Abstract
Both sperm typing and linkage disequilibrium patterns from large population genetic data sets have demonstrated that recombination hotspots are responsible for much of the recombination activity in the human genome. Sperm typing has also revealed that some hotspots are heterogeneous in the population; and linkage disequilibrium patterns from the chimpanzee have implied that hotspots change at least on the separation time between these species. We propose a population genetics model, inspired by the double-strand break model, which features recombination hotspots that are heterogeneous across the population and whose population frequency changes with time. We have derived a diffusion approximation and written a coalescent simulation program. This model has implications for the “hotspot paradox.”
Analysis of the Seattle SNP, Perlegen, and HapMap data sets has suggested that the fine-scale recombination rate varies with position across the chromosome (1–3). Indeed 80% of the recombination activity is believed to occur in as little as 10–20% of the sequence. So-called recombination hotspots, narrow regions generally 1 kb wide with elevated recombination rates, have been estimated approximately every 100 kb across the human genome. Sperm typing (4–6) (for reviews, see refs. 7 and 8) has confirmed the presence of recombination hotspots. Although more recent population genetic modeling efforts (9–11) have included recombination hotspots, these methods, like their predecessors (12–16), assume that the recombination rate is (i) homogeneous across the population, and (ii) constant throughout time. However, sperm typing has also demonstrated that some hotspots are heterogeneous in the population (17). Moreover, analysis of linkage disequilibrium patterns in the human and chimpanzee populations (18, 19) has shown little congruence between the location of hotspots in the two species, implying that recombination rates change at least on the order of the separation time between these species.
The predominant mechanistic model of recombination is the double-strand break model illustrated in Fig. 1(20) (for a review see ref. 21). In this model, there is a break through both strands of one of the chromosomes. On this chromosome there is a loss of several hundred base pairs around the break. This loss is then replaced by copying from the other chromosome. The break can be resolved as either a crossover or a conversion event. In most population genetic models, this loss and copy of DNA has been ignored: in these models there is an exchange of DNA between chromosomes, but, despite this rearrangement, all sections of the chromosome are assumed to retain their original quantities. Because the length of this loss is relatively small, this simplification had seemed benign.
Fig. 1.
Double-strand break model. Each red and blue line represents the same chromosomal region; the model shown is during meioses so there are two copies of both the red and blue chromosomes. The short black line represents a double-strand break on the red chromosome; some surrounding DNA has been lost and is then copied from the blue chromosome. The break can be resolved either through a conversion or a crossover event. The colored triangles and ovals represent potential locations for the hotspot motif: in this example, a motif at the triangle location loses one red copy and gains one blue one, whereas a motif at the oval location retains the original quantities.
However, this loss is the key factor in the “hotspot paradox” (e.g., refs. 22 and 23). This paradox states that if the hotspot is caused by some motif in the DNA sequence that elevates the local double-strand break rate, then this motif will often be lost in a recombination event; and, thus, all hotspots will be so short-lived that they will never be observed. Researchers have considered positive selection on the hotspot or multiple hotspots at different nearby positions, without successfully resolving the paradox (22, 23). Another suggestion (24) is that perhaps there is some distance between the motif and the break position: after a break event, then, whether or not the motif is transmitted depends on this distance and the amount of lost DNA. In yeast, researchers have observed double-strand breaks as far as 1.3 kb away from a known motif (25). In humans, researchers have measured gene conversion tract lengths averaging less than this distance, namely several hundred base pairs (26). In prokaryotes (27, 28) and yeast (24, 29), such hotspot-causing motifs have been identified. In humans, there is compelling evidence that some hotspot-causing motifs are found in retrotransposons; however, these motifs explain <20% of the inferred human hotspots (2, 30–32). The cause of most human hotspots is unclear; for the purposes of this article, a motif is anything genetically inheritable that elevates the local double-strand break rate on a chromosome harboring it, thus increasing the chance that it will not be transmitted to the next generation. Therefore our model applies not just to simple DNA sequence motifs, but also to possible epigenetic factors (33) or even motifs comprised of multiple interacting DNA sequence patterns.
In this article, we incorporate the double-strand break model into the standard population genetics model (e.g., ref. 34). The model has parameters governing the random amount of DNA lost and copied from the other chromosome after a double-strand break. In addition, we include a recombination hotspot model. We assume that, in a given chromosomal region, a motif originated once in the past, and since then it has been transmitted genetically. In chromosomes harboring this motif, the double-strand break rate is elevated at a specified distance from the motif. Whether an individual heterozygous for the motif transmits the motif is random and depends on this distance, the amount of lost and copied DNA, and the hotspot recombination rate.
