Abstract
Cells responding to dramatic environmental changes or undergoing a developmental switch typically change the expression of numerous genes. In bacteria, σ factors regulate much of this process, whereas in eukaryotes, four RNA polymerases and a multiplicity of generalized transcription factors (GTFs) are required. Here, by using a systems approach, we provide experimental evidence (including protein-coimmunoprecipitation, ChIP-Chip, GTF perturbation and knockout, and measurement of transcriptional changes in these genetically perturbed strains) for how archaea likely accomplish similar large-scale transcriptional segregation and modulation of physiological functions. We are able to associate GTFs to nearly half of all putative promoters and show evidence for at least 7 of the possible 42 functional GTF pairs. This report represents a significant contribution toward closing the gap in our understanding of gene regulation by GTFs for all three domains of life and provides an example for how to use various experimental techniques to rapidly learn significant portions of a global gene regulatory network of organisms for which little has been previously known.
Keywords: archaea, regulatory networks, systems biology
Transcriptional regulation by general transcription factors (GTFs) is an important control point for large-scale regulation of physiology across all three domains of life. It is generally understood that in bacteria, σ factors help to modulate large changes in gene expression in response to environmental stimuli. Meanwhile, in eukaryotes, families of large multisubunit general transcription complexes initiate large-scale changes in transcription from various promoters. Relatively little is known, however, about how GTFs in archaea help confer fitness across a broad range of environments, including hostile ones, such as thermal vents and hypersaline ponds, to milder environments, such as the oceans and the human oral cavity and gut (1–4).
In archaea, two GTFs orthologous to eukaryotic transcription factor IIB (TFB) and TATA-binding proteins (TBPs) are necessary and sufficient for initiating basal transcription (5). These GTFs are present in multiple copies in several archaeal species [supporting information (SI) Table 2]. Halobacterium NRC-1 is particularly complex, because its genome encodes six TBP and seven TFB proteins (6). Because we know that a TBP must pair with a TFB to recruit RNA polymerase to the promoter to form an active preinitiation complex the presence of six TBPs and seven TFBs in Halobacterium NRC-1 theoretically encodes 42 possible TFB–TBP pairs, which may drive transcription from an equally diverse set of promoters in Halobacterium NRC-1
Here, we investigate the following: (i) the degree to which members of the two families interact with one another; (ii) whether the promoter specificities of the individual TFBs and TBPs have diverged significantly from one another, or whether they have retained considerable overlap in their genomic binding sites; (iii) whether a higher-order regulatory network architecture has emerged by interactions among these factors; and (iv) if so, what are the consequences of such a network on global transcriptional control? We address these questions through an integrated analysis of protein–DNA interactions (evidence for direct regulation of gene expression), protein–protein interactions (evidence for pairwise interactions), and environmental and genetic (nonnative expression and knockouts of GTFs) perturbation-induced global transcriptional changes to provide a rare view into global transcriptional regulation in archaea. Further, we present several testable hypotheses regarding specific mechanistic principles of gene regulation in archaea.
Results and Discussion
Preliminary Analysis of Existing Data.
Phylogenetic analysis.
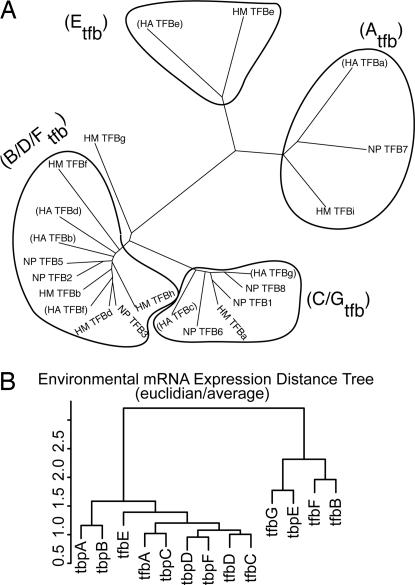
We show that the haloarchaeal TFB family can be roughly divided into four phylogenetic clades on the basis of their amino acid sequence similarities (Fig. 1A and SI Fig. 6). A similar analysis for TBP is presented in SI Fig. 7. These evolutionary groupings represent a line of experimental evidence regarding the putative relationships among GTFs in Halobacterium NRC-1 (6). The patterns seen in the evolutionary analysis suggest certain GTF interactions may have coevolved to occur preferentially over others.
Fig. 1.
