Abstract
Mitochondria are not produced de novo but are maintained by division. Mitochondrial division is a coordinated process of positioning and constriction of the division site and fission of double membranes, in which dynamin-related protein is believed to mediate outer membrane fission. Part of the mitochondrial division machinery was purified from M phase-arrested Cyanidioschyzon merolae cells through biochemical fractionation. The dynamin-related protein Dnm1 was one of the two major proteins in the purified fraction and was accompanied by a newly identified protein CMR185C, named Mda1. Mda1 contained a predictable coiled-coil region and WD40 repeats, similarly to Mdv1 and Caf4 in yeasts. Immunofluorescence and immunoelectron microscopy showed that Mda1 localizes as a medial belt or ring on the mitochondrial outer surface throughout the division. The ring formation of Mda1 followed the plane of the ring of FtsZ, a protein that resides in the matrix. Dnm1 consistently colocalized with Mda1 only in the late stages of division. Mda1 protein was expressed through S to M phases and was phosphorylated specifically in M phase when Mda1 transformed from belt into foci and became colocalizing with Dnm1. Dephosphorylation of Mda1 in vitro increased its sedimentation coefficient, suggesting conformational changes of the macromolecule. Disassembly of the purified mitochondrial division machinery was performed by adding GTP to independently release Dnm1, suggesting that Mda1 forms a stable homo-oligomer by itself as a core structure of the mitochondrial division machinery.
Keywords: dynamin, FtsZ, mitochondrial division, plastid
Eukaryotic cells must properly control the number, morphology, position, and distribution of mitochondria for their specialized functions (1). Mitochondrial division is a major process in this control, as mitochondria cannot be produced de novo. Besides maintaining the number and morphology of mitochondria, recent studies have reported that mitochondrial division has significant roles in apoptotic cell death (2). The mitochondrial division apparatus was first described as a mitochondrion dividing (MD) ring, observed as a pair of electron-dense deposits at the equatorial region of the dividing mitochondria in Cyanidioschyzon merolae under transmission electron microscopy (3), led by the discovery of the plastid dividing (PD) ring for plastid or chloroplast division (4). Similar structures were also found in other algae, slime molds, and higher plants (5, 6). Electron micrographs in recent studies in humans (7) and yeast (8) showed electron-dense structures at the mitochondrial constriction site, although they were not mentioned as representing the MD ring. It is at present entirely unknown which molecules are responsible for the MD ring.
Mitochondrial and chloroplast division implicate two types of self-assembling GTPases. The first, FtsZ, is a bacterial cell-division protein, and had been diverted to mitochondrial and plastid division by means of endosymbiosis (9). Chloroplast or plastid FtsZ can be found in most photosynthetic eukaryotes, whereas mitochondrial FtsZ is retained only in relatively primitive eukaryotes (10–13). FtsZ forms ring or belt at the division site of plastids or chloroplasts (9) and mitochondria (10, 11) from the inside of the inner membrane. The second GTPase is dynamin-related protein (DRP). The involvement of DRP in mitochondrial fission was first reported in budding yeasts as Dnm1 (14), and was later found to be widespread in a diverse variety of eukaryotes. The necessity and function of DRPs in mitochondrial division is likely to be universal, as all of them act in a similar manner in that the DRPs accumulate at the cytosolic side of the mitochondrial tip or constriction site. However, several lines of evidence suggested that unlike FtsZ, DRPs are required only in the late stages of mitochondrial division (15–17), suggesting that FtsZ and DRP have distinct roles (16). The findings of DRPs involved in chloroplast division (18, 19) further strengthen the idea that chloroplast/plastid and mitochondrial division employ a basically similar mechanism, that involves FtsZ, the PD/MD ring, and dynamin (16, 19). However, there are undoubtedly more molecules involved, and the kinetics and regulation of the division machinery of the organelles need to be elucidated. In the budding yeast Saccharomyces cerevisiae, the outer membrane protein Fis1 (20, 21) and two WD40 repeat-containing proteins Mdv1 (21–23) and Caf4 (24) are implicated in Dnm1-dependent mitochondrial fission pathway. Fis1 mediates Dnm1 localization to mitochondrial surface through Mdv1 or Caf4 (21, 23, 24). The modes of their interaction are still open to discussion, and additional or distinctive roles for Mdv1 and Caf4 are also suggested (24–27). Orthologs for Mdv1 or Caf4 have not been found in other organisms whereas potent Fis1 orthologs have been characterized in human (7, 28) and plants (29).
Although isolation would be a direct way to understand how the molecular machineries are assembled, isolation of the mitochondrial division machinery has never been achieved, presumably because it is believed to be a dynamic and temporal structure and the quantities involved were considered to be too low in comparison with the other cellular components. With regard to this problem, C. merolae provides an ideal experimental system, as the alga has a single mitochondrion and a single chloroplast in a small cell of 1–2 μm in diameter. Furthermore, cell and organelle division can be synchronized. Indeed, isolation and analysis of the chloroplast division machinery have been explored exclusively in this alga (30, 31).
