Abstract
Through the analysis of hundreds of full-length cDNAs from fifteen species representing all major orders of dinoflagellates, we demonstrate that nuclear-encoded mRNAs in all species, from ancestral to derived lineages, are trans-spliced with the addition of the 22-nt conserved spliced leader (SL), DCCGUAGCCAUUUUGGCUCAAG (D = U, A, or G), to the 5′ end. SL trans-splicing has been documented in a limited but diverse number of eukaryotes, in which this process makes it possible to translate polycistronically transcribed nuclear genes. In SL trans-splicing, SL-donor transcripts (SL RNAs) contain two functional domains: an exon that provides the SL for mRNA and an intron that contains a spliceosomal (Sm) binding site. In dinoflagellates, SL RNAs are unusually short at 50–60 nt, with a conserved Sm binding motif (AUUUUGG) located in the SL (exon) rather than the intron. The initiation nucleotide is predominantly U or A, an unusual feature that may affect capping, and hence the translation and stability of the recipient mRNA. The core SL element was found in mRNAs coding for a diverse array of proteins. Among the transcripts characterized were three homologs of Sm-complex subunits, indicating that the role of the Sm binding site is conserved, even if the location on the SL is not. Because association with an Sm-complex often signals nuclear import for U-rich small nuclear RNAs, it is unclear how this Sm binding site remains on mature mRNAs without impeding cytosolic localization or translation of the latter.
Keywords: gene regulation, mRNA, Alveolata, Sm-binding site
Dinoflagellates are unicellular eukaryotes that contribute significantly to marine primary production, coral reef growth, and marine toxins. They are members of the Alveolata, which also include ciliates and apicomplexa (1). Dinoflagellate genomes are enormous (3–200 pg of DNA per cell) and lack typical histones, with chromosomes permanently condensed, nuclear membranes remaining intact in mitosis, and mitotic spindle being extranuclear (for review see ref. 2). The mechanism of gene regulation is largely unknown. Sporadic investigations have shown that a relatively small fraction of genes are under transcriptional control (3–6) and that introns are not common (7, 8). The few comprehensive studies of dinoflagellate gene structures reveal genes with high copy number and arrangement in polycistronic or otherwise tandem arrays (e.g., refs. 7–10).
Spliced leader (SL) trans-splicing has been found in a disjointed group of eukaryotes, in which a short RNA fragment (i.e., SL, ≈15–50 nt) from a small noncoding RNA (SL RNA) is transplanted to the 5′ end of independently transcribed pre-mRNAs to yield mature mRNAs. This process converts a polycistronic transcript into translatable monocistronic mRNAs. SL trans-splicing has been well studied in Euglenozoa. It has been detected in nematodes, Platyhelminthes, cnidarians, rotifers, ascidians, and appendicularia (for review, see 11–13). SL RNA contains two functional domains: an exon (i.e., SL) that is spliced to an mRNA and an intron that contains a spliceosomal (Sm) binding site believed to facilitate splicing. Although SL RNA bears low sequence similarity across phyla, features of its secondary structure are conserved (11). For example, in trypanosomes, the SL carries a 5′ cap structure consisting of a 7-methylguanosine followed by four methylated nucleotides (cap 4), whereas in worms, the SL carries a 5′ cap structure containing 2,2,7-trimethylguanosine (11, 13, 14). In both groups of organisms, the SL RNAs interact with the Sm-complex via a binding site in the introns. The Sm complex forms on several U-rich small nuclear ribonucleoprotein particles consisting of small nuclear RNA (snRNA) and Sm proteins and is involved in both cis- and trans-splicing (11, 14). Here, we document the widespread use of SL trans-splicing by nuclear mRNAs throughout major dinoflagellate orders. Further, we characterize dinoflagellate SL RNA genes and transcripts and model the RNA secondary structure. The results provide a critical advancement in understanding dinoflagellate gene expression and help future genetic and evolutionary studies of splicing.
Results
SL Addition Is Common in Dinoflagellate mRNA.
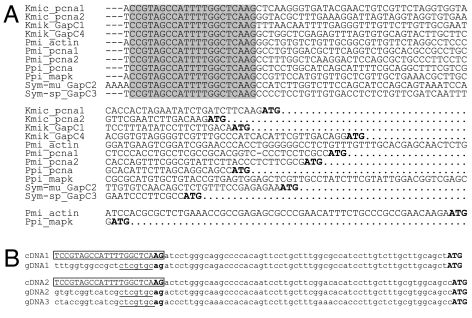
In organisms that use SL trans-splicing, mRNAs contain a common 5′ leader. BLAST analyses indicated that the first 22 nt of the 5′ end of the sequence observed in the cDNAs of pcna (9) and mapk (6) from Pfiesteria piscicida and actin from Prorocentrum minimum (GenBank accession no. AF512889) also occurred in GAPDH cDNAs from Karenia mikimotoi (AB164183, AB164186), Symbiodinium sp. (AB106686), and Symbiodinium muscatinei (AY314974). This 22-nt sequence was used to query existing EST datasets from Alexandrium tamarense, Amphidinium carterae, Heterocapsa triquetra, Karenia brevis, Lingulodinium polyedrum, and Pyrocystis lunula. SL was detected in all but the last species. In each positive case, only a small number of ESTs contained the SL, most likely because of incomplete 5′ end sequence. The 22-nt motif, DCCGTAGCCATTTTGGCTCAAG (D = T, A, or G), was conserved and located at the beginning of the 5′ UTRs (Fig. 1A). In a total of 21 full-length cDNAs obtained by using the GeneRacer kit (for actin, mapk and pcna) from P. piscicida, Karlodinium micrum, and Pr. minimum, variations in the 22-nt motif were observed only at the first position: 11 started with T, 9 started with A, and 1 started with G. Sequence alignment of P. piscicida pcna cDNA with genomic DNA showed that the 22-nt sequence was absent in the genomic pcna sequence (Fig. 1B). PCR, using P. piscicida genomic DNA as template and the dinoflagellate SL (DinoSL) and a pcna-specific primer, yielded no product under conditions that successfully amplified the cDNA templates (9). These results are consistent with the hypothesis that the 22-nt sequence is a SL sequence trans-spliced to the mRNA. Dinoflagellate SL shared no similarity with other phyla except at the last two nucleotides, AG, which are largely conserved (e.g., 12, 15, 16).