Evolutionary and gene expression relationships among GTFs (A) Haloarchaeal TFB sequence analysis reveals four distinct groups. TFB sequences from three fully sequenced haloarchaeal genomes were initially aligned by using ClustalW (24) and subsequent dendograms generated with Phylip (25). Groups are named in parentheses according to the membership of Halobacterium NRC-1 TFBs. Sequences were accessed from the National Center for Biotechnology Information database and renamed according to organism as follows: HA, Halobacterium sp. NRC-1; HM, Haloarcula marismortui; and NP, Natronomonas pharaonis DSM 2160. There are four clear groups of TFBs, with the exception of HM-TFBg. (B) Dendogram of the expression profiles of GTFs across >300 microarray conditions summarized in SI Table 3.
GTF gene expression profiles across numerous conditions.
We investigated the broad-scale patterns in transcriptional responses of GTFs in Halobacterium NRC-1 from >300 microarrays reflecting a number of environmental perturbations (Fig. 1B, SI Tables 3–5, and SI Fig. 8) by calculating a dendogram of expression profile distances of GTFs relative to themselves. This revealed two large groupings of expression profiles. One contains only a single TBP (TBPe) and TFBs -b, -f, and -g. The second group contains all other GTFs. Finer divisions in expression profiles are also evident. We also note similarity of expression for TBPc and TFBa and the TFB/TBP pair TBPe and TFBg, suggesting that these specific pairs of GTFs may function together. Moreover, this analysis, in combination with the phylogenetic analysis in Fig. 1A, shows that the transcriptional regulation of several structurally related GTFs, those from similar phylogenetic groupings (specifically pairs TFBc and -g and TFBb, -g, and -d) have clearly diverged from one another.
Systematic Gene Knockouts Reveal Likely Essential GTFs.
A systematic effort to knock out each of the GTFs has revealed that two TFBs (-f and -g) and three TBPs (-b, -e, and -f) appear to be essential for growth under standard laboratory conditions. Although three TBPs (TBPa, -c, and -d) that are present in at least two copies in the genome were successfully deleted, TBPb presents a particular challenge, because it is present in four copies. Although this result implies that only 5 of the 13 GTFs in Halobacterium are absolutely necessary for life under standard laboratory conditions, it does not exclude the possibility that the other GTFs provide added fitness under various stressful or nonstandard laboratory growth conditions.
Genome-Wide Localization of TFB-Binding Sites.
ChIP followed by microarray chip analysis (ChIP-Chip) of C-terminally myc-tagged TFBs and TBPs was performed as described in Methods. An analysis of putative binding sites shows a significant bias toward binding regions that overlap intergenic sequence (Table 1). Because such a description of a system-wide architecture of an archaeal GTF network is previously uncharacteized, we used stringent criteria to select what most likely represent high occupancy and/or high binding-affinity sites.
Table 1.
Intergenic bias in ChIP-Chip experiments
| GTF | Number of tiles | Fraction intergenic | Random intergenic | P value |
|---|---|---|---|---|
| tbpB | 317 | 0.74 | 0.50 | 1.1E-20 |
| tfbA | 322 | 0.57 | 0.50 | 1.5E-3 |
| tfbB | 279 | 0.85 | 0.50 | 1.0E-37 |
| tfbC | 228 | 0.82 | 0.50 | 3.1E-25 |
| tfbD | 271 | 0.87 | 0.50 | 1.1E-40 |
| tfbE | 253 | 0.43 | 0.50 | 0.986 |
| tfbF | 324 | 0.86 | 0.50 | 4.4E-46 |
| tfbG | 396 | 0.79 | 0.50 | 8.5E-37 |
Using this approach, we mapped binding sites for all seven TFBs and TBPb to 1,048 promoters in the Halobacterium NRC-1 genome (SI Dataset 1). When this set is expanded by putatively cotranscribed operon structures, it represents 1,266 genes (heretofore operon-expanded; SI Dataset 2). This represents 51% of the 2,445 unique genes in the Halobacterium NRC-1 genome and is well correlated with the maximum number of genes we have observed to be differentially expressed during some single environmental perturbation experiments.
Although this approach resulted in a low false-positive discovery rate, it does not exclude the possibility of binding sites with lower affinity. In addition, because of the dynamic condition-dependent nature of biological networks, the complete set of protein–DNA interactions elucidated from this ChIP-Chip represents a portion of all possible interactions. Despite these stipulations, this data set is comprised of real high-affinity/-occupancy physical interactions that can occur in vivo and can be interrogated for functional insight.