In this study, we developed a procedure to purify at least part of the mitochondrial division machinery in C. merolae, revealed its composition and subcellular localization, and assayed its biochemical properties.
Results
Purification of the Organelles Division Proteins.
The results of previous studies have indicated that inhibition of nuclear DNA synthesis uncouples the orderly division of chloroplast and mitochondria in C. merolae, suggesting that organelle division is under the control of cell cycle checkpoints (32, 33). In accordance with the observation that mitochondrial constriction occurs during S/G2 phase whereas final severance is suppressed until entering into M phase, a dynamin-related protein Dnm1 specifically accumulates in M phase, being otherwise dispersed among the cytoplasm (34). To purify such accumulated Dnm1 in M phase as part of mitochondrial division machinery, we developed a biochemical fractionation procedure using cell cycle-arrested cells. Cells were lysed with detergent and fractionated by several steps of differential and iodixanol-density centrifugations, followed by NaCl extraction. The residual pellet was further extracted with heptylthioglucoside.
Dnm1 was enriched in the insoluble fractions of the 35% and 20% iodixanol pellets (35P and 20P), as one of two major proteins only when M-arrested cells were used (Fig. 1A, open arrowheads), subsequently confirmed by immunoblot (data not shown). The other major protein, at ≈100 kDa (Fig. 1A, arrows), was subjected to time-of-flying mass spectrometry. Peptide mass fingerprint identified the protein as being encoded by CMR185C in the C. merolae genome. In the S/G2-arrested fractions two proteins were enriched (Fig. 1A, arrows and arrowheads) and were identified as CMR185C and Dnm2 (the other dynamin-related protein involved in chloroplast division) (19) by mass spectrometry and immunoblot (data not shown), respectively. The primary features of the deduced 783-aa sequence of the CMR185C protein were a putative coiled-coil domain in the central region and seven WD40 repeat units at the carboxyl terminal (Fig. 1B). Similar domain arrangements could be seen in Mdv1 and Caf4 from yeasts although the N-terminal region of CMR185C shares little homology with that of either Mdv1 or Caf4 [supporting information (SI) Fig. 6]. Overall sequence identities between CMR185C and Mdv1, CMR185C and Caf4, and Mdv1 and Caf4 were 18.0%, 19.2%, and 32.7%, respectively.
Fig. 1.
Fractionation and identification of organelle division proteins. (A) All fractions were separated with 10% SDS/PAGE and stained with high-sensitivity CBB. Each set of three lanes corresponds to the fractions from nonarrested (DMSO-treated), S/G2-arrested (camptothecin-treated), and M-arrested (oryzalin-treated) cells, from left to right. Each lane contained the total indicated fraction from the starting cell lysate of ≈8 mg of protein except for the (S) fraction, of which only 1/20th was loaded. The cell lysate was centrifuged on 40% iodixanol bedding to give the top layer of the soluble fraction (S), the bottom pellet (40P) and the middle layer at the interface, which was subsequently centrifuged on 35% iodixanol bedding to give the bottom pellet (35P), and on 20% iodixanol bedding to give the bottom pellet (20P) and upper layers (20U). The pellets 35P and 20P were subsequently further extracted with 1 M NaCl (NaCl) and 2% heptylthioglucoside (HTG), to give an insoluble pellet (P) in which the organelle division proteins were concentrated. The filled arrowheads indicate Dnm2 and open arrowheads indicate Dnm1. Arrows indicate bands subjected to mass spectrometry. The molecular weight marker (m) is indicated on the left. (B) Predicted domain architecture of CMR185C Mda1. From the bands indicated by arrows in A, a protein was identified by mass spectrometry as being encoded by CMR185C in the C. merolae genome; this protein was later named Mda1. (C) Characterization of the affinity-purified antibody to CMR185C. C. merolae total protein was separated and immunoblotted by using antibody raised against the recombinant N-terminal region (1–297) of CMR185C. (D) Protein expression throughout the cell cycle and in arrest. Equal culture volumes of cells were harvested at indicated times (hours) after the onset of synchronization and immunoblotted for Dnm1, α-tubulin (αTub), and Mda1. Cells were also arrested with camptothecin (Cpt) at S/G2 phase or with oryzalin (Orz) in M phase.
Subcellular Localization of CMR185C Mda1.