This combined model, explained in more detail in Materials and Methods, relaxes the two assumptions from the first paragraph in a natural way. Other researchers have considered similar models (22, 23, 31), but we derive a diffusion approximation rather than just relying on computer simulations. This same diffusion arises as a model for gene conversion (35, 36); for a coalescent description of gene conversion see refs. 37 and 38. This equivalence makes sense because the transmission of the motif is analogous to an allele experiencing gene conversion. The diffusion approximation allows us to analytically study the evolution of the hotspot's frequency in the population and to better understand how this evolution depends on the model parameters. To study the effect of the model on linkage disequilibrium patterns, we have written a coalescent simulation program. We investigate the range of parameters for which the Hotspotter (10) recombination rate estimation program, which assumes the recombination rate is homogeneous across the population, detects the simulated heterogeneous hotspots. The simulation code is available at our website, www.cmb.usc.edu/people/petercal.
Results
The only parameter in the diffusion approximation is α. α is a scaled parameter proportional to the probability that the hotspot motif both causes a double-strand break, and the motif is then lost in the subsequent loss and copying of DNA, see Eq. 2. To interpret α, we find it useful to consider a hotspot with 1-kb width and a per-base recombination rate elevated h1 times above the genome average. Substituting 10−8 for this average and N = 10,000 for the diploid population size, then α = h1fh/10, where fh is the probability that the motif is lost after a double-strand break caused by the hotspot. Therefore, for example, α = 1 can be parameterized as a 1-kb hotspot with recombination rate elevated h1 = 100 times, and probability fh = 0.1.
Fig. 2 shows various diffusion solutions with different colors representing different α values. Fig. 2A shows the normalized probability that a hotspot achieves a specified frequency as a function of that frequency. The probability is normalized by dividing by the probability that a neutral mutation achieves this same frequency; the neutral mutation solution is the case α = 0. The solutions are u(1/2N, b) from Eq. 4. Fig. 2B shows the normalized probability that a hotspot currently at a set frequency eventually fixes in the population. The solution is u(x, 1) from Eq. 4. For Fig. 2 A and B, for the 0.1 dark and light blue solutions, the probabilities are similar to the probabilities for a neutral mutation. For the α = 1 green solution, the probabilities, which depend on the current frequency, are not less than a factor of 3 lower than for the neutral mutation. For the α ≥ 10 orange and red solutions, the probabilities are much smaller than for a neutral mutation. Fig. 2C shows the expected time for a hotspot currently at a specified frequency to be either fixed or lost in the population; the solution is Eq. 5. Fig. 2D shows the mean age of a hotspot as a function of its current frequency; the solution is Eq. 7. In both parts, the α ≤ 0.1 dark and light blue solutions nearly coincide both with each other and the neutral mutation solution (α = 0, not shown). The α = 1 green solution is similar. The α ≥ 10 orange and red solutions have smaller mean times and ages.
Fig. 2.
Diffusion solutions. Different colors represent different values of the model parameter α: red, 100; orange, 10; green, 1; light blue, 0.1; dark blue, 0.01. (A) The normalized probability that a hotspot achieves a specified frequency. The probability is normalized by dividing by the probability a neutral mutation (α = 0) achieves the same frequency. (B) The normalized probability that a hotspot currently at a set frequency eventually fixes in the population. (C) The expected time that a hotspot currently at a specified frequency takes to either be fixed or lost from the population. (D) The mean age of a hotspot as a function of its current frequency.
The coalescent simulation program has more parameters than the diffusion approximation. In Table 1, we vary the probability fh and the current hotspot frequency (same frequency in the population and the sample). The other parameters and their values are discussed in Materials and Methods. The simulation program produces a sample of 100 haplotypes that are randomly combined into 50 genotypes and input to the Hotspotter (10) recombination rate estimation program. If the hotspot's recombination estimate exceeds the flanking regions 95% credibility region, the hotspot is counted as detected. When the current hotspot frequency is 50% (or higher), a sufficient signal has been left in the linkage disequilibrium patterns so that the hotspot is detected with high frequency for all tested fh values. When the current hotspot frequency is 30%, whether the hotspot is detected depends on the fh parameter. As expected, lower fh values imply older hotspots and, therefore, increased frequency of detection. When the current hotspot frequency is 10% (or lower), there is little chance of detecting the hotspot.