Distribution of TFB-Binding Events.
Unique associations.
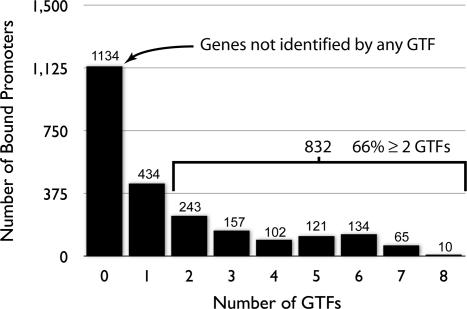
There are two components of the binding distribution that are of functional interest. The first are the genomic locations (promoters) that are bound by only a single GTF in our experiment (Fig. 2). The second are the remaining genomic regions bound by multiple GTFs (Fig. 2). In this section, we discuss the 37% of all identified promoters that were bound by only a single GTF. Although we do not explicitly exclude the possibility that other GTFs could bind to these regions under different conditions, our data strongly suggest that some unique promoter–GTF combinations have indeed evolved. Notwithstanding yet-uncharacterized influences on gene expression, this GTF promoter selectivity strongly implies that functional segregation of transcriptional regulation can be mediated by GTFs in archaea.
Fig. 2.
Distribution of ChIP-Chip interactions across the genome. This histogram summarizes the number of putative genes that have been associated with none, one, or multiple TFBs or TBPb by the ChIP-Chip method described elsewhere. The number of promoters in each category is written above each bar. Nearly 66% of all identified promoters are associated with more than one GTF. This suggests there is likely some mechanistic redundancy in the gene regulatory network specified by these factors and from the standpoint of high-level regulatory processes that many cellular processes have a high degree of functional overlap.
To extend this idea, we filtered gene sets selected by a single GTF by expression correlation to that particular GTF (SI Fig. 9). This allows us to reduce our list of candidate genes to those that have the greatest probability of being directly regulated by a given GTF (SI Table 6). This also yields an assessment of functional association and segregation of gene expression by GTFs. One interesting conclusion from these filtered data suggests that TFBf binds in a functionally relevant manner to promoters of genes encoding proteins involved in ribosome biogenesis. Meanwhile, TFBg likely uniquely regulates genes encoding phosphate transport and phototrophy proteins. These two observations are indirectly supported by our inability to knock out TFBf and -g, because deleting either would seriously impact the regulation of functions that are key for survival. A similar analysis is provided for all TFBs and TBPb in SI Table 6. We also note that segments of the GTF regulatory network that are poorly represented in Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) databases or other functional association databases will not be amenable to this sort of analysis but are nevertheless important components of cellular physiology, for which we now present recently discovered associations.
Shared associations.
More than half (832, operon-expanded) of the target genes in the protein–DNA interaction map are associated with more than one GTF. This suggests that at least 34% of all genes in Halobacterium NRC-1 may be regulated by more than one TFB. This high degree of multiple protein–DNA associations mechanistically offers at least two distinct physiological benefits for the organism. First, genes putatively under the control of more than one GTF may be less susceptible to changes in the relative concentrations of GTFs within the cytosol that, as observed in our microarray data (7–9), are expected to occur as a consequence of environmental perturbations. This scheme can allow for the uninterrupted transcription of genes whose products are required for distinct but overlapping physiological states in a variety of environmental conditions. Alternatively, competitive binding of multiple GTFs with different binding affinities for the same promoter (a process that may be modulated by other factors) may alter promoter activity, which can further contribute to the regulation of physiology. Although further evidence for this idea is presented later, we note that examples of such processes are well characterized in bacteria by using alternate sigma factors (10–12). Each of these proposed functions adds a measure of complexity to the possible modes of gene regulation. Second, multiple GTF associations contribute resilience to core biological processes in the face of random damaging events such as loss of GTF through mutation. This hypothesis is supported by the observation that many of the GTFs could be singly removed without compromising viability.
Our inability to localize binding sites for TBPs aside from TBPb is curious but may reflect a consequence of epitope-tagging TBPs, functional attenuation by dimerization, condition-specific TBPb recruitment, or the requirement of other factors. Nevertheless, the significant influence of TFBs on transcriptional segregation (particularly their abundant segregation along global gene expression profile groups) suggests that, although useful, knowledge of TBP-binding site distributions would simply serve to refine the current regulatory divisions delineated by the existing TFB network presented herein.