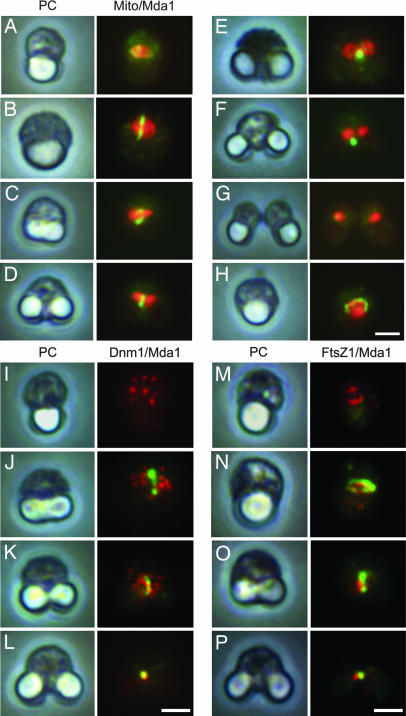
To characterize the cellular functions of CMR185C, polyclonal antibodies were raised. The affinity-purified antibody as well as the antisera specifically recognized a broad, ≈95- to 100-kDa band from C. merolae total protein (Fig. 1C), roughly consistent with the size of the bands in the purified fraction. Immunoblot on total protein of synchronized cultures showed that Dnm1 does not change throughout cell division whereas α-tubulin increases from S to M phase (Fig. 1D) as reported (34). CMR185C appears specifically in dividing cells and in cells arrested at S/G2 and M phase (Fig. 1D). To identify the subcellular localization of CMR185C, immunofluorescence was performed at several stages of cell division (Fig. 2, detailed in SI Fig. 7). In the early stage, CMR185C was to some extent concentrated around mitochondria (Fig. 2A), then formed a medial belt on mitochondria (Fig. 2B), which seemed like an arch along the mitochondrial surface when a cell at a similar stage was viewed from another angle (Fig. 2H). In the next stage when mitochondria and chloroplasts began to elongate, CMR185C formed an apparently enclosing ring or belt at an equatorial region of the mitochondria (Fig. 2 C and D). The medial ring of CMR185C was also constricted so as to be focused as mitochondrial constriction progressed (Fig. 2E). After mitochondrial division was completed, the CMR185C focus was affixed to one side of the divided mitochondria, and gradually disappeared (Fig. 2 F and G). Based on these observations, we named the protein Mda1 (mitochondrial division apparatus 1). To reveal the spatial relationship between Mda1 and Dnm1, both proteins were simultaneously immunostained. As reported (16), Dnm1 was found as several patches throughout the cytoplasm before and in the early phase of mitochondrial division, when Mda1 began to form a medial ring or belt (Fig. 2 I and J). As mitochondrial constriction proceeded, some of the Dnm1 patches became “attached” to the Mda1 belt (Fig. 2K). In the later stage of division, Mda1 and Dnm1 accumulated almost in the same focus (Fig. 2L). Next, we compared the order and placement of ring formation of Mda1 with that of FtsZ, which resides in the matrix and may be involved in the placement of the division plane (16). In most cases, at the earliest stage we could observe the FtsZ ring without consistently observing the Mda1 ring (Fig. 2M), however, the reverse case was rarely seen. Both rings always formed in the same plane of the mitochondria (Fig. 2 N and O), whereas their localization did not completely overlap and their respective intensities were not correlated. In the final stage of mitochondrial division, the FtsZ ring split into two when Mda1 formed the single focus (Fig. 2P). The ultrastructural localization of Mda1 was examined by immunoelectron microscopy. Each of the five thin serial sections contained parts of the Mda1 localization at the mitochondrial outer surface, which altogether showed the belt localization of Mda1 around the mitochondrion which was not yet constricted (Fig. 3 A and E). In the middle and final stages of mitochondrial division, Mda1 localizations were detected at or around the electron-dense image of MD ring on division site (Fig. 3 B–D).
Fig. 2.
Immunofluorescence for Mda1 and mitochondria, Dnm1 or FtsZ1. (A–H) Images of cells fixed and immunostained for mitochondria (red) and Mda1 (green) are aligned (A–G) according to the predicted order of cell cycle progression. (H) Cells viewed from another angle, considered to correspond to the stages of the cell cycle of cells in B. Images of fixed and immunostained cells are aligned vertically for Mda1 (green) and Dnm1 (red) (I–L) or FtsZ1 (red) (M–P) according to the predicted order of cell cycle progression except for (N) and (O), both of which are from almost the same stage and show views from different angles. PC, phase contrast image; Mito, images for mitochondria stained with anti-CmEF-Tu(mt) antibody. (Scale bars: 1 μm.) Independent fluorescence images and additional cells were shown in SI Fig. 7.
Fig. 3.