Table 1:
Detection frequency for simulated hotspots as a function of fh and the current hotspot frequency
| fh | Current hotspot frequency, % |
||
|---|---|---|---|
| 10 | 30 | 50 | |
| 0.99 | 0 | 30 | 90 |
| 0.90 | 10 | 40 | 90 |
| 0.10 | 20 | 50 | 100 |
Discussion
We propose a population genetics model with recombination hotspots that are heterogeneous in the population and whose population frequency changes with time. These complications have been shown to exist, and the double-strand break model provides a natural way to model them. We have derived a diffusion approximation for the evolution of the hotspot in the population. α is a scaled parameter proportional to the product of the probability that the motif causes a double-strand break and fh, the probability that this motif is lost in the subsequent loss and copying of DNA. For α ≤ 1, the hotspot behaves similarly to a neutral mutation, thus possibly resolving the hotspot paradox. For this parameter range, hotspots are not automatically eliminated from the population, but evolve much like neutral polymorphisms. One of the advantages of the diffusion approach is that it allows one to study how the parameter values affect the model without waiting for ever more computer simulations. Our conclusion is different from some others who have considered this paradox (22, 23); one reason may be that, presumably because of computational constraints, these researchers simulated the model under, in our opinion, unrealistic parameter values. For all α, the mean ages and mean times to fixation or loss considered in Fig. 2 are on the order of the effective population size [N = 10,000 (39)] in generations and are thus much less than the species separation time between humans and chimpanzees [6–7 million years (40)]. This result is consistent with the inferred incongruity between human and chimpanzee hotspots (18, 19). For different human populations, however, because the time of the last great out-of-Africa migration is estimated to be ≈100,000 years ago (41), some hotspots are predicted to be population-specific, whereas others will be present in all populations.
To study the effect of this model on linkage disequilibrium patterns, we have written a coalescent simulation program. We varied the current frequency of the hotspot and the probability fh and then measured the frequency at which the hotspot was detected. Hotspots with current frequency 50% and above left a sufficient linkage disequilibrium pattern to almost always be detected, whereas hotspots with current frequency <10% were rarely detected. Thus, recombination estimation programs such as Hotspotter (10) and LDHat (11), which assume that the recombination rate is homogeneous across the population, will detect most of the high-frequency hotspots, few of the low-frequency hotspots, and some of the intermediate-frequency ones.
The ideas presented in this article could be applied to model multiple recombination hotspots. However, for an intermediate or large number of hotspots, we would advocate developing a new forward-simulation algorithm rather than modifying the presented coalescent program; otherwise, one would have to consider not just one group with the hotspot and one without but many groups for all of the different combinations of the multiple hotspots.
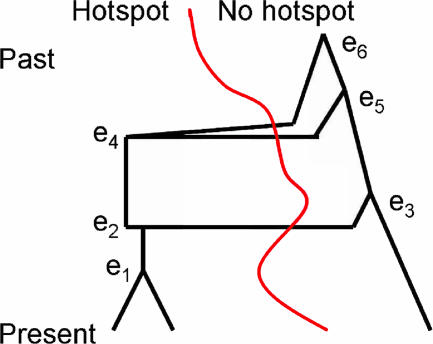
In this article, we have assumed that the single hotspot in the chromosomal region of interest originated once in the past. It is unclear how hotspots originate: they may be due to a de novo mutation, repeatedly introduced by transposable elements (2, 30), or some epigenetic factor (33). Fig. 3 shows the genealogy of a sample. An interesting observation is that, in some sense, it appears that the hotspot is created multiple times, whenever a recombination event causes a descendent to possess the motif even though its ancestor did not (when the black genealogy line crosses the red population curve). These instances, however, are due to the loss of DNA surrounding a double-strand break, and the subsequent copying of the existing motif from the other chromosome and not to recurrent creations of the motif.
Fig. 3.
Coalescent model. The sample has three chromosomes. The red curve represents the changing hotspot population frequency. Events e1, e3, e5, and e6 are coalescent events. Event e2 is a recombination event: one region with the hotspot motif is broken into two sections, only one of which has the motif. Event e4 is another recombination event: one region with the hotspot motif is broken into two sections, neither of which has the motif.
Materials and Methods
Diffusion Approximation.