Protein–Protein Interactions Among TFBs and TBPs.
The presence of multiple GTFs (seven TFBs and six TBPs) in Halobacterium NRC-1 allows 42 different potential combinations of TFB–TBP pairs to drive transcriptional initiation from presumably as many promoter subsets across the genome (6). To assess whether the organism functionally explores this theoretically possible combinatorial repertoire and if so, in what fashion, we analyzed the protein composition of transcription complexes enriched by immunoprecipitation with antibodies targeting each of the individual proteinA-tagged GTFs expressed in Halobacterium NRC-1. Using the method described in Methods, control immunoprecipitations did not identify any GTF peptides, thus all statistically significant identifications of either TFBs or TBPs in noncontrol experiments were interpreted as evidence of association with the epitope-tagged GTF. Fig. 3 and SI Dataset 3 show the resulting interaction network among families of TFBs and TBPs.
Fig. 3.
GTF protein–protein and protein–DNA self interactions. The GTF protein–protein association network reveals inter- and intraspecies interactions. Protein–protein associations were determined by coimmunopreciptation experiments of protein A-tagged basal transcription factors followed by tandem mass spectrometry. All experiments were conducted under 4.5 M salinity to preserve native interactions. The source node from which a directed edge leaves is the bait protein, whereas the target node is that which was immunopreciptated with the bait. All edges were selected with a peptide probability (19) of ≥0.9 and a minimum of two unique identifying peptides (20).
Seven TFB–TBP pairs were detected in the GTF protein–protein interaction map, and at least five of the seven TFBs (TFBb, -c, -d, -e, and -g) interact with TBPe. In addition, there are two unique TFB–TBP interactions; TFBc with TBPd and TFBa with TBPc. High similarity between mRNA level changes for TFBa and TBPc across all assayed environmental conditions reinforce the selective and likely cofunctional relationship between these two factors (Fig. 1B). These two unique TFB–TBP interactions do not preclude alternative pairings that may form under physiological conditions different from those assayed in this experiment. They illustrate, however, that some physiologically important TFB–TBP pairings are favored under selected conditions and thus, that the evolutionary expansion of transcription regulation through selective pairings among TBPs and TFBs has indeed occurred in archaea.
Meanwhile, the observation that TBPe interacts with numerous TFBs recalls the results of a proteomic survey of Halobacterium NRC-1 in which TBPe was the only detected TBP (13). Taken together, protein abundance of TBPe and its numerous interactions with TFB, although perhaps linked, suggest a dominant role among TBPs for TBPe. This view may also be related to the observation that most archaea have a single TBP and multiple TFBs and highlights that, whereas the set of Halobacterium NRC-1 GTF is highly expanded with respect to other sequenced archaea, the general principles learned in this study are likely applicable to other archaea, both simpler and the not-yet-discovered more sophisticated. In such a system, the partitioning of basal transcription must be largely accomplished by the TFBs. Intriguingly, Halobacterium NRC-1 has, nevertheless, taken advantage of the opportunities for the unique combinatorial pairings of TFBs and TBPs leading to a mechanism for increasing the partitioning of transcription by GTFs.
Integration of ChIP Data with TFB Perturbation Transcriptome Profiles.
To further refine our understanding of GTF behavior in Halobacterium, we applied a complementary approach to investigate the influence of directed perturbation of individual GTFs on the response of the global transcriptome to environmental changes. In this experiment, all TFBs and TBPs were perturbed by expression from a nonnative promoter and, if possible, by gene knockout. Transcriptomes from each of these perturbed strains were collected in duplicate during three different growth phases (late lag, exponential, and early stationary). One clear example of the power of combining transcriptome profiles of genetically perturbed strains with physical interaction data is shown in Fig. 4.
Fig. 4.