Immunoelectron microscopy for Mda1. Ultra-thin sections were labeled with an anti-Mda1 rabbit antibody and a 10 nm gold particle-conjugated secondary antibody, and were observed under transmission electron microscopy. (A) One of the five serial sections showing the entire cell at the early stage of mitochondrial division. (B) Cross-sectioned dividing mitochondria with electron-dense images of sectioned MD ring pinching the constriction site. (C) Section showing a cell at the final stage of mitochondrial division. (D) Three-fold magnification of C, showing Mda1 signals around a constricted MD ring. (E) The five panels on the Left are images of the five serial sections showing the belt localization of Mda1 around the mitochondrion, one of which is in A. The Right end panel shows signals in the five superimposed left images. N, nucleus; m, mitochondrion; mb, microbody; cp, chloroplast. [Scale bars: 200 nm (A and B) and 500 nm (C).]
Disassembly of the Purified Proteins.
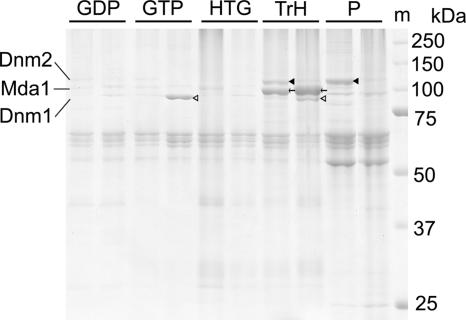
To characterize the biochemical properties of the purified proteins as well as to evaluate their intactness, the combined fraction of 35P and 20P after NaCl treatment was further extracted. By adding 2 mM GTP but not GDP, large part of Dnm1 was released from the M-arrested fraction (Fig. 4, GDP and GTP). The remaining Mda1 and the smaller amount of Dnm1 were then solubilized in a buffer containing 1 M Tris·HCl and 5% heptylthioglucoside (Fig. 4, TrH). In the S/G2-arrested fraction, Mda1 was also solubilized whereas a large fraction of the Dnm2 remained insoluble (Fig. 4, TrH and P).
Fig. 4.
Disassembly of the mitochondrial division machinery. The sets of two lanes are fractions from S/G2-arrest and M-arrest. Pellets 35P and 20P in Fig. 1 were combined and subsequently extracted with 2 mM GDP (GDP), 2 mM GTP (GTP), 2% heptylthioglucoside (HTG), and 1 M Tris·HCl plus 5%heptylthioglucoside (TrH). Proteins from extracts and pellets (P) were resolved by 10% SDS/PAGE. The molecular weight marker (m) is indicated on the right. Filled arrowheads indicate Dnm2; open arrowheads indicate Dnm1. Arrows indicate Mda1.
Phosphorylation of Mda1.
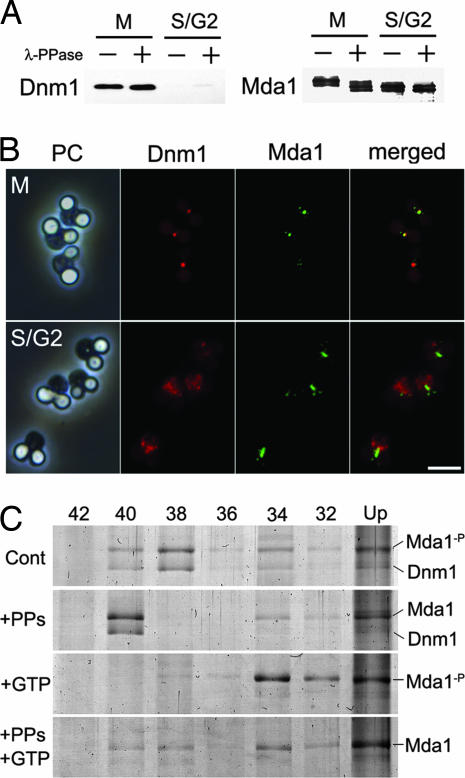
Mda1 in the S/G2-arrested and M-arrested fractions, respectively, seemed slightly different in its molecular size in SDS/PAGE. Because phosphorylation is one of the most potent modifications correlating to the cell cycle, we applied phosphatase treatment in combination with low-bis-acrylamide gel electrophoresis, which efficiently separates phosphorylated and nonphosphorylated peptides. As expected, the band for Mda1 in M-arrest was shifted down by phosphatase treatment to the level in S/G2-arrest (Fig. 5A). Treatment with phosphatase did not affect the disassembly of Mda1 and Dnm1 in the conditions tested. The subcellular localization of Mda1 and Dnm1 in S/G2- or M-arrest was also checked. In M-arrested cells Dnm1 and Mda1 localized largely in the same foci, whereas in S/G2-arrested cells Mda1 formed belts and Dnm1 was dispersed among the cytoplasm (Fig. 5B).
Fig. 5.