To derive a diffusion approximation (e.g., refs. 42 and 43), we consider a Moran model (e.g., ref. 43). There are 2N chromosomes, each of which may have the recombination hotspot motif. At rate 2N2, a randomly selected chromosome dies and is replaced by the product of two randomly selected chromosomes. If both of these two chromosomes have the motif, then so does the replacement chromosome; if neither of these two chromosomes has the motif, then neither does the replacement. We refer the reader to Fig. 1 for the case when one of these two chromosomes has the hotspot motif and the other does not. In this case, each of the chromosomes suffers a double-strand break with probability r. With independent probability fr, the DNA at the motif's position is lost and copied from the other chromosome. Chromosomes with the hotspot motif suffer a double-strand break with an additional probability h and with independent probability fh: the motif sequence is lost and replaced with DNA from the other chromosome. Because of the low probabilities, we ignore the possibility of multiple breaks near the location of the motif. The probability that the hotspot motif is transmitted when the two chromosomes are heterozygous for the motif is,
 |
The rfr term comes from the case when the double-strand break affects the motif location on the chromosome without the motif, causing there to be two copies of the motif, one of which will be transmitted to the replacement chromosome; the term (1/2)(1 − hfh − 2rfr) comes from the case when there are no breaks on either chromosome near the motif location, so the motif will be transmitted to the replacement chromosome with probability 1/2. We take the usual diffusive limit as N increases to infinity. Define
and assume α is positive and finite. α is a scaled parameter proportional to the probability that a hotspot motif both causes a double-strand break, and the motif is then lost in the subsequent loss and copying of DNA. Let Xt be the fraction of chromosomes with the hotspot motif at time t. Then,
This same diffusion arises as a model for gene conversion (36) and for the Wright-Fisher model with selection (e.g., ref. 42). The case α = 0 models a neutral mutation, where a heterozygous individual transmits the mutation with probability 1/2. In the Results, we use this case for comparisons.
We are now able to use diffusion theory (e.g., refs. 42 and 43), to calculate many quantities of interest. The probability that a hotspot at frequency x < b achieves frequency b is
Eq. 4 can then be used to find the probability that a hotspot achieves frequency b: set x = ε, where ε is small, representing the single chromosome where the motif originated. Likewise, to find the probability that a hotspot currently at frequency x eventually fixes in the population, set b = 1. The expected time for a hotspot to go from frequency x to either fixation or loss is, in units of 2N generations,
 |
We consider the diffusion conditional on the hotspot frequency Yt eventually reaching zero,
 |
This diffusion is time-reversible (e.g., ref. 43), so we can use it to study the hotspot frequency going backwards in time. Then, conditioning on the hotspot originating at an arbitrarily small frequency, the mean age of a hotspot currently at frequency x is, in units of 2N generations,
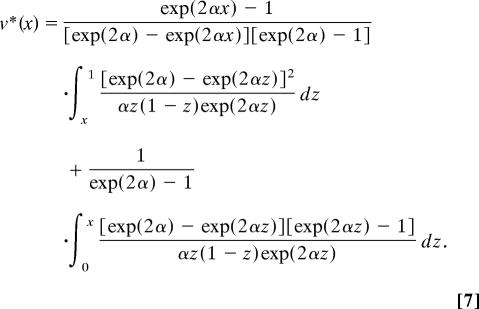 |
Coalescent Simulation.
We consider an equivalent coalescent model looking backwards in time (e.g., ref. 44). We refer the reader to Fig. 3. The population has a constant number 2N of chromosomes. We are interested in modeling the genealogy of a small sample s of chromosomes. We specify the initial number of chromosomes in the population Nh,t= 0 that possess the hotspot motif and the initial number of chromosomes in the sample sh,t= 0 with the motif. Note the number of chromosomes in the population without the motif Nr,t = 2N − Nh,t for all times t; the number of chromosomes in the sample without the motif is sr,t. This model is similar to some selection models (45) in the way it separates those chromosomes with and without the motif and in the way it uses the population frequency to govern the sample's genealogy. An ancestor possesses the hotspot motif if and only if its descendent does, unless a recombination event has possibly affected this inheritance. Going backwards in time, at each generation, two samples with the hotspot motif coalesce with probability
and two samples without the motif coalesce with probability
We use the diffusion equation (Eq. 6) to simulate the hotspot's frequency in the population in the previous generation, conditioned on the number of hotspots eventually decreasing to one at the point in the past when the motif originated. At each generation, each chromosome in the sample is paired with a chromosome from the population; whether this second chromosome has the hotspot motif is randomly determined based on the hotspot population frequency. The only genetic information of interest from this second chromosome is whether or not it possesses the motif. For each of these two chromosomes, with probability r, there is a double-strand break, and the location of the break is uniform in the region. For chromosomes harboring the motif, there is an additional probability h of a double-strand break due to the motif, and this break is always located in the middle of the region. Because of the small probabilities, we assume that there is, at most, one break per chromosome pair per generation. After a break, the chromosome with the break loses a random amount of DNA: a uