with pFLAG vectors expressing TIP60 (32) or HAT-deficient TIP60(pFLAGMUTTIP60) (35) in 293T cells. Full-length FOXP3a, when expressed as a Gal4 fusion protein, acted as a repressor of the pG5 luciferase reporter (Fig. 4B, lane 2). Overexpression of wild-type TIP60 (Fig. 4B, lane 3), but not the HAT-deficient TIP60 (Fig. 4B, lane 4) specifically promotes Gal4-FOXP3-mediated transcriptional repression. Therefore the HAT activity of TIP60 is important for this transcription repressive activity.
Fig. 4.
FOXP3 mediates transcriptional repression via the forkhead domain as part of an ensemble with HDAC7 and TIP60. (A) Schematic representation of GAL4-FOXP3 binding to 5x GAL4-binding sites luciferase reporter construct (pG5Luc) used in luciferase reporter assay. (B) Overexpression of TIP60 promotes FOXP3-mediated transcriptional repression. 293T cells were transfected with the control pBIND empty vector (pBIND), pBIND-FOXP3a, pBIND-FOXP3a and pFLAG-TIP60 or, pBIND-FOXP3qa and the HAT-deficient TIP60 expressing construct (pFLAG-MUT-TIP60), plus pG5Luc luciferase reporter and control MSV-β-Gal as indicated, then analyzed by means of luciferase assay normalized with β-Gal activity. Results are means of three separate experiments with SD. (C) Knockdown of endogenous TIP60 relieves FOXP3-mediated transcriptional repression. 293T cells were transfected with indicated vectors and cell lysates were analyzed by means of luciferase assay normalized with β-Gal activity. Results are means of 3 separated experiments with SD. The knockdown efficiency of TIP60 shRNA was evaluated by Western blotting with Rabbit anti-TIP60, and reprobed with anti-α-tubulin (Inset). (D) A role of FOXP3-TIP60-HDAC7 ensemble in the repression of IL-2 production. Transfected Jurkat E6.1 T cells with vectors as indicated were stimulated respectively with plate-bound TCR Vβ 8.1 plus soluble anti-CD28. IL-2 production in cultured medium was measured with IL-2 ELISA kit (eBioscience). The repression efficiency of the empty vector transfected sample was defined as zero, and the one with 10 μg each of FOXP3a, FOXP3b, TIP60, and HDAC7 plasmids transfected was defined as 100%. The result is the average ± standard error by mean of three independent experiments. (Inset) One representative result of three independent experiments showing the actual amount of IL-2 production after TCR plus CD28 stimulation in Jurkat T cells, which were cotransfected with either 10 μg each of FOXP3a, TIP60 and HDAC7 expressing plasmids, or equal amounts of empty vectors. Transfection of the HAT-deficient TIP60 or HDAC deficient HDAC7 species led to less inhibition of IL2 production.
We next established that the endogenous TIP60 expression in 293T cells contributes to FOXP3-mediated transcriptional repression. We tested whether knockdown of endogenous TIP60 expression would affect FOXP3-mediated repression. We found that inhibition of the endogenous TIP60 expression dramatically relieved FOXP3-mediated transcriptional repression (Fig. 4C, lane 3). The nontargeted short hairpin RNA (shRNA) construct (Sigma) was used as a negative control (Fig. 4C, lane 2). We also performed endogenous TIP60 knockdown by lentiviral based shRNA transduction into in vitro expanded human CD4+CD25+ regulatory T cells. However, the TIP60 shRNA transduction dramatically reduced proliferation and viability of the lentiviral transduced CD4+CD25+ regulatory T cells compared with the nontargeted shRNA transduced cells (data not shown) preventing appropriate biochemical analyses. This observation is consistent with the known early lethal effect of TIP60 knockout in mice.
Based on the knockdown data of endogenous TIP60, we conclude that TIP60 is an essential subunit of the FOXP3 repression complex, and the HAT enzymatic activity of TIP60 plays an important defining role in repression mediated by the FOXP3 complex.
A FOXP3 Ensemble Is Necessary for IL-2 Production Regulation.
We extended our analysis of the FOXP3–TIP60–HDAC7 ensemble, to study its effects on IL-2 production. Coexpression of both FOXP3 isoforms was used to mimic the physiological expression pattern of human FOXP3 in vivo. Jurkat E6.1 T cells were cotransfected with both FOXP3a and FOXP3b, together with or without TIP60 alone, HDAC7 alone or the titrated amounts of the combination of both TIP60 and HDAC7. As depicted in Fig. 4D, the combination of TIP60 and HDAC7 with FOXP3a and FOXP3b led to the maximal repression in a dose dependent manner (lanes 2, 3, and 4), and the combination of either TIP60 or HDAC7 alone with FOXP3a and FOXP3b reduced the total repression efficiency (lanes 5 and 6). FOXP3a and FOXP3b alone also repressed IL-2 production to a degree as a consequence of recruitment of endogenous HAT–HDAC complexes (Fig. 4D, lane 7). The Inset in Fig. 4D shows the actual amount of IL-2 produced after transfections with different forms of Tip60 or HDAC7. Clearly optimal suppression of IL2 production by Foxp3 requires both intact enzymes. Therefore, our data indicates that FOXP3 recruitment of a functional HAT/HDAC complex is essential for its repression of cytokine production and that expression of wild-type HATs or HDACs can directly affect the stoichiometric and functional features of the ensembles needed for regulation.
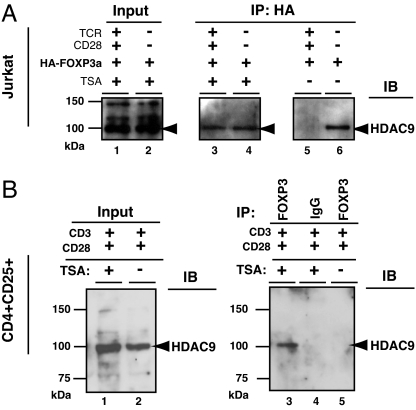
TCR Stimulation Disrupts FOXP3 and HDAC9 Interaction, Which Can Be Restored by Trichostatin A (TSA) Treatment.
Another class II HDAC expressed in T cells, namely HDAC9 (31), also exists in the FOXP3 complex under different physiological conditions. We found that, in the absence of T cell stimulation (Fig. 5A, lanes 4 and 6), HDAC9 coimmunoprecipitates with FOXP3. However, TCR plus CD28 stimulation is sufficient to antagonize the FOXP3 complex from its association with endogenous HDAC9 (Fig. 5A, comparing lanes 5 and 6). Interestingly, the disruption of the FOXP3–HDAC9 complex can be reversed by treating these activated T cells with the HDAC inhibitor TSA (Fig. 5A, lanes 3 and 4). To confirm that protein HDAC function plays a role in the dynamic assembly of endogenous FOXP3–HDAC9 complex, we immunoprecipitated endogenous FOXP3 complex from in vitro anti-CD3/CD28 activated and expanded human CD4+CD25+ T cells, with or without pretreatment with TSA for 4 h, followed by Western blotting for HDAC9.
Fig. 5.
T cell stimulation antagonizes FOXP3 recruiting HDAC9. (A) HA-FOXP3a transfected Jurkat E6.1 T cells (10 × 106) were not stimulated, or stimulated with plate-bound TCR Vβ 8.1 plus soluble anti-CD28 for 4 h (with or without 400 nM TSA, indicated above lanes), lysed, and equal nuclear extracts were immunoprecipitated with anti-HA probe (F-7), then analyzed by immunoblotting with anti-HDAC9 H-45 (lanes 3–6). The input nuclear extracts of TSA treated cells, with or without stimulation, were also immunoblotted with anti-HDAC9 H-45 (lanes 1 and 2). (B) In vitro activated and expanded human CD4+CD25+ T cells were treated with or without 400 nM TSA and lysed, and equal nuclear extracts were immunoprecipitated with anti-FOXP3 mAb 221D or control IgG, then analyzed by immunoblotting with anti-HDAC9 H-45 (Right, lanes 3, 4, and 5). The input nuclear extracts of TSA treated or untreated cells were also immunoblotted with anti-HDAC9 H-45 (Left, lanes 1 and 2).
Consistent with the result in Jurkat T cells, we also did not detect the endogenous association of FOXP3-HDAC9 in the in vitro anti-CD3/CD28 activated and expanded primary human CD4+CD25+ T cells (Fig. 5B, lane 5). However, TSA treatment is sufficient to promote endogenous FOXP3-HDAC9 association (Fig. 5B, lane 3). Together, these data indicate that in addition to a T cell receptor plus CD28 stimulation signal, HDAC activity may also play a determining role on the stability of the dynamic ensembles of the FOXP3–HDAC9 complex. We suggest that HDAC9 subserves a function distinct from HDAC7 in FOXP3-mediated regulation and that disabling HDACs may alter regulatory T cell functions in vivo.