Phosphorylation of Mda1. (A) The purified proteins from M-arrest or S/G2-arrest were treated with (+) or without (−) λ-protein phosphatase (λ-PPase), subjected to low-bis-acrylamide gel electrophoresis and immunoblotted with anti-Dnm1 or anti-Mda1. (B) Cells in M-arrest or S/G2-arrest were immunostained with anti-Dnm1 and anti-Mda1 antibodies and detected with Alexa Fluor 555- or Alexa Fluro 488-conjugated secondary antibody, respectively. (Scale bar: 2 μm.) (C) Narrow-ranged differential density centrifugation of Mda1-containing macromolecules in various states. Partly purified fraction from M-arrest cells was incubated in the presence of phosphatase and/or GTP, then fractionated by differential density centrifugation, and analyzed by SDS/PAGE. Each lane contains pellets through a certain concentration of iodixanol (%) as indicated above. Cont, incubated in control buffer; +PPs, incubated in the presence of λ-PPase; +GTP, incubated in the presence of GTP; Mda1-p, band for natively phosphorylated Mda1; Mda1, band for Mda1 dephosphorylated in vitro.
To investigate how phosphorylation of Mda1 affects the property of the division apparatus, a purified fraction from M-arrested cells was incubated in the presence or absence of phosphatase and/or GTP, and separated by narrow-ranged differential density centrifugations (Fig. 5C). Under any conditions, the upper fraction contained proteins with higher background, and thought to be inefficiently resolved or aggregated (Fig. 5C, Up). In the absence of phosphatase and GTP, Mda1 and Dnm1 was mainly partitioned in F38, whereas they sharply shifted to F40 by phosphatase treatment (Fig. 5C, Cont and +PPs). By GTP addition, Dnm1 was largely released and Mda1 appeared in F34 whereas it was partly shifted to heavier fractions in the presence of phosphatase (Fig. 5C, +GTP and +PPs+GTP).
Discussion
Major Components of the Mitochondrial Division Machinery.
Use of our procedure led to the purification of Dnm1 and phosphorylated Mda1 in the same fraction from M-arrested cells, and Dnm2 and Mda1 in the same fraction from S/G2-arrested cells. Although it seems unlikely that Dnm2 and Mda1 are in the same complex, because Dnm2 localizes as chloroplast division machinery (19, 31) whereas Mda1 was revealed to localize on mitochondria, it is still possible that Dnm2 and Mda1 are physically bound because the chloroplast and mitochondrial division apparatuses are in close association in C. merolae (35). Alternatively, the chloroplast and mitochondrial division machineries may have similar biochemical properties. Immunofluorescence as well as immunoelectron microscopy showed that Mda1 begins to localize as a belt form around the outer surface of mitochondria before mitochondrial constriction occurs, but after FtsZ ring formation in the matrix. It is further suggested that FtsZ is involved in the determination of the division plane from the inside of mitochondria, and that the placement information is passed through the inner and outer membranes by as yet unknown mechanisms, then Mda1 localization follows this plane. Mda1 remained in the form of a medial ring or belt throughout mitochondrial division, whereas Dnm1 accumulated to colocalize with Mda1 only in the late stage of division when cells were entering into M phase. Combined with the in vitro results showing that Dnm1 could be independently released by GTP addition, it is likely that Mda1 forms a stable homo-oligomer by itself. Mda1 was also shown to be phosphorylated during M phase. In M-arrested cells Mda1 forms foci with Dnm1 presumably after completion of mitochondrial division, whereas in S/G2-arrested cells Mda1 exists as a longer medial belt of the mitochondria, devoid of Dnm1. Narrow-ranged differential centrifugation showed that Mda1-containing macromolecules decrease their sedimentation coefficient by Dnm1 release with GTP, whereas dephosphorylation of Mda1 does not separate Dnm1 from Mda1 but increases the sedimentation coefficient. This suggested that phosphorylation of Mda1 does not contribute to the stability of Dnm1 association, but induced certain changes in the conformation or organization of the Mda1-containing macromolecule. This phosphorylation may mediate changes in the arrangement of the Mda1 oligomer, or in the mode of interaction with other unknown molecules, whereas we do not exclude the possibility that phosphorylation of Mda1 facilitate the incorporation of Dnm1 into division apparatus. Such possible changes in organization might induce the constriction of the division apparatus within M phase. Our purification procedure outlined here was performed in harsh conditions to determine the core structure, which we revealed to include Mda1. Therefore, there is the potential for modifications in future studies to preserve mildly associated molecules.
Similar Domain Arrangement with Distinct Behaviors.
Mda1 shares significant characteristics with the yeast proteins Mdv1 and Caf4, in that they all contain a central coiled-coil region and C-terminal WD40 repeats, and are all involved in mitochondrial division in association with DRPs. Supposing that the yeast Mdv1 and Caf4 and algal Mda1 share a common evolutionary origin, it would have arisen before the eukaryotic divergence of Bikonta and Unikonta, so its descendants would have been rather more widespread than has been found. Interestingly, the proteins also show differences in their functional behaviors. Mda1 appears as a medial belt in the early stage of mitochondrial division, before Dnm1 accumulation. In yeasts, exogenously expressed dsRed-Mdv1 follows GFP-Dnm1 accumulation (25), whereas genetic studies indicated that Mdv1 and Caf4 function upstream of Dnm1 (24). Our biochemical experiments as well as localization studies suggested that Mda1 exists as a stable homo-oligomer independently of Dnm1. In addition, Mda1 is under the control of the cell cycle, in that Mda1 is expressed during S to M phase and is phosphorylated in M phase. Such characteristics may be due to a specialized feature of C. merolae, or perhaps possible corresponding characteristics of Mdv1 and Caf4 are not yet sufficiently explored in yeast. Taking into consideration the fact that the timing and frequency of organelle division differs between cell types, we propose that eukaryotic cells are capable of organizing organelle division with several layers of control in the cell cycle as well as in other systems of control. Higher similarity between Mdv1 and Caf4 in primary sequences than those between Mda1 and each of them implies that they occurred by duplication after evolutional divergence, or that it may represent their functional similarity and/or binding partners. The primal function of the WD40 proteins is assumed to be an adopter between Dnm1 and Fis1. In C. merolae, a potent Fis1 homolog CMQ197C can be found. Our present study, however, did not detect the protein in the purified fraction. This is not inconsistent because Fis1 is evenly distributed on mitochondrial surface and does not accumulate at fission site (20). Fis1 homologs in plants as well as human have been shown to function in mitochondrial morphogenesis, although no WD40 proteins have yet been reported to be involved in this pathway. In human, direct and transient interaction between hFis1 and DRP is proposed (7). It is possible that WD40 proteins were lost in such organisms in the course of evolution, as was mitochondrial FtsZ in yeasts, animals and higher plants. Even so, an evolutional origin of eukaryote probably once had used DRP, Fis1, FtsZ, and the WD40 protein for organizing the mitochondrial division machinery.
Materials and Methods
Cell Preparation.
Cell culture, drug treatment, and fixation were performed as described (21) with minor modifications. For details, see SI Text.
Fractionation Procedure and Electrophoresis.
Cells were pelleted by centrifugation at 2,300 × g for 10 min at room temperature, washed with the basal buffer (50 mM Hepes-KOH/200 mM NaCl/2 mM MgSO4/1 mM EGTA-Na/1 mM DTT/10% wt/vol glycerol, pH 7.1) and suspended in the lysis buffer (basal buffer containing 5% wt/vol Triton X-100/2% wt/vol sodium cholate/protease inhibitor mixture) in a concentration of ≈10 mg protein/ml. The cell suspension was frozen with liquid nitrogen and stored at −80°C. The lysate was thawed on ice. Then, DNase I was added to give a final concentration of 200 μg/ml, and the mixture was incubated on ice for 30 min. From this point, procedures were performed on ice or at 4°C, in 1.5 ml of microcentrifuge tubes, and with fixed angle rotors for centrifugation. The lysate was centrifuged at 1,000 × g to remove cell debris. The supernatant was saved, and the pellet was resuspended in the lysis buffer and centrifuged again at 1,000 × g, and the supernatants were combined. The supernatant was then layered on basal buffer containing 40% iodixanol, 2% Triton X-100, and 0.5% sodium cholate, centrifuged at 30,000 × g for 5 min, changing the side of angle rotor, then centrifuged again to form the upper layer supernatant (S), bottom pellet (40P), and middle layer at the interface. The middle layer was suspended in basal buffer, layered on basal buffer containing 35% iodixanol, and centrifuged at 30,000 × g to form the bottom pellet (35P) and upper layers. The upper layers were suspended, layered on basal buffer containing 20% iodixanol, and centrifuged at 30,000 × g to form the bottom pellet (20P) and upper layers (20U). The pellets 35P and 20P were suspended in basal buffer containing 1 M NaCl, 2% Triton X-100 and 0.5% sodium cholate for 30 min, and then centrifuged at 30,000 × g for 10 min. The pellets were washed with basal buffer, suspended in 2% heptylthioglucoside in basal buffer, and incubated for 30 min and then centrifuged at 30,000 × g. For the disassembling assay, the 35P and 20P fractions were combined and sequentially extracted with 1 M NaCl, 2 mM GDP at 37°C for 30 min, 2 mM GTP at 37°C for 30 min, 2% heptylthioglucoside, and finally 1 M Tris·HCl (pH 7.4) plus 5% heptylthioglucoside, and centrifugation was performed at 30,000 × g for 10 min in each extraction step. For SDS/PAGE, extracts were precipitated with trichloroacetic acid followed by acetone rinse, and the pellets were resolved in SDS sample buffer containing 50 mM DTT, 8 M urea, and 1 mM EDTA. After electrophoresis, the gels were stained with the high-sensitivity Coomassie brilliant blue (CBB), GelCode blue stain reagent, or Imperial protein stain (Pierce Biotechnology, Rockford, IL). Protein bands were identified by immunoblot or subjected to time-of-flying mass spectrometry (Shimazu Biotech, Ibaraki, Japan). For low-bis-acrylamide gel electrophoresis, a ratio of 99:1 acrylamide:bisacrylamide was used. For phosphatase treatment, samples were incubated in basal buffer containing 4,000 units/ml λ-protein phosphatase (λ-PPase) (New England Biolabs, Beverly, MA) and 2 mM MnCl2 at 30°C for 30 min.
For narrow-ranged differential density centrifugation, we used every 2% step of iodixanol concentration from 42% to 32% in the basal buffer containing 1% TritonX-100. The partly purified fraction from M-arrest cells was prepared as described above, whereas 42% instead of 40% iodixanol was used for bedding. The resultant layer upon the bedding was washed twice with basal buffer and suspended in basal buffer containing 2 mM MnCl2, 1 mM DTT, and a protease inhibitor mixture, and incubated at 30°C for 30 min in the presence or absence of 4,000 units/ml λ-protein phosphatase λ-PPase and/or 5 mM GTP. Samples were then loaded on layer containing certain concentrations of iodixanol and centrifuged at 30,000 × g for 5 min, changing the side of angle rotor, then centrifuged again. Resultant pellet (F42 to F32) was collected whereas the layer at the interface was suspended and carried to the next step by using a lower concentration of iodixanol. The final upper fraction was pelleted in 16% iodixanol (Up). All pellets were washed with 1 M NaCl and then 2% heptylthioglucoside as described above, and analyzed by SDS/PAGE.
Antibodies, Immunofluorescence, Immunoblotting, and Electron Microscopy.
For generation of anti-CMR185C rabbit antiserum, a bacterial recombinant protein of the amino acid residues from 1 to 297 of the predicted 783-aa sequence of CMR185C protein with 6 histidine tag at N-terminal was used for immunization and affinity purification. For generation of anti-CMR185C mouse antiserum, a bacterial recombinant of the total protein with 6 histidine tag at N-terminal was used to immunize. Immunofluorescence and immunoblotting were performed by conventional methods. For electron microscopy, rapid frozen, acetone-fixed cells were used. For details, see SI Text.
Supplementary Material
Acknowledgments
This work was supported by Grant 7472 from the Japan Society for the Promotion of Science Fellowships (to K.N.), Grant 17051029 from Scientific Research on Priority Areas (to T.K.), the Frontier Project “Adaptation and Evolution of Extremophiles” from the Ministry of Education, Culture, Sports, Science and Technology of Japan, and the Program for the Promotion of Basic Research Activities for Innovative Biosciences (PROBRAIN) (to T.K.).
Abbreviations
- MD
mitochondrion dividing
- DRP
dynamin-related protein.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0609364104/DC1.
References
- 1.Yaffe MP. Science. 1999;283:1493–1497. doi: 10.1126/science.283.5407.1493. [DOI] [PubMed] [Google Scholar]
- 2.Youle RJ, Karbowski M. Nat Rev Mol Cell Biol. 2005;6:657–663. doi: 10.1038/nrm1697. [DOI] [PubMed] [Google Scholar]
- 3.Kuroiwa T, Suzuki K, Kuroiwa H. Protoplasma. 1993;175:173–177. [Google Scholar]
- 4.Mita T, Kanbe T, Tanaka T, Kuroiwa T. Protoplasma. 1986;130:211–213. [Google Scholar]
- 5.Hashimoto H. Cytologia. 2004;69:323–326. [Google Scholar]
- 6.Kuroiwa T, Nishida K, Yoshida Y, Fujiwara T, Mori T, Kuroiwa H, Misumi O. Biochim Biophys Acta. 2006;1763:510–521. doi: 10.1016/j.bbamcr.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 7.Yoon Y, Krueger EW, Oswald BJ, McNiven MA. Mol Cell Biol. 2003;23:5409–5420. doi: 10.1128/MCB.23.15.5409-5420.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ingerman E, Perkins EM, Marino M, Mears JA, McCaffery JM, Hinshaw JE, Nunnari J. J Cell Biol. 2005;170:1021–1027. doi: 10.1083/jcb.200506078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Osteryoung KW. Curr Opin Microbiol. 2001;4:639–646. doi: 10.1016/s1369-5274(01)00263-6. [DOI] [PubMed] [Google Scholar]
- 10.Beech PL, Nheu T, Schultz T, Herbert S, Lithgow T, Gilson PR, McFadden GI. Science. 2000;287:1276–1279. doi: 10.1126/science.287.5456.1276. [DOI] [PubMed] [Google Scholar]