Fig. 1.
The conserved SL sequence from dinoflagellate mRNAs. (A) Alignment of the 5′ end of several full-length dinoflagellate cDNAs. The 21-nt identical sequence is shaded. ATG in bold indicates the predicted start codon. Kmic, K. micrum; Kmik, Kar. mikimotoi; Pmi, Pr. minimum; Ppi, P. piscicida; Sym-mu, S. muscatinei; Sym-sp, Symbiodinium sp. (B) Alignment of the 5′ UTRs of P. piscicida pcna cDNAs and corresponding genomic (gDNA) clones revealing the 22-nt trans-spliced SL (boxed). cDNA1, GenBank accession no. DQ239852; gDNA1, DQ239839; cDNA2, DQ239853; gDNA2, DQ239842; gDNA3, DQ239843. The dinucleotide AG at the SL-mRNA boundary is in bold. Underlined are the 5′ end conserved sequence in different genomic clones, ended with the 3′ acceptor splice site AG.
Wide Taxonomic Distribution of SL and Functional Diversity of Trans-Spliced mRNAs.
To examine the hypothesis that the paucity of detected SL-containing cDNAs in the existing EST datasets was due to incomplete 5′ ends of most of the sequences, we set out to retrieve full-length or 5′ end cDNAs from 15 species of dinoflagellates representing all major taxonomic orders, from ancestral (e.g., Oxyrrhis marina) to derived lineages (e.g., Alexandrium spp.). cDNA that was synthesized by using GeneRacer oligo(dT) primer was PCR-amplified, using the 22-nt DinoSL as the forward primer and a gene-specific primer or the GeneRacer 3′ primer (GR3) as the reverse primer. First, we isolated the 5′ UTR and the coding region of pcna from Akashiwo sp., Alexandrium affine, Alexandrium fundyense, K. micrum, Katodinium rotundatum, Peridinium foliaceum, Pr. minimum, Prorocentrum micans, and Symbiodinium goreaui to yield pcna sequence from a total of ten species, including P. piscicida [supporting information (SI) Table 1]. Sequence alignment indicated that the 5′ UTRs of the pcna transcripts shared little similarity except for the SL (Fig. 1A). This survey indicated that SL trans-splicing of pcna was widespread throughout the phylum.
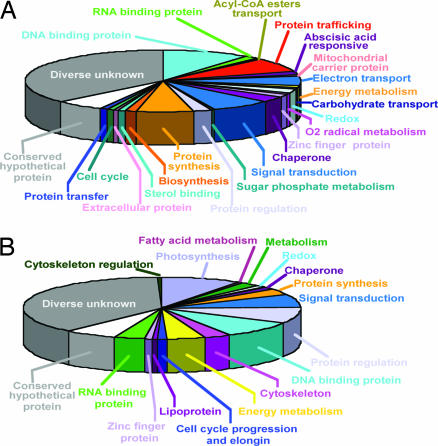
To determine whether SL trans-splicing is limited to certain functional groups of genes, we amplified SL-containing cDNAs for seven dinoflagellate species, and randomly sequenced 35–226 clones for each species (SI Tables 1 and 2). These cDNAs encoded proteins of diverse function (Fig. 2) along with numerous proteins of unknown function, indicating no preferred SL recipients.
Fig. 2.
The mRNA targets for trans-splicing encompass a wide variety of cellular functions. (A) Full-length cDNAs (226) from P. piscicida. (B) 165 cDNAs from K. micrum.
SL Trans-Splicing Is Restricted to Nuclear mRNAs.
We examined whether nuclear genes of plastids or other origin have adopted SL trans-splicing for mRNA processing. Genes encoding chloroplast light harvesting proteins, ferredoxin, plastocyanin, O-acetyl-serine lyase, and ATP synthase subunit C (atpH) have presumably transferred to the nuclear genome from chloroplast (17). The nuclear gene for Rubisco (form II) in peridinin-containing dinoflagellates was likely horizontally transferred to the nucleus from a proteobacterium (18). These transcripts possess SL at their 5′ ends, as shown by cDNA-templated PCR that used DinoSL and gene-specific primers (SI Table 1), demonstrating that the mRNAs of nuclear-encoded chloroplast-targeted genes are SL trans-spliced.
We further investigated whether SL is added to organelle-encoded mRNAs. P. piscicida mitochondrial cytochrome c oxidase 1 (cox1) and cytochrome b (cob) were chosen as the representatives of mitochondrion-encoded genes (19). Form I Rubisco large subunit gene (rbcL) of K. micrum was chosen as a representative chloroplast-encoded gene. The K. micrum chloroplast is thought to be of haptophyte origin through tertiary replacement (20). psbA, encoded in chloroplast minicircular DNA in lineages characterized by peridinin as the major accessory pigment (2), was examined for Heterocapsa and Amphidinium. PCR, using DinoSL with gene-specific reverse primers for these three genes, did not amplify cDNA-templated product. Control PCRs, using gene-specific primers for psbA and cDNA templates, yielded correct fragments. Using the GeneRacer technique, we successfully isolated the full-length cDNA of form I rbcL from K. micrum and found no SL addition at its 5′ end. Thus, all queries for the presence of SL in organellar transcripts were negative (SI Table 1). Similarly, no product was obtained in PCRs that used DinoSL and small subunit ribosomal RNA (18S rRNA)-specific primer sets for cDNA synthesized with random hexamers, indicating that SL is not added to 18S rRNA, a result consistent with previous reports that SL is not present at the 5′ end of the transcripts of 5.8S rRNA (i.e., RNA pol I) (21), 5S rRNA (i.e., RNA pol III) (22), or mitochondrial mRNAs in Trypanosoma brucei (23), although trans-splicing of the gene-internal variety occurs in mitochondrial genes of some organisms (24). The presence of SL on nuclear-encoded mRNAs of proteins destined for organellar import supports the hypothesis that trans-splicing is generally required for nuclear gene expression; alternatively, adoption of the trans-splicing pathway may have allowed an easier transition for gene relocation into the nuclear genome.
Dinoflagellate SL RNA Genes Are in Tandem Arrays.
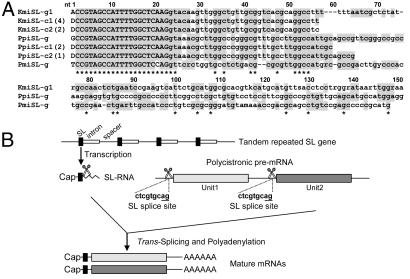
SL RNA genes in kinetoplastids are arranged in multiple head-to-tail tandem repeats (25). Because most dinoflagellate genes studied to date are also arranged in arrays (e.g., refs. 7, 9, 26, 27), we examined whether dinoflagellate SL RNA genes occur in tandem. An overlapping but not self-priming set of dinoflagellate core SL primers was used for PCR to amplify adjacent SL RNA gene repeats from dinoflagellate DNA, adapted from the strategy used in kinetoplastids (25). In 10 clones obtained from K. micrum, eight had one repeat unit, and two had dimers with an intact SL RNA gene in the center, flanked by upstream and downstream noncoding regions and adjacent gene fragments. The full-gene repeat ranged from 354 to 365 bp (GenBank accession nos. EF143070–EF143079). Three one-unit clones (179 bp; EF143080–EF143082) were recovered from P. piscicida. From Pr. minimum, one two-unit and one seven-unit clone, with 143 bp per unit, were obtained (EF143083–EF143084). Each unit consists of the 22-bp SL (i.e., exon), followed by an intron of varying size (see next section) and an intergenic spacer sequence (predicted to be 298–309 bp in K. micrum, 115–120 bp in P. piscicida and 89 bp in Pr. minimum). All repeats in the Pr. minimum SL RNA clones were identical in SL sequence, with the exception of an A→C substitution at position 20. The nucleotide sequence of SL RNA in different clones or different units of the same clone in the same species were conserved with few substitutions or insertions/deletions. The first 90 bp were moderately conserved, but within the 90 bp, the splice donor dinucleotide GT was strictly conserved as observed in all SL RNAs (Fig. 3A). Trypanosome SL RNA genes use a polyT tract to terminate transcription (28). No such motif was evident in the examined dinoflagellate sequences.
Fig. 3.
Conservation of SL RNAs (SL donor) in dinoflagellates. (A) Alignment of SL RNA and genomic sequences. Nucleotides identical in least two species are shaded, and those identical in all species are denoted by asterisks. Exon (SL) is shown in uppercase, and intron is shown in lowercase. D = T, A, or G; r = a or g; v = a, c or g; m = a or c; k = t or g. Kmi, K. micrum; Ppi, P. piscicida; Pmi, Pr. minimum; g, gene sequence; c, cDNA, with the number immediately after it indicating different forms. Numbers in parentheses are the number of identical sequences retrieved. For cDNAs of P. piscicida SL RNA, only the longest two clones are shown. (B) Schematic diagram illustrating how SL (exon) in the SL RNA is trans-spliced to pre-mRNA to yield mature mRNA with SL at the 5′ end. A putative 3′ acceptor site (i.e., SL splice site), ag, is underlined.
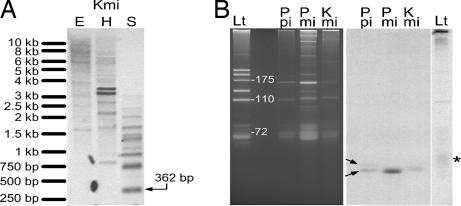
To further study the genomic structure of the SL RNA in dinoflagellates, Southern blot hybridization was carried out. Multiple bands were detected with different restriction enzymes for P. piscicida, K. micrum, and Pr. minimum. Fig. 4A shows an example for K. micrum. In addition to the predicted 362-bp band, numerous other bands were present for Sau3AI-digested DNA, suggesting the existence of polymorphism in dinoflagellate SL RNA gene arrays as reported in kinetoplastids (29).
Fig. 4.
The SL RNA gene is multicopy but produces a consistent transcript size. (A) Southern blot, using K. micrum SL RNA gene clone1 as the probe. K. micrum genomic DNA (equivalent to 4 × 105 cells) was digested by EcoRI (E), HindIII (H), and Sau3AI (S). Arrow indicates the band with predicted size of one unit of SL RNA gene. (B) Northern blot. (Left) Ethidium bromide stain of total RNA extracted from L. tarentolae (Lt), P. piscicida (Ppi), Pr. minimum (Pmi), and K. micrum (Kmi). Size standards are 5.8S rRNA (175 nt), 5S rRNA (110 nt), and tRNAGly (72 nt). (Right) Blot hybridized consecutively with oligonucleotides DinoSLa/s and S-255. Arrows indicate the two bands in P. piscicida RNA, and the asterisk indicates the cytochrome oxidase subunit III gRNA-1 (in lane Lt).
Minimal Sizes of Dinoflagellate SL RNA.
To estimate the size of the SL RNA in dinoflagellates, Northern blot analysis was carried out for K. micrum, P. piscicida, and Pr. minimum. The reverse-complement of 14-nt SL and 4-nt intron was used as the probe to favor SL RNA (SL plus intron) over mRNA (containing only SL at the 5′ end). Distinct bands of hybridization were detected for K. micrum and Pr. minimum that migrated slightly faster than the heterogeneous 55–60 nt cytochrome oxidase subunit III guide RNA (30) of Leishmania tarentolae (Fig. 4B). For P. piscicida, a second band with slightly larger size was also detected.
To map the 3′ ends of the dinoflagellate SL RNA, we used the 3′ end Racer technique (15). SL RNAs in other organisms are not polyadenylated, with two developmentally regulated exceptions (31, 32). The small and homogeneous size of the dinoflagellate SL RNAs indicated an absence of poly(A) tails. We removed the poly(A) RNA from total RNA, polyadenylated the remaining RNA pool and performed 3′ RACE to retrieve the SL RNA cDNA from K. micrum (six clones; GenBank accession nos. EF143085–EF143090) and P. piscicida (15 clones; EF143091–EF143105). Several nucleotide substitutions were detected in the SL RNA intron in both species (Fig. 3A). Intron length in all K. micrum clones was identical (34 nt). However, in P. piscicida clones it ranged from 20 to 42 nt (Fig. 3A). The shorter clones did not correspond to truncated products as judged by Northern blot analysis (Fig. 4B), but may represent minor subpopulations, sample degradation, or amplification artifacts. The introns of dinoflagellate SL RNAs were slightly longer than that in Ciona [30 nt (16)] but shorter than those in other organisms (typically 50–100 nt). The total length of SL RNA was 56 nt in K. micrum, and 59 and 64 nt in P. piscicida, consistent with the size estimates from the Northern blot analyses. Our attempt to isolate SL RNA cDNA from Pr. minimum was unsuccessful. The intron length of the Pr. minimum SL RNA was predicted to be 32-nt within an SL RNA of 54 nt. From a poly(A)-selected library of P. piscicida we also obtained SL RNA clones, which shared the same sequences as the poly(A)-depleted RNA library (data not shown), suggesting that in dinoflagellates some of the SL RNA may be polyadenylated.
Unusual SL RNA Structures and Sm Binding Site Locale.
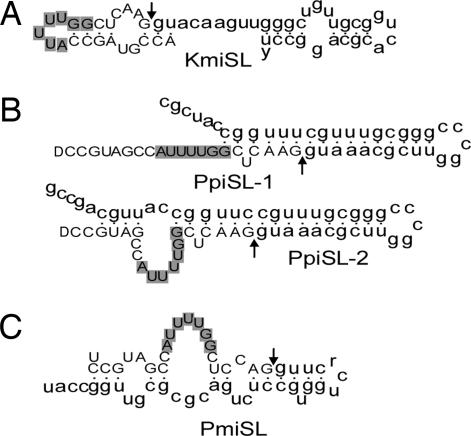
With few exceptions (16, 33), the known SL RNAs share conserved structures. The exon and the beginning of the intron form a stem-loop that contains the splice donor dinucleotide GU, followed by two additional stem-loops that flank a single-stranded region containing a binding site for the Sm-protein complex (34). The Sm binding site sequence is conserved in different organisms, with RAU4–6GR in the kinetoplastids, freshwater planarians and Caenorhabditis, RAUUUUCGG in Hydra, AGCUUUGG in Ciona, AGCUUUUCUUUGG in Schistosoma, and AAYUYUGA in Rotifera (12, 15, 16, 33, 35–38). Surprisingly, in dinoflagellate SL RNA, no Sm binding site was found in the intron. Instead, a common Sm variant, AUUUUGG, was detected within the exon. We speculate that this sequence acts as the Sm-complex binding site.
Modeling analysis yielded a single structural prediction of SLRNA for K. micrum, P. piscicida, and Pr. minimum (Fig. 5). Under the constraint that the splice-donor dinucleotide [GU in GGUA (U)C (a)] is double-stranded and the putative Sm binding site (AUUUUGG) single-stranded, all predicted SL structures were thermodynamically stable except for K. micrum. Without the constraint (default setting of the modeling program), a stable structure was generated for K. micrum. As a result, 2 nt of the Sm binding site were located in the double-stranded region (Fig. 5A). The same models were produced when folds were computed at 10°C and 30°C, temperatures at which these three algae could be found in the natural marine environment. Similar to the predicted structure of SL RNA in Ciona and Schistosoma (16, 33), the dinoflagellate SL RNAs have one (for P. piscicida and Pr. minimum) or two (for K. micrum) stem-loops as a result of their short sequence. The predicated structures differ from one dinoflagellate species to the other, whereas two P. piscicida SL genes exhibited similar structures.
Fig. 5.
Predicted secondary structures of SL RNAs from dinoflagellates. Shown are K. minimum (A), P. piscicida (B), and Pr. minimum (C). Exons are shown in uppercase, and introns are shown in lowercase; arrows indicate the exon–intron boundaries. Potential Sm binding sites are shaded.
Conservation of Sm Complex Subunits.
The Sm proteins form a heptameric ring around the Sm binding site and facilitate assembly of the core small nuclear ribonucleoprotein responsible for RNA splicing (39). Five major types of snRNAs (U1, U2, U4, U5, and U6) involved in cis-splicing have been identified in eukaryotes. All but one (U1) of these snRNAs are believed to function in trans-splicing (11, 13). Sm-D1 is necessary for SL RNA biogenesis in T. brucei (35, 40). Several cDNAs for Sm binding proteins have been detected in dinoflagellate cDNA libraries. In P. piscicida, three cDNA clones were obtained, one most similar to Sm-G (GenBank accession no. DQ864835) and two most similar to Sm-D2 (GenBank accession nos. DQ864832–DQ864833). Analysis of a K. micrum cDNA library yielded two different sequences (DQ867062 and EF134109) highly similar to Sm-D1 (e value = 4e −29). The presence of these subunits indicates that the Sm binding site interactions are conserved in dinoflagellates.
Discussion
Cis-splicing is common in eukaryotes. It transforms primary transcripts into mature mRNAs by removing intervening region (introns) and joining protein-coding regions (exons) (11) using ribosomes or spliceosomal complexes. The latter consists of snRNAs (U1, U2, U4, U5, and U6) and Sm proteins (11). Two forms of trans-splicing exist in eukaryotes. One form joins sequences from two separate protein coding transcripts. The other form (SL trans-splicing) splices a short noncoding leader RNA to the 5′ end of an independently transcribed pre-mRNA. Trans-splicing is believed to share machinery with cis-splicing because both involve the Sm complex (11, 13). In kinetoplastids, cis-splicing occurs in few genes and SL trans-splicing affects most genes (41). Similarly, introns have been detected in a small fraction of dinoflagellate genes (7, 8), and trans-splicing occurs in many if not all of the dinoflagellate nuclear protein coding genes as demonstrated in this study.
This is the first systematic analysis of SL trans-splicing within one phylum. The detection of SL trans-splicing in the major orders of dinoflagellates, even in early branches like O. marina and Noctiluca scintillans, suggest that SL trans-splicing was present early in dinoflagellate evolution and remains common in extant dinoflagellates. SL trans-splicing has not been detected for ciliates and apicomplexa. However, our preliminary analysis of Perkinsus marinus (ATCC 50439) and Perkinsus chesapeaki (ATCC PRA-65) cDNAs, using the 22-nt SL sequence, revealed the usage of SL trans-splicing in these organisms (data not shown). Because Perkinsus spp. are phylogenetically intermediate between Apicomplexa and dinoflagellates (42), this finding indicates that the SL trans-splicing machinery emerged in early ancestors of dinoflagellates.