Integrating multiple data sources to reveal nested regulatory relationships between TFB. (A) Transcriptome data for a subset of TFB perturbations. Transcriptome data, displayed as a heat map, for a plasmid-only control strain (cmyc-only), a strain expressing TFBa-cmyc from a nonnative promoter (TFBa-cmyc), a TFBa knockout strain (TFBa-KO), and similarly constructed strains TFBc-cmyc, TFBc-KO, TFBe-cmyc, and TFBe-KO. The gene number corresponding to each profile is indicated on the far right. A legend displaying colors corresponding to log10ratios is located below the heat map. These data indicate functional influences of the GTFs on the expression of the selected genes and allows for grouping of genes based on GTF influence. (B) An influence network derived from ChIP-Chip data and inference from transcriptome data. Red solid arrows indicate direct interactions from ChIP-Chip experiments, and blue dashed arrows indicate putative regulatory interactions inferred from transcriptome data. Genes with the red color label appear to have some additional regulatory input from other regulators. The simplest testable hypothesis that can account for most of these data is that TFBe represses all genes presented by direct competition to the promoter with TFBa (the likely activator). The group of genes in the 6400 series is likely indirectly regulated by TFBa through direct regulation of TFBc expression, which subsequently up-regulates this small cluster. This analysis shows how we can simultaneously analyze transcriptome data (indirect evidence of regulation) with ChIP-Chip data (direct measure of regulatory influence) to formulate very specific hypotheses for gene regulation by GTFs in Halobacterium NRC-1.
These data provide evidence for multiple TFBs influencing the expression of groups of genes on the replicon pNRC200 and how these possible influences may be logically nested with respect to one another (Fig. 4). We look for differences in gene expression with respect to the cmyc-only control for either GTF knockout or nonnative expression perturbations. For instance, less expression in a knockout strain implies that the perturbed GTF either directly or indirectly regulates expression of these genes. Additional perturbation experiments and ChIP-Chip data (evidence for direct influence) can be combined to help resolve direct vs. indirect regulatory influences or provide clear testable hypotheses.
The data in Fig. 4 reveal that TFBs likely compete for common promoters, and that this competition can lead to negative regulation of groups of genes. This picture is consistent with the general observation from the entire set of ChIP-Chip data that suggests that many promoters can be bound by multiple TFBs. Moreover, this integrated analysis has suggested that regulatory interactions among TFBs (TFBs regulating the expression of other TFBs) are also a likely important component of global gene regulation. Associated hypotheses raised from these data concern the influence of other “missing” regulators and how quantitative differences in promoter-binding affinity can lead to differential gene expression. Although this experimental data set does not fully answer these final questions, it does provide a firm foundation on which we are building future investigations to refine our understanding of the architecture and functional role of this regulatory circuit.
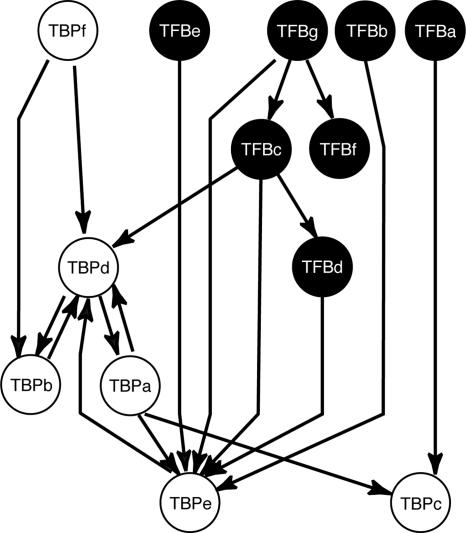
Architecture of the Intra-TFB Gene Regulatory Circuit Mirrors Transcriptome Profile Patterns.
Integrating ChIP-Chip, coimmunoprecipitation (including protein–protein interactions among TFBs), and GTF perturbation transcriptome analysis, we construct a hypothesis for a high-level component of the gene regulatory network. Fig. 5 shows some of the likely regulatory relationships among the TFBs themselves. In principle, this regulatory structure should influence large-scale patterns of gene expression, admitting the likely participation of other regulators. We broadly examine the general trends in organization between the TFBs in ChIP-Chip [i.e., interaction of a TFB and a tfb promoter (TFB-Ptfb)] and their expression relative to other genes across relatively large numbers of transcriptome conditions. For instance, the similar promoter structures for TFBb and -g based on ChIP-Chip data are consistent with their expression-based segregation (Figs. 1B and 5). In addition, TFBa, -c, -d, and -e appear to organize into a second large functional group of GTFs based on the dearth of regulatory inputs into their promoters. Again, this is reflective of their own segregation from TFBb, -f, and -g in their patterns of gene expression (Fig. 1B). We should also note that the multiple binding events indicated for TFBb and -g do not likely occur simultaneously. Rather, they represent an integrated picture drawn from multiple experiments and likely reflect the idea that multiple TFBs can independently associate with these promoters, adding to the robustness of these nodes in the regulatory network.