- 11.Takahara M, Takahashi H, Matsunaga S, Miyagishima S, Takano H, Sakai A, Kawano S, Kuroiwa T. Mol Gen Genet. 2000;264:452–460. doi: 10.1007/s004380000307. [DOI] [PubMed] [Google Scholar]
- 12.Gilson PR, Yu XC, Hereld D, Barth C, Savage A, Kiefel BR, Lay S, Fisher PR, Margolin W, Beech PL. Eukaryot Cell. 2003;2:1315–1326. doi: 10.1128/EC.2.6.1315-1326.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kiefel BR, Gilson PR, Beech PL. Protist. 2004;155:105–115. doi: 10.1078/1434461000168. [DOI] [PubMed] [Google Scholar]
- 14.Bleazard W, McCaffery JM, King EJ, Bale S, Mozdy A, Tieu Q, Nunnari J, Shaw JM. Nat Cell Biol. 1999;1:298–304. doi: 10.1038/13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Labrousse AM, Zappaterra MD, Rube DA, van der Bliek AM. Mol Cell. 1999;4:815–826. doi: 10.1016/s1097-2765(00)80391-3. [DOI] [PubMed] [Google Scholar]
- 16.Nishida K, Takahara M, Miyagishima SY, Kuroiwa H, Matsuzaki M, Kuroiwa T. Proc Natl Acad Sci USA. 2003;100:2146–2151. doi: 10.1073/pnas.0436886100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Legesse-Miller A, Massol RH, Kirchhausen T. Mol Biol Cell. 2003;14:1953–1963. doi: 10.1091/mbc.E02-10-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao H, Kadirjan-Kalbach D, Froehlich JE, Osteryoung KW. Proc Natl Acad Sci USA. 2003;100:4328–4333. doi: 10.1073/pnas.0530206100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miyagishima SY, Nishida K, Mori T, Matsuzaki M, Higashiyama T, Kuroiwa H, Kuroiwa T. Plant Cell. 2003;15:655–665. doi: 10.1105/tpc.009373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mozdy AD, McCaffery JM, Shaw JM. J Cell Biol. 2000;151:367–380. doi: 10.1083/jcb.151.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tieu Q, Nunnari J. J Cell Biol. 2000;151:353–366. doi: 10.1083/jcb.151.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fekkes P, Shepard KA, Yaffe MP. J Cell Biol. 2000;151:333–340. doi: 10.1083/jcb.151.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cerveny KL, McCaffery JM, Jensen RE. Mol Biol Cell. 2001;12:309–321. doi: 10.1091/mbc.12.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Griffin EE, Graumann J, Chan DC. J Cell Biol. 2005;170:237–248. doi: 10.1083/jcb.200503148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Naylor K, Ingerman E, Okreglak V, Marino M, Hinshaw JE, Nunnari J. J Biol Chem. 2006;281:2177–2183. doi: 10.1074/jbc.M507943200. [DOI] [PubMed] [Google Scholar]
- 26.Bhar D, Karren MA, Babst M, Shaw JM. J Biol Chem. 2006;281:17312–17320. doi: 10.1074/jbc.M513530200. [DOI] [PubMed] [Google Scholar]
- 27.Schauss AC, Bewersdorf J, Jakobs S. J Cell Sci. 2006;119:3098–3106. doi: 10.1242/jcs.03026. [DOI] [PubMed] [Google Scholar]
- 28.James DI, Parone PA, Mattenberger Y, Martinou JC. J Biol Chem. 2003;278:36373–36379. doi: 10.1074/jbc.M303758200. [DOI] [PubMed] [Google Scholar]
- 29.Scott I, Tobin AK, Logan DC. J Exp Bot. 2006;57:1275–1280. doi: 10.1093/jxb/erj096. [DOI] [PubMed] [Google Scholar]
- 30.Miyagishima S, Itoh R, Aita S, Kuroiwa H, Kuroiwa T. Planta. 1999;209:371–375. doi: 10.1007/s004250050645. [DOI] [PubMed] [Google Scholar]
- 31.Yoshida Y, Kuroiwa H, Misumi O, Nishida K, Yagisawa F, Fujiwara T, Nanamiya H, Kawamura F, Kuroiwa T. Science. 2006;313:1435–1438. doi: 10.1126/science.1129689. [DOI] [PubMed] [Google Scholar]
- 32.Itoh R, Takahashi H, Toda K, Kuroiwa H, Kuroiwa T. Eur J Cell Biol. 1996;71:303–310. [PubMed] [Google Scholar]
- 33.Itoh R, Takahashi H, Toda K, Kuroiwa H, Kuroiwa T. Eur J Cell Biol. 1997;73:252–258. [PubMed] [Google Scholar]
- 34.Nishida K, Yagisawa F, Kuroiwa H, Nagata T, Kuroiwa T. Mol Biol Cell. 2005;16:2493–2502. doi: 10.1091/mbc.E05-01-0068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miyagishima S, Kuroiwa H, Kuroiwa T. Planta. 2001;212:517–528. doi: 10.1007/s004250000426. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.