The unique sequence of SL and its ubiquity in mRNAs offers a convenient tool for isolation of 5′ end cDNAs from dinoflagellates. The SL sequence is particularly useful when only a single conserved region is known for the target genes. The 22-nt SL combined with RACE techniques has led to the isolation of 682 full-length transcripts from various dinoflagellate species (SI Tables 1 and 2), the largest set of full-length cDNA data reported for dinoflagellates. Transcriptome studies of heterotrophic species that account for 50% of the phylum will benefit from the use of the SL marker, because the separation of algal prey mRNA from dinoflagellate mRNA is otherwise impossible. Using SL, we have isolated ≈200 full-length P. piscicida transcripts without any prey-derived contaminants. Transcriptome profiling for natural dinoflagellate assemblages will thus be possible despite the coexistence of other organisms. SL is also a marker for determining whether organelle-targeted gene products are encoded in the nuclear genome. Most of the plastid genes in peridinin-containing dinoflagellates have been transferred from plastids to the nucleus (2). Currently, nuclear-encoded plastid-targeted genes are identified by a poly(A) tail at the 3′ end of their mRNA along with N-terminal transit and signal peptides on the proteins (e.g., ref. 17). Use of the 22-nt SL in cDNA construction simplifies the process of isolating full-length or 5′ end mRNAs, and its exclusive presence on nuclear-encoded transcripts serves as a marker.
SL trans-splicing may confer mechanisms to modulate gene expression in dinoflagellates, in which transcriptional regulation is known only for a small fraction of genes studied so far (3–6). SL trans-splicing enables translation in kinetoplastids (43, 44) and nematodes (13). In addition, SL trans-splicing may serve as a strategy to generate mature monocistronic mRNAs from polycistronic pre-mRNA transcripts (11). There is increasing evidence for polycistronic or otherwise tandem-repeated gene transcripts in dinoflagellates (e.g., refs. 7–10, 26, 27). For example, pcna transcripts in P. piscicida contain at least two coding units in a tandem array (9), suggesting that the pre-mRNA transcript is processed, likely through trans-splicing, to monocistronic mRNA (Fig. 3B).
Comparison of genomic DNA and cDNA from P. piscicida pcna gives hints as to how the splice sites are recognized. Different pcna genomic clones have variable 5′ noncoding regions. However, at the junction between SL sequence and the 5′ UTR of pcna, the genomic DNA sequence bears a dinucleotide AG consistent with the common canonical cis-splicing acceptor boundary, apparently serving as the acceptor site of SL. The conserved sequence (CTCGTGC) immediately upstream of the boundary in different forms of pcna genomic clones may also be important for recognizing the splice site (Figs. 1B and 3B). Furthermore, this conserved sequence with the AG acceptor site is present in the intergenic region, evidence that SL trans-splicing generates monocistronic mRNA from polycistronic transcripts. Interestingly, no apparent splice site can be identified in the intergenic region of the form II Rubisco in Pr. minimum, consistent with the fact that the tandem-repeated gene copies are cotranscribed and cotranslated (10). This mode of gene expression apparently allows more effective transport of Rubisco to the chloroplast. In kinetoplastids, the conserved “AG” genomic acceptor site is preceded by a dinucleotide pattern that includes a TC-rich region just before the AG (41). As more genomic sequences of dinoflagellate genes become available, prediction tools for genomic trans-splicing acceptor sites can be built, as they have for trypanosomes (44) and Leishmania (41), enabling further computational and experimental investigation into the way in which common polycistronic transcripts are converted to monocistronic mRNA by SL trans-splicing in dinoflagellates.
The use of SL allows for a multitude of variations on the standard eukaryotic mRNA rubric. The ubiquitous presence of the SL on mRNAs in kinetoplastids has allowed some unusual variants in gene expression, such as the use of RNA polymerase I to transcribe genes coding for variant surface glycoproteins (45). Because SL donates the 5′ cap structure required for recognition by the translation machinery, these noncanonical mRNA transcripts can be translated despite their origin. A similar mechanism may be at play in dinoflagellates. As the major purveyors of cap structure, SLs in most of the studied organisms are initiated with a purine. Thus, the predominant use of U or A in the dinoflagellate SL may signal the presence of additional novel enzymes, specifically those catalyzing the efficient capping of pyrimidines. Dinoflagellate SL may convey 7-methylguanosine caps with additional 5′ methylations, as in kinetoplastids (11), or a trimethylguanosine cap as in worms (13).
The structural organization of dinoflagellate SL RNAs is unique and poses a molecular dilemma. The only Sm binding-site consensus found in dinoflagellate SL RNA is located in the exon, whereas in all other trans-splicing organisms this element occurs in the intron and falls between two hairpin loops to facilitate targeting of the Sm proteins, with the exception of Ciona and Schistosoma where the intronic site has just one stem loop upstream (16, 33). The importance of the Sm binding site on small RNAs has been demonstrated by mutagenesis in L. tarentolae (46) and Leptomonas collosoma (47). The 2–7 nt mutations in the Sm binding site affected SL RNA cap 4 methylation, transcription termination and 3′ end processing, thereby abolishing trans-splicing. Because components of the heptametric Sm ring (39) are present in dinoflagellates, the splicing machinery is likely to assemble through a pathway similar to other organisms. Studies have provided evidence of a spliceosome-based RNA processing system in dinoflagellates (48). Whether the Sm proteins assemble on SL to form a splicing-competent ribonucleoprotein particle that carries out the trans-splicing and whether they stay bound to the trans-spliced mRNA remains to be determined. Because Sm proteins bound to snRNAs comprise part of the signal for nuclear import, the presence of an Sm complex on the mRNA, whose final destination is presumably the cytosol, could lead to a localization tug-of-war. The Sm-complex might be displaced by the initiation of translation, or alternatively could impede ribosomal scanning. It is also possible that the dinoflagellate SL is too short to allow effective binding of Sm proteins to the exon, thereby eliminating the danger of a misplaced nuclear targeting signal.
Materials and Methods
Database Searches and Sequence Comparison.
The common 22-nt sequence was observed in the 5′ UTR for pcna and mapk from P. piscicida (6, 9) and actin from Pr. minimum (AF512889). This sequence was used to query GenBank databases by using BLAST and compared with newly obtained full-length cDNA sequences.
Selection of Species and DNA/cDNA Preparation.
Fifteen species representing all major taxonomic orders of dinoflagellates were selected to examine the distribution of the 22-bp SL sequence: Akashiwo sp., Am. carterae, and K. micrum in the order of Gymnodiniales; A. affine, A. fundyense, and A. tamarense in Gonyaulacales; H. triquetra, Kat. rotundatum, Peridinium foliaceum, and P. piscicida in Peridiniales; Pr. micans and Pr. minimum in Prorocentrales; S. goreaui in Suessiales; N. scintillans in Noctilucales; and O. marina in Oxyrrhinaceae (for strain information, see SI Table 1). Total RNA was isolated, and first-strand cDNA was synthesized by using GeneRacer oligo(dT) primer (Invitrogen) (9) or random hexamers (for testing on 18S rRNA and the chloroplast-encoded psbA). Full-length first-strand cDNA libraries for K. micrum, P. piscicida, and Pr. minimum were synthesized by using a GeneRacer kit (6). Genomic DNA from K. micrum, P. piscicida, and Pr. minimum were isolated according to ref. 19.
PCR Amplification and Sequencing of Dinoflagellate pcna and Other Polyadenylated mRNAs with 5′ End SL Addition.
To isolate pcna, first-strand, poly(dT)-based cDNAs of Akashiwo sp., A. affine, A. fundyense, K. micrum, Kat. rotundatum, P. foliaceum, Pr. minimum, P. micans, and S. goreaui were subjected to PCR amplification by using ExTaq (Takara Mirus Bio, Madison, WI) with the dinoflagellate SL sequence (DinoSL: 5′-TCCGTAGCCATTTTGGCTCAAG-3′) and DinoPCNA3c and DinoPCNA3d (9) as the primers. To study taxonomic and functional distribution of SL-containing mRNA, first-strand cDNAs of A. fundyense, Am. carterae, K. micrum, N. scintillans, P. piscicida, Pr. minimum, and O. marina were used as the templates for PCR amplification with DinoSL and GR3 (Invitrogen) under a touch-down PCR program as follows: 95°C for 20 s, 72°C for 2.5 min for 5 cycles; 95°C for 20 s, 65°C for 30 s, 72°C for 2 min for 5 cycles; 95°C for 20 s, 60°C for 30 s, 72°C for 2 min for 5 cycles; and 95°C for 20 s, 58°C for 30 s, 72°C for 2 min for 15 cycles. PCR products, obtained here and in following sections, were cloned and sequenced by following ref. 9.
Examination of Organelle-Encoded mRNA and Nuclear-Encoded rRNA.
Reverse primers were designed for dinoflagellate cox1 (5′-TAGAAAAATTATTDACTCTAGGATATACMACTTC-3′ and 5′-AATCCTCCAAABAAKCCDGGCATTACTA-3′), cob (5′-ATTGGCATAGGAAATACCATTCAGG-3′ and 5′-CTTCTAAKGCATTATCTGGATGTGA-3′), the chloroplast-encoded rbcL (5′-ATGGAACTCTAGCAACCCTATAAGC-3′ and 5′-GCTGCTGTAAGTAAATCTGTCCATA-3′ for K. micrum) and psbA (5′-TGACCGATAGGATAAACAATGAACAC-3′ and 5′-AACGACAGCACCAGAGATGATG-3′ for Am. carterae; 5′-TACCACCGTTGTATAACCATTCATC-3′ and 5′-AGCACCTGAGATGATGTTGTTAC-3′ for H. triquetra). For 18S rRNA, Dino18S R2 and Dino18S R1 (49) were used. Two rounds of touch-down PCR were performed under the same cycles as described above with 30S extension time. First-strand cDNAs of P. piscicida (for cob, cox1, and 18S rRNA), K. micrum (for rbcL and 18S rRNA), Pr. minimum (for 18S rRNA), and Am. carterae and H. triquetra (for psbA), synthesized by using oligo(dT) for cob and cox1, and the remaining random hexamers for were used as the template with DinoSL primer paired with the first of the two gene-specific reverse primers. The cDNA-PCR products were diluted 100-fold and used as the template for the second round of PCR with DinoSL paired with the second (nested) reverse gene-specific primer.
Examination of Nuclear-Encoded Chloroplast-Targeted mRNA.
Reverse primers for nuclear-encoded chloroplast targeted genes included the form II Rubisco in Pr. minimum (5′-GGTCAGCACGGAACACATCAT-3′ and 5′-TTGTGAAGTCGTCGGTGGTG-3′) and ATP synthase subunit C (atpH) for A. tamarense, A. affine, and H. triquetra (5′-GCTCACTTGATSAGNGGGTTNGC-3′ and 5′-AGGGACTCCATGAAGGCAAGC-3′). Two rounds of touch-down PCR were performed.
Amplification of SL RNA Gene from K. micrum, P. piscicida, and Pr. minimum.
The SL RNA gene encompassing at least two copies in tandem was amplified by using primers derived from SL modified to reduce the likelihood of self-priming: 5′-cgagagtatcAGCCATTTTGGCTCAAG-3′ (forward) and 5′-acagaacaAGCCAAAATGGCTACGG-3′ (reverse), where the uppercase letters indicate SL and the lowercase letters indicate random nucleotides. DNA from 104 to 105 cells of K. micrum, P. piscicida, and Pr. minimum was used in PCR as described above, with a 1-min extension time.
Southern and Northern Blots for K. micrum, P. piscicida, and Pr. minimum SL RNA.
Genomic DNA from K. micrum, P. piscicida, and Pr. minimum were digested with EcoRI, HindIII, and Sau3AI, electrophoresed, and transferred to Hybond+ membranes (GE Healthcare, Little Chalfont, U.K.). Cloned SL RNA gene sequences from K. micrum (GenBank accession no. EF143078, two units), P. piscicida (EF143080, one unit) and Pr. minimum (EF143084, seven units) were labeled by using AlkPhos Direct Labeling and Detection Systems (GE Healthcare) and used as probes for hybridization. Membranes were hybridized, washed, and exposed to x-ray film (Fuji, Tokyo, Japan) for 2 or 20 h.
Total RNA from 106 cells of K. micrum, P. piscicida, and Pr. minimum were electrophoresed in an 8% acrylamide/8 M urea gel and transferred to nylon membranes (28). This gel, with a medium resolution, was optimal for RNAs smaller than 350 nt and usually did not indicate the SL-containing mRNA bands for kinetoplastids (28). A dinoflagellate SL RNA oligonucleotide (DinoSLa/s; 5′-TGTACCTTGAGCCAAAATG-3′) was labeled with 32P for hybridization (28). Total RNA from L. tarentolae cells was included as size marker. The cytochrome oxidase subunit III transcript was detected with the oligonucleotide S-255 (GAATAGTGTTTTCATCTCTC) (30).
Rapid Amplification of cDNA 3′ End of K. micrum, P. piscicida, and Pr. minimum SL RNA.
Poly(A) mRNA was depleted from total RNA by using PolyATtract mRNA Isolation System IV (Promega, Madison, WI). A poly(A) tail was added to the non-poly(A) RNA by using Escherichia coli Poly(A) Polymerase (Takara Mirus Bio). The resulting RNA (total RNA as well in the case of P. piscicida) was used to synthesize the first-strand cDNA with GeneRacer oligo(dT) primer (Invitrogen). Two rounds of touch-down PCR were carried out as described above. The first round of PCR was performed by using DinoSL and GR3 for all three species; the second round of PCR was conducted by using a 100-fold dilution of the first-round PCR products as a template and GR3 paired with the following nested primers: 5′-GCTCAAGGTACAAGTTGGGCTG-3′ for K. micrum; 5′-CTCAAGGTAAACGCTTGGCCC-3′ for P. piscicida; and 5′-TGGCTCAAGGTTCACTGGTG-3′ for Pr. minimum.
Modeling of RNA Structure.
Secondary structures of K. micrum, P. piscicida, and Pr. minimum SL RNAs were modeled by using MFOLD Version 3.1.2: Prediction of RNA secondary structure modeling program (http://bioweb.pasteur.fr/seqanal/interfaces/mfold-simple.html). Folding was performed at 20°C, the culture temperature of the three species, with the constraint that the splice-donor dinucleotide [GU in GGUA (U)C (a)] remained double-stranded and that the putative Sm binding site (AUUUUGG) remained single-stranded. These are conserved structural features predicted in previous SL RNA models (34). For K. micrum, because the constraints led to an unstable structure, default setting was used in the modeling. Folding was also performed at 10°C and 30°C to examine effects of temperature.
Supplementary Material
Acknowledgments
We thank D. M. Anderson (Woods Hole Oceanographic Institution) for A. fundyense and A. tamarense, E. J. Buskey (University of Texas) for N. scintillans, and C. F. Dungan (Maryland Department of Natural Resources) for Perkinsus. This work was supported by National Science Foundation Grants DEB-0344186, OCE0452780, and EF-0626678; National Oceanic and Atmospheric Administration Ecology and Oceanography of Harmful Algal Blooms Grant NA05NOS4781196 (to S.L. and H.Z.); and National Institutes of Health Grant AI056034 (to D.A.C. and N.R.S.).
Abbreviations
- PCNA
proliferating cell nuclear antigen
- SL
spliced leader
- Sm
spliceosomal
- snRNA
small nuclear RNA.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AF512889, DQ864761–DQ864971, DQ867053–DQ867070, DQ884413–DQ884451, EF133854–EF133905, EF133961–EF134003, EF134083–EF134402, EF141835, and EF143070–EF143105).
This article contains supporting information online at www.pnas.org/cgi/content/full/0700258104/DC1.
References
- 1.Cavalier-Smith T. Biol Rev. 1998;73:203–266. doi: 10.1017/s0006323198005167. [DOI] [PubMed] [Google Scholar]
- 2.Hackett JD, Yoon HS, Soares MB, Bonaldo MF, Casavant TL, Scheetz TE, Nosenko T, Bhattacharya D. Curr Biol. 2004;14:213–218. doi: 10.1016/j.cub.2004.01.032. [DOI] [PubMed] [Google Scholar]
- 3.Taroncher-Oldenburg G, Anderson DM. Appl Environ Microbiol. 2000;66:2105–2112. doi: 10.1128/aem.66.5.2105-2112.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Okamoto OK, Robertson DL, Fagan TF, Hastings JW, Colepicolo P. J Biol Chem. 2001;276:19989–19993. doi: 10.1074/jbc.M101169200. [DOI] [PubMed] [Google Scholar]
- 5.Okamoto OK, Hastings JW. J Phycol. 2003;39:519–526. [Google Scholar]
- 6.Lin S, Zhang H. Appl Environ Microbiol. 2003;69:343–349. doi: 10.1128/AEM.69.1.343-349.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rowan R, Whitney SM, Fowler A, Yellowlees D. Plant Cell. 1996;8:539–553. doi: 10.1105/tpc.8.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Okamoto OK, Liu L, Robertson DL, Hastings JW. Biochemistry. 2001;40:15862–15868. doi: 10.1021/bi011651q. [DOI] [PubMed] [Google Scholar]
- 9.Zhang H, Hou Y, Lin S. J Eukaryot Microbiol. 2006;53:142–150. doi: 10.1111/j.1550-7408.2005.00085.x. [DOI] [PubMed] [Google Scholar]
- 10.Zhang H, Lin S. J Phycol. 2003;39:1160–1171. [Google Scholar]
- 11.Mayer MG, Floeter-Winter LM. Mem Inst Oswaldo Cruz. 2005;100:501–513. doi: 10.1590/s0074-02762005000500010. [DOI] [PubMed] [Google Scholar]
- 12.Pouchkina-Stantcheva NN, Tunnacliffe A. Mol Biol Evol. 2005;22:1482–1489. doi: 10.1093/molbev/msi139. [DOI] [PubMed] [Google Scholar]
- 13.The C. elegans Research Community, editor. Blumenthal T. WormBook. 2005 Jun 25; doi: 10.1895/wormbook. 1.5.1. www.wormbook.org. [DOI]
- 14.Hastings KEM. Trends Genet. 2005;21:240–247. doi: 10.1016/j.tig.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 15.Stover NA, Steele RE. Proc Natl Acad Sci USA. 2001;98:5693–5698. doi: 10.1073/pnas.101049998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vandenberghe AE, Meedel TH, Hastings KEM. Genes Dev. 2001;15:294–303. doi: 10.1101/gad.865401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Patron NJ, Waller RF, Archibald JM, Keeling PJ. J Mol Biol. 2005;348:1015–1024. doi: 10.1016/j.jmb.2005.03.030. [DOI] [PubMed] [Google Scholar]
- 18.Morse D, Salois P, Markovic P, Hastings JW. Science. 1995;268:1622–1624. doi: 10.1126/science.7777861. [DOI] [PubMed] [Google Scholar]
- 19.Lin S, Zhang H, Spencer DF, Norman JE, Gray MW. J Mol Biol. 2002;320:727–739. doi: 10.1016/s0022-2836(02)00468-0. [DOI] [PubMed] [Google Scholar]
- 20.Tengs T, Dahlberg OJ, Shalchian-Tabrizi K, Klaveness D, Rudi K, Delwiche CF, Jakobsen KS. Mol Biol Evol. 2000;17:718–729. doi: 10.1093/oxfordjournals.molbev.a026350. [DOI] [PubMed] [Google Scholar]
- 21.Dorfman DM, Lenardo MJ, Reddy LV, Van der Ploeg LH, Donelson JE. Nucleic Acids Res. 1985;13:3533–3549. doi: 10.1093/nar/13.10.3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lenardo MJ, Dorfman DM, Donelson JE. Mol Cell Biol. 1985;5:2487–2490. doi: 10.1128/mcb.5.9.2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Volloch V, Schweitzer B, Rits S. Nature. 1990;343:482–484. doi: 10.1038/343482a0. [DOI] [PubMed] [Google Scholar]
- 24.Bonen L. FASEB J. 1993;7:40–46. doi: 10.1096/fasebj.7.1.8422973. [DOI] [PubMed] [Google Scholar]
- 25.Murthy VK, Dibbern KM, Campbell DA. Mol Cell Probes. 1992;6:237–243. doi: 10.1016/0890-8508(92)90022-p. [DOI] [PubMed] [Google Scholar]
- 26.Le QH, Markovic P, Hastings JW, Jovine RVM, Morse D. Mol Gen Genet. 1997;255:595–604. doi: 10.1007/s004380050533. [DOI] [PubMed] [Google Scholar]
- 27.Li L, Hastings JW. Plant Mol Biol. 1998;36:275–284. doi: 10.1023/a:1005941421474. [DOI] [PubMed] [Google Scholar]
- 28.Sturm NR, Yu MC, Campbell DA. Mol Cell Biol. 1999;19:1595–1604. doi: 10.1128/mcb.19.2.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thomas S, Westenberger SJ, Campbell DA, Sturm NR. Gene. 2005;352:100–108. doi: 10.1016/j.gene.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 30.Sturm NR, Simpson L. Cell. 1990;61:871–878. doi: 10.1016/0092-8674(90)90197-m. [DOI] [PubMed] [Google Scholar]
- 31.Pelle R, Murphy NB. Mol Biochem Parasitol. 1993;59:277–286. doi: 10.1016/0166-6851(93)90225-m. [DOI] [PubMed] [Google Scholar]
- 32.Lamontagne J, Papadopoulou B. J Biol Chem. 1999;274:6602–6609. doi: 10.1074/jbc.274.10.6602. [DOI] [PubMed] [Google Scholar]
- 33.Rajkovic A, Davis R, Simonsen J, Rottman F. Proc Natl Acad Sci USA. 1990;87:8879–8883. doi: 10.1073/pnas.87.22.8879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bruzik JP, Doren KV, Hirsh D, Steitz JA. Nature. 1988;335:559–562. doi: 10.1038/335559a0. [DOI] [PubMed] [Google Scholar]
- 35.Zeiner GM, Foldynova S, Sturm NR, Lukes J, Campbell DA. Eukaryot Cell. 2004;3:241–244. doi: 10.1128/EC.3.1.241-244.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zayas RM, Bold TD, Newmark PA. Mol Biol Evol. 2005;22:2048–2054. doi: 10.1093/molbev/msi200. [DOI] [PubMed] [Google Scholar]
- 37.Dassanayake RS, Chandrasekharan NV, Karunanayake EH. Gene. 2001;269:185–193. doi: 10.1016/s0378-1119(01)00438-3. [DOI] [PubMed] [Google Scholar]
- 38.Davis RE. Parasitol Today. 1996;12:33–40. doi: 10.1016/0169-4758(96)80643-0. [DOI] [PubMed] [Google Scholar]
- 39.Stark H, Dube P, Luehrmann R, Kastner B. Nature. 2001;409:539–542. doi: 10.1038/35054102. [DOI] [PubMed] [Google Scholar]
- 40.Mandelboim M, Barth S, Biton M, Liang XH, Michaeli S. J Biol Chem. 2003;278:51469–51478. doi: 10.1074/jbc.M308997200. [DOI] [PubMed] [Google Scholar]
- 41.Gopal S, Awadalla S, Gasterland T, Cross GAM. Genome Biol. 2005;6:R95. doi: 10.1186/gb-2005-6-11-r95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leander BS, Keeling PJ. J Phycol. 2004;40:341–350. [Google Scholar]
- 43.Zeiner GM, Sturm NR, Campbell DA. J Biol Chem. 2003;278:38269–38275. doi: 10.1074/jbc.M304295200. [DOI] [PubMed] [Google Scholar]
- 44.Gopal S, Cross GAM, Gaasterland T. Nucleic Acids Res. 2003;31:5877–5885. doi: 10.1093/nar/gkg798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kooter JM, Borst P. Nucleic Acids Res. 1984;12:9457–9472. doi: 10.1093/nar/12.24.9457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sturm NR, Campbell DA. J Biol Chem. 1999;274:19361–19367. doi: 10.1074/jbc.274.27.19361. [DOI] [PubMed] [Google Scholar]
- 47.Mandelboim M, Estrano CL, Tschudi C, Ullu E, Michaeli S. J Biol Chem. 2002;277:35210–35218. doi: 10.1074/jbc.M201910200. [DOI] [PubMed] [Google Scholar]
- 48.Alverca E, Franca S, Diaz de la Espina SM. Bio Cell. 2006;98:709–720. doi: 10.1042/BC20050083. [DOI] [PubMed] [Google Scholar]
- 49.Lin S, Zhang H, Hou Y, Miranda L, Bhattacharya D. Appl Environ Microbiol. 2006;72:5626–5630. doi: 10.1128/AEM.00586-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.