Fig. 5.
The structure of the TFB self-association network. If we take into account the behavior of TFB gene expression (Fig. 1B) with the ChIP-Chip data, we can infer the TFB self-association network as shown here in which lines represent interactions between TFBs and the promoters of other TFBs. Colored circles represent TFB protein products, and the thick gray lines connecting them represent putatively repressing protein–protein interactions derived from Fig. 3. This protein–protein regulation represents a second layer of regulation by GTFs among themselves and may help provide both regulation and stability to the regulatory module. The structure of this network reveals a highly connected central group, TFBb and -g. The remaining TFBs are much less connected and appear to influence the expression of TFBb and/or -g directly or indirectly. One edge, inferred from perturbation transcriptome data (Fig. 4) between TFBa and -c (dashed line) coupled with the edge between TFBe and –c, highlights the idea that TFBc may act as a separate layer in this regulatory network. The differential expression of TFBf from that of TFBa, -c, -d, and -e (Fig. 1) also suggests it may act as a unique component of the regulatory network. Therefore, we have annotated the network as having four putative regulatory components, C1–C4.
Although these protein–DNA interactions imply direct regulatory control at the level of RNA polymerase (RNAP) recruitment, the protein–protein interactions among the TFBs (Fig. 3) most likely occur away from the DNA and may represent another mechanism to modulate RNAP recruitment. These TFB–TFB interactions are similar to those known to occur among TBPs. If functional, these would represent a regulatory layer that needs to be further investigated and integrated into our understanding of archaeal and eukaryotic gene regulatory networks. In this network, however, these interactions provide a direct means of communication between the expression-segregated TFB and may constitute a mechanism of regulatory feedback during the switch between states defined by individual sets of GTFs.
Conclusion
We have provided evidence, like numerous σ factors in bacteria and four different RNA polymerases and multiple GTFs in eukaryotes, that a multiplicity of GTFs in archaea also accomplishes large-scale regulation of transcription. Specifically, our study provides insights into how an expanded family of TFBs controls transcription in Halobacterium NRC-1 to functionally segregate genes while also providing some resistance to environmental or genetic perturbation. In addition, like most archaea whose genomes encode a single TBP, this organism favors interactions of most TFBs with a single TBP (TBPe). The use of at least some of the 42 possible TFB–TBP pairs by Halobacterium NRC-1 represents an instance in evolution wherein expansion of families of interacting transcription factors has been exploited for selective combinatorial use to regulate transcription. In light of our study, a particularly relevant example of a similar mechanism in eukaryotes is the use of developmental and tissue-specific TBP and TFIIB homologues (14, 15).
Finally, this study has demonstrated that the integration of various systems-level data types (protein–DNA and –protein interactions, differential gene regulation in response to environmental and genetic perturbations) can provide relatively rapid and unprecedented insight into the organization of global gene regulatory circuits, in this case specified by two expanded families of GTFs. We expect that this type of systems approach can easily be adapted to uncover additional gene regulatory mechanisms adopted by other relatively poorly studied organisms from each of the three domains of life. We further expect that Halobacterium NRC-1 will continue to serve as a model organism in which to test a large number of hypotheses raised in this study, such as role of TFB–TFB interactions in transcription regulatory mechanisms in archaea.
Methods
Strain Culturing and Plasmid Construction.
All Halobacterium NRC-1 strains were cultured under standard conditions (16). pNBPA vector was constructed by replacing the GFP coding sequence (CDS) with a proteinA (pA) CDS downstream to the ferredoxin promoter in pKJ419 (a generous gift from John Spudich, University of Texas, Houston, TX). For vector pMTFcmyc, the pA CDS was replaced with a Cmyc CDS. GTF genes were cloned into the NdeI restriction sites of the pMTFcmyc and pNBPA vectors for Cmyc and pA tagging, respectively, and transformed by standard protocols. Transformants were selected on CM agar containing 20 μg/ml mevinolin (A.G. Scientific, San Diego, CA).
Immunoprecipitaion and Mass Spectrometry Analyses of Protein Complexes.