Discussion
Specific recruitment of a HAT and an HDAC complex to target genes by transcription factors is relevant for transcriptional activation as well as transcriptional repression (36, 37). We previously found that FOXP3 existed as a large complex independent of FOXP3 oligomerization (11), which led us to explore the possible role of FOXP3 as a positive transcriptional repressor. The repressor function is clearly further developed by recruitment of transcription corepressors such as HAT and HDAC.
Here, we have identified HAT TIP60 and HDAC7 and HDAC9 associated in a dynamic ensemble with FOXP3 in vivo. TIP60 is the only detectable HAT coimmunoprecipitated with FOXP3 which suggests that TIP60 is the principal HAT responsible for FOXP3 acetylation and FOXP3-mediated transcriptional regulation in vivo. Moreover, we have discovered that FOXP3 is acetylated and that this modification is linked to its function in regulatory T cells.
HATs can directly interact with HDACs in vivo, although the molecular mechanisms and consequences of such an arrangement are currently unclear (37, 38). Notably the TIP60 and HDAC7-associated N-terminal 106–190 aa of FOXP3 is required for FOXP3-mediated transcriptional repression and should be considered as a repression domain.
Bray and Lay (39) have analyzed multimeric ensembles and found suppression of protein complex formation by disproportionately high concentrations of their individual components. We have also identified a general stoichiometric relationship of the constituents of the FOXP3–TIP60–HDAC7 complex that is needed to obtain the maximal repression of IL-2 production in T cells after TCR plus CD28 stimulation.
Overexpression of the wild-type TIP60, but not the HAT-deficient TIP60, promotes FOXP3-mediated repression, whereas knockdown of endogenous TIP60 relieved FOXP3-mediated repression. These studies suggest that stimuli signals which promote TIP60 function or decrease HDAC activity may modify T cell-mediated suppression. Finally, these studies extend our notion that modifications by distinct acetyltransferases and deacetylases may occur on the same transcription factor perhaps at distinct sites, times, and after different signaling events. At this point we have not resolved the totality of activities that TIP60-mediated FOXP3 acetylation effects. Which sites of FOXP3 that are acetylated also remain undefined. Based on our observation that inhibition of endogenous TIP60 expression by shRNA knockdown relieved FOXP3-mediated transcriptional repression, we speculate that the association of FOXP3 with HAT/HDACs occurs to facilitate the preferential transcription of FOXP3-targeted genes and serves as a mechanism whereby cellular repression is established and regulated. Moreover, the transcriptional repression effect of GAL4-FOXP3 is reduced but still detected even when the endogenous TIP60 expression had been completely knockdown by shRNA transduction. This finding indicates that other unknown transcriptional corepressors could also contribute to FOXP3-mediated repression.
Based on the biochemical analysis of FOXP3 complex in human CD4+CD25+ T cells, we hypothesize that FOXP3 ensembles are composed of multiple protein subunits, and the dynamic assembly of the FOXP3 complex may be responsible for the transcriptional repression of IL-2 in regulatory T cells. HAT-deficient TIP60 mutant mice may help to further clarify certain aspects of Tip60's physiological role in regulatory T cells in vivo.
Whereas endogenous FOXP3 consistently associated with HDAC7 in human in vitro activated and expanded regulatory T cells, we also found that FOXP3 disassociated from another class II deacetylase, HDAC9, after TCR plus CD28 stimulation. The observation suggests a dynamic quality to FOXP3 complex ensembles that occurs in response to T cell receptor signals. Moreover, the dynamic association of FOXP3 and HDAC9 is promoted by the protein deacetylase inhibitor TSA. It is noteworthy that HDAC9 can function as a signal-responsive repressor independently of its HDAC catalytic domain (40, 41).
Our initial description of the dynamic ensembles of FOXP3 with HAT/HDAC complexes provides a molecular explanation of how FOXP3 mediates transcriptional repression in regulatory T cells and identifies pharmaceutical approaches such as altering the enzymatic activity of HATs or HDACs to modify regulatory T cell functions.
Experimental Procedures
For additional details, see supporting information (SI) Methods.
Human CD4+CD25+ T Cells.
Human FOXP3+CD4+CD25+ T cells were obtained by in vitro expansion as follows: 200 million PBLs were stained for CD4 and CD25, and by using a Mo Flo high speed sorter, the brightest (top 1%) CD4+CD25+ cells were purified. These cells were stimulated with anti-CD3, anti-CD28 coated beads by using a three-bead to one-cell ratio or by using a cell-based αAPC expressing CD64 and CD86 loading with anti-CD3 Ab (42) in the presence of high levels of IL-2 (300 units/ml) and cultured in RPMI medium 1640 with 10% FCS for the next 20–25 days.
Cloning of Human FOXP3 cDNA.
Based on the nucleotide sequence of FOXP3 in the human genome (www.ensembl.org/Homo_sapiens), we cloned two FOXP3 cDNAs, one corresponding to the full length named as FOXP3a, as well as a splice variant lacking exon 2 named as FOXP3b.
Preparation of Nuclear Extracts.
Nuclear extracts were prepared according to Nishiya et al. (43) with some modification.
Plasmids, Reagents, and Antibodies.
We are grateful to Alison H. Banham for anti-FOXP3 mAb (44), Eric Verdin for pcDNA-FLAG-HDAC7 (30), He-Jin Lee for pCMV2-FLAG-wild-type TIP60 (32), and Craig N Robson for pcDNA-FLAG-HAT-deficient TIP60 (Q377E/G380E, MutTIP60) (20).
shRNA Vectors and Reagent.
TRC shRNAs (Lenti) targeting human TIP60, TRCN0000020315 (sh15), and the Arrest-In transfection reagent (cat. no. ATR1741) were purchased from Open Biosystem. The non-target shRNA control vector was purchased from Sigma (St. Louis, MO) (cat. no. SHC002).
Dual Luciferase Assay.
Jurkat transfections and all luciferase assays were performed as described in Shapiro et al. (45) with some modification.
Supplementary Material
Acknowledgments
We thank G. R. Crabtree (Department of Pathology, Stanford University, Stanford, CA) for providing human IL-2 promoter reporter; W. H. Biggs III (Ludwig Institute for Cancer Research, La Jolla, CA) for 8xFK1tk-luciferase reporter; E. E. Morrisey (Department of Medicine, University of Pennsylvania, Philadelphia, PA) for MSV-β-gal reporter; E. Verdin (Gladstone Institute of Virology and Immunology, University of California, San Francisco, CA) for pcDNA-FLAG-HDAC7; H.-J. Lee (Parkinson's Institute, Sunnyvale, CA) for pCMV2-FLAG-wild-type TIP60 and C. N. Robson (University of Newcastle Upon Tyne, Newcastle Upon Tyne, U.K.) for pcDNA-FLAG-HAT deficient TIP60 (Q377E/G380E, MutTip60) constructs; A. H. Banham (University of Oxford, John Radcliffe Hospital, Oxford, U.K.) for anti-FOXP3 mAb 221D; and V. S. Shapiro, T. Golovina, Y. Hirohashi, H. Zhang, Q. Wang, M. Tone, and R. Murali for discussion and advice. M.I.G. is the John Eckman Professor of Medical Science at University of Pennsylvania.
Abbreviations
- HAT
histone acetyltransferase
- TIP60
Tat-interactive protein, 60 kDa
- HDAC
histone deacetylase
- TSA
trichostatin A
- shRNA
short hairpin RNA.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0700298104/DC1.
References
- 1.Schwartz RH. Nat Immunol. 2005;6:327–330. doi: 10.1038/ni1184. [DOI] [PubMed] [Google Scholar]
- 2.Schubert LA, Jeffery E, Zhang Y, Ramsdell F, Ziegler SF. J Biol Chem. 2001;276:37672–37679. doi: 10.1074/jbc.M104521200. [DOI] [PubMed] [Google Scholar]
- 3.Hori S, Nomura T, Sakaguchi S. Science. 2003;299:1057–1061. [PubMed] [Google Scholar]
- 4.Fontenot JD, Gavin MA, Rudensky AY. Nat Immunol. 2003;4:330–336. [PubMed] [Google Scholar]
- 5.Khattri R, Cox T, Yasayko SA, Ramsdell F. Nat Immunol. 2003;4:337–342. [PubMed] [Google Scholar]
- 6.Cowell IG. Trends Biochem Sci. 1994;19:38–42. doi: 10.1016/0968-0004(94)90172-4. [DOI] [PubMed] [Google Scholar]
- 7.Thiel G, Lietz M, Hohl M. Eur J Biochem. 2004;271:2855–2862. doi: 10.1111/j.1432-1033.2004.04174.x. [DOI] [PubMed] [Google Scholar]
- 8.Bettelli E, Dastrange M, Oukka M. Proc Natl Acad Sci USA. 2005;102:5138–5143. doi: 10.1073/pnas.0501675102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu Y, Borde M, Heissmeyer V, Feuerer M, Lapan AD, Stroud JC, Bates DL, Guo L, Han A, Ziegler SF, et al. Cell. 2006;126:375–387. doi: 10.1016/j.cell.2006.05.042. [DOI] [PubMed] [Google Scholar]
- 10.Earle KE, Tang Q, Zhou X, Liu W, Zhu S, Bonyhadi ML, Bluestone JA. Clin Immunol. 2005;115:3–9. doi: 10.1016/j.clim.2005.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li B, Samanta A, Song X, Furuuchi K, Iacono KT, Kennedy S, Katsumata M, Saouaf SJ, Greene MI. Immunol Rev. 2006;212:99–113. doi: 10.1111/j.0105-2896.2006.00405.x. [DOI] [PubMed] [Google Scholar]
- 12.Avni O, Lee D, Macian F, Szabo SJ, Glimcher LH, Rao A. Nat Immunol. 2002;3:643–651. doi: 10.1038/ni808. [DOI] [PubMed] [Google Scholar]
- 13.Fields PE, Kim ST, Flavell RA. J Immunol. 2002;169:647–650. doi: 10.4049/jimmunol.169.2.647. [DOI] [PubMed] [Google Scholar]
- 14.Tsuda E, Goto M, Mochizuki S, Yano K, Kobayashi F, Morinaga T, Higashio K. Biochem Biophys Res Commun. 1997;234:137–142. doi: 10.1006/bbrc.1997.6603. [DOI] [PubMed] [Google Scholar]
- 15.Sobulo OM, Borrow J, Tomek R, Reshmi S, Harden A, Schlegelberger B, Housman D, Doggett NA, Rowley JD, Zeleznik-Le NJ. Proc Natl Acad Sci USA. 1997;94:8732–8737. doi: 10.1073/pnas.94.16.8732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gray SG, De Meyts P. Diabetes Metab Res Rev. 2005;21:416–433. doi: 10.1002/dmrr.559. [DOI] [PubMed] [Google Scholar]
- 17.Cosio BG, Tsaprouni L, Ito K, Jazrawi E, Adcock IM, Barnes PJ. J Exp Med. 2004;200:689–695. doi: 10.1084/jem.20040416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang XJ. Nucleic Acids Res. 2004;32:959–976. doi: 10.1093/nar/gkh252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barnes PJ, Adcock IM, Ito K. Eur Respir J. 2005;25:552–563. doi: 10.1183/09031936.05.00117504. [DOI] [PubMed] [Google Scholar]
- 20.Gaughan L, Logan IR, Cook S, Neal DE, Robson CN. J Biol Chem. 2002;277:25904–25913. doi: 10.1074/jbc.M203423200. [DOI] [PubMed] [Google Scholar]
- 21.Kamine J, Elangovan B, Subramanian T, Coleman D, Chinnadurai G. Virology. 1996;216:357–366. doi: 10.1006/viro.1996.0071. [DOI] [PubMed] [Google Scholar]
- 22.Squatrito M, Gorrini C, Amati B. Trends Cell Biol. 2006;16:433–442. doi: 10.1016/j.tcb.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 23.Sapountzi V, Logan IR, Robson CN. Int J Biochem Cell Biol. 2006;38:1496–1509. doi: 10.1016/j.biocel.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 24.Sun Y, Jiang X, Chen S, Fernandes N, Price BD. Proc Natl Acad Sci USA. 2005;102:13182–13187. doi: 10.1073/pnas.0504211102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patel JH, Du Y, Ard PG, Phillips C, Carella B, Chen CJ, Rakowski C, Chatterjee C, Lieberman PM, Lane WS, et al. Mol Cell Biol. 2004;24:10826–10834. doi: 10.1128/MCB.24.24.10826-10834.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sykes SM, Mellert HS, Holbert MA, Li K, Marmorstein R, Lane WS, McMahon SB. Mol Cell. 2006;24:841–851. doi: 10.1016/j.molcel.2006.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tang Y, Luo J, Zhang W, Gu W. Mol Cell. 2006;24:827–839. doi: 10.1016/j.molcel.2006.11.021. [DOI] [PubMed] [Google Scholar]
- 28.Xiao H, Chung J, Kao HY, Yang YC. J Biol Chem. 2003;278:11197–11204. doi: 10.1074/jbc.M210816200. [DOI] [PubMed] [Google Scholar]
- 29.Yuan ZL, Guan YJ, Chatterjee D, Chin YE. Science. 2005;307:269–273. doi: 10.1126/science.1105166. [DOI] [PubMed] [Google Scholar]
- 30.Dequiedt F, Kasler H, Fischle W, Kiermer V, Weinstein M, Herndier BG, Verdin E. Immunity. 2003;18:687–698. doi: 10.1016/s1074-7613(03)00109-2. [DOI] [PubMed] [Google Scholar]
- 31.Fontenot JD, Rasmussen JP, Williams LM, Dooley JL, Farr AG, Rudensky AY. Immunity. 2005;22:329–341. doi: 10.1016/j.immuni.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 32.Lee HJ, Chun M, Kandror KV. J Biol Chem. 2001;276:16597–16600. doi: 10.1074/jbc.C000909200. [DOI] [PubMed] [Google Scholar]
- 33.Biggs WH, 3rd, Meisenhelder J, Hunter T, Cavenee WK, Arden KC. Proc Natl Acad Sci USA. 1999;96:7421–7426. doi: 10.1073/pnas.96.13.7421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li S, Weidenfeld J, Morrisey EE. Mol Cell Biol. 2004;24:809–822. doi: 10.1128/MCB.24.2.809-822.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Halkidou K, Logan IR, Cook S, Neal DE, Robson CN. Nucleic Acids Res. 2004;32:1654–1665. doi: 10.1093/nar/gkh296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roth SY, Denu JM, Allis CD. Annu Rev Biochem. 2001;70:81–120. doi: 10.1146/annurev.biochem.70.1.81. [DOI] [PubMed] [Google Scholar]
- 37.Nusinzon I, Horvath CM. Sci STKE 2005. 2005:re11. doi: 10.1126/stke.2962005re11. [DOI] [PubMed] [Google Scholar]
- 38.Yamagoe S, Kanno T, Kanno Y, Sasaki S, Siegel RM, Lenardo MJ, Humphrey G, Wang Y, Nakatani Y, Howard BH, Ozato K. Mol Cell Biol. 2003;23:1025–1033. doi: 10.1128/MCB.23.3.1025-1033.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bray D, Lay S. Proc Natl Acad Sci USA. 1997;94:13493–13498. doi: 10.1073/pnas.94.25.13493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang CL, McKinsey TA, Olson EN. Proc Natl Acad Sci USA. 2001;98:7354–7359. doi: 10.1073/pnas.131198498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang CL, McKinsey TA, Chang S, Antos CL, Hill JA, Olson EN. Cell. 2002;110:479–488. doi: 10.1016/s0092-8674(02)00861-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maus MV, Thomas AK, Leonard DG, Allman D, Addya K, Schlienger K, Riley JL, June CH. Nat Biotechnol. 2002;20:143–148. doi: 10.1038/nbt0202-143. [DOI] [PubMed] [Google Scholar]
- 43.Nishiya N, Yamamoto K, Imaizumi Y, Kohno T, Matsuyama T. Mol Immunol. 2004;41:855–861. doi: 10.1016/j.molimm.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 44.Roncador G, Brown PJ, Maestre L, Hue S, Martinez-Torrecuadrada JL, Ling KL, Pratap S, Toms C, Fox BC, Cerundolo V, et al. Eur J Immunol. 2005;35:1681–1691. doi: 10.1002/eji.200526189. [DOI] [PubMed] [Google Scholar]
- 45.Shapiro VS, Mollenaver MN, Weiss A. J Immunol. 1998;161:6455–6458. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.