Culture pellets (1.5 liters) of epitope-tagged GTF strains were lysed in 50 ml of basal salt solution (BSS -CM without peptone) containing Complete Protease Inhibitors (CH; Roche, Indianapolis, IN) by using a microfluidizer model 110 S (Microfluidics, Newton, MA), clarified by centrifugation, and incubated overnight at 4°C with IgG Sepharose 6 Fast Flow gel matrix (Amersham, Piscataway, NJ) preequilibrated in BSS. IgG-Sepharose-bound protein complexes were washed with 5 ml of BSS, transferred to Micro Bio-Spin columns (BioRad, Hercules, CA), and washed thrice with 1× PBS. Protein complexes were eluted with 150 ml of 0.5 M acetic acid, dried, dissolved in 100 μl of H2O, trypsinized, and analyzed with microliquid chromatography–electrospray ionization tandem MS, as described (17, 18). SEQUEST (ThermoFinnigan, Waltham, MA), PeptideProphet (19), and ProteinProphet were used to assign spectra to peptides and peptides to proteins (20).
HaloSpan Array Construction, ChIP, and TFB Localization.
PCR products representing 99.9% of the entire genome were generated by amplifying successive 500-bp regions of the Halobacterium genome (21). PCR products were verified for purity on an agarose gel, cleaned by using a 96-well PCR vacuum filtration plate, mixed 50:50 with DMSO (Sigma, St. Louis, MO), and spotted in quadruplicate on GAPSII slides (Corning, Corning, NY) with a VersArray (BioRad) spotter instrument (HaloSpan array).
ChIP of Cmyc-tagged protein complexes was conducted as described by Ren et al. (22). Amplified DNA from both ChIP complexes and non-IP DNA were each directly labeled by using either Alexa532/Alexa546 and Alexa647 (Invitrogen, Carlsbad, CA) or Dyomics-547 and -647 (Kreatech, Leiden, The Netherlands). Hybridization, washing, scanning, and spot-finding were conducted by using standard microarray procedures (7).
Statistical Analysis to Identify Significant GTF–Promoter Associations.
Each ChIP-Chip consisted of at least two independent biological replicates, with at least 16 replicate spots in each. The significance of DNA fragment enrichment in raw ChIP-chip microarray data was estimated by maximum-likelihood analysis, which yielded log10 ratios and the λ significance statistic (23). Plots of log10 ratios against λ demonstrated even distributions of log10 ratios in the absence of tagged transcription factors. In the presence of transcription factors, the log10 ratio distribution was significantly skewed toward the cross-linked sample relative to control, indicating specific nonrandom enrichment of DNA fragments. Therefore, features (tiles) associated with a log10 ratio in the experimental sample greater than the fifth-highest λ statistic in the control were considered representative of true enrichment attributable to transcription factor binding (SI Fig. 10). Finally, genes 50 bp downstream of tiles associated with transcription factor binding were considered to be putatively under control of the transcription factor.
Transcriptome Profiling.
RNA preparation, labeling, and microarray analysis were conducted as described (7). For environmental data presented in Fig. 1, data collection procedures are described in SI Table 2. For GTF perturbation experiments (Fig. 4), duplicate biological samples were harvested during three (late-lag, exponential, and early-stationary) growth phases (for 13 nonnatively expressed GTF strains (seven TFB and six TBP) and all possible knockouts (five TFB and three TBP) and processed as noted above.
Construction of In-Frame Gene Deletion Strains.
Gene knockouts were constructed as described (7) to chromosomally delete all but the first 21 bp of each gene.
Supplementary Material
Acknowledgments
We thank S. Proper, M. Slota, A. Weston, B. Marzolf, K. Whitehead, A. Schmid, J. Ranish, J. Roach, and D. Galas for help preparing versions of this manuscript and J. Aitchison and M. Marelli for advice and help with the protein A immunoprecipitation strategy. This work was supported by National Science Foundation Postdoctoral Fellowship DBI 0400598 (to M.T.F.); and National Science Foundation Grants EIA-0220153, MCB-0425825, and EF-0313754; Department of Energy Grants DE-FG02-04ER63807 and DE-AC02-05CH11231; and National Institutes of Health Grant P050 GM076547. (to N.S.B.).
Abbreviations
- GTF
general transcription factor
- TFB
archaeal transcription factor IIB ortholog
- TBP
TATA-binding protein.
Footnotes
The authors declare no conflict of interest.
Data deposition: The microarray and proteomic data reported in this paper have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GEO) database (GEO accession nos. GSE6776 and GSE7045).
This article contains supporting information online at www.pnas.org/cgi/content/full/0611663104/DC1.
References
- 1.DeLong EF, Karl DM. Nature. 2005;437:336–342. doi: 10.1038/nature04157. [DOI] [PubMed] [Google Scholar]
- 2.Lepp PW, Brinig MM, Ouverney CC, Palm K, Armitage GC, Relman DA. Proc Natl Acad Sci USA. 2004;101:6176–6181. doi: 10.1073/pnas.0308766101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, Wu D, Paulsen I, Nelson KE, Nelson W, et al. Science. 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- 4.Robertson CE, Harris JK, Spear JR, Pace NR. Curr Opin Microbiol. 2005;8:638–642. doi: 10.1016/j.mib.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 5.Geiduschek EP, Ouhammouch M. Mol Microbiol. 2005;56:1397–1407. doi: 10.1111/j.1365-2958.2005.04627.x. [DOI] [PubMed] [Google Scholar]
- 6.Baliga NS, Goo YA, Ng WV, Hood L, Daniels CJ, DasSarma S. Mol Microbiol. 2000;36:1184–1185. doi: 10.1046/j.1365-2958.2000.01916.x. [DOI] [PubMed] [Google Scholar]
- 7.Baliga NS, Bjork SJ, Bonneau R, Pan M, Iloanusi C, Kottemann MC, Hood L, DiRuggiero J. Genome Res. 2004;14:1025–1035. doi: 10.1101/gr.1993504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaur A, Pan M, Meislin M, Facciotti MT, El-Gewely R, Baliga NS. Genome Res. 2006;16:841–854. doi: 10.1101/gr.5189606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Whitehead K, Kish A, Pan M, Kaur A, Reiss DJ, King N, Hohmann L, DiRuggiero J, Baliga NS. Mol Syst Biol. 2006;2:47. doi: 10.1038/msb4100091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hengge-Aronis R. Curr Opin Microbiol. 1999;2:148–152. doi: 10.1016/S1369-5274(99)80026-5. [DOI] [PubMed] [Google Scholar]
- 11.Huang X, Fredrick KL, Helmann JD. J Bacteriol. 1998;180:3765–3770. doi: 10.1128/jb.180.15.3765-3770.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wade JT, Roa DC, Grainger DC, Hurd D, Busby SJ, Struhl K, Nudler E. Nat Struct Mol Biol. 2006;13:806–814. doi: 10.1038/nsmb1130. [DOI] [PubMed] [Google Scholar]
- 13.Goo YA, Yi EC, Baliga NS, Tao WA, Pan M, Aebersold R, Goodlett DR, Hood L, Ng WV. Mol Cell Proteomics. 2003;2:506–524. doi: 10.1074/mcp.M300044-MCP200. [DOI] [PubMed] [Google Scholar]
- 14.Chen X, Hiller M, Sancak Y, Fuller MT. Science. 2005;310:869–872. doi: 10.1126/science.1118101. [DOI] [PubMed] [Google Scholar]
- 15.Hansen SK, Takada S, Jacobson RH, Lis JT, Tjian R. Cell. 1997;91:71–83. doi: 10.1016/s0092-8674(01)80010-6. [DOI] [PubMed] [Google Scholar]
- 16.Baliga NS, DasSarma S. J Bacteriol. 1999;181:2513–2518. doi: 10.1128/jb.181.8.2513-2518.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yi EC, Lee H, Aebersold R, Goodlett DR. Rapid Commun Mass Spectrom. 2003;17:2093–2098. doi: 10.1002/rcm.1150. [DOI] [PubMed] [Google Scholar]
- 18.Gygi SP, Rochon Y, Franza BR, Aebersold R. Mol Cell Biol. 1999;19:1720–1730. doi: 10.1128/mcb.19.3.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Anal Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 20.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. Anal Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 21.Ng WV, Kennedy SP, Mahairas GG, Berquist B, Pan M, Shukla HD, Lasky SR, Baliga NS, Thorsson V, Sbrogna J, et al. Proc Natl Acad Sci USA. 2000;97:12176–12181. doi: 10.1073/pnas.190337797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ren B, Robert F, Wyrick JJ, Aparicio O, Jennings EG, Simon I, Zeitlinger J, Schreiber J, Hannett N, Kanin E, et al. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- 23.Ideker T, Thorsson V, Siegel AF, Hood LE. J Comput Biol. 2000;7:805–817. doi: 10.1089/10665270050514945. [DOI] [PubMed] [Google Scholar]
- 24.Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Felsenstein J. Phylogeny Inference Package. Seattle: Department of Genome Science, University of Washington; 2005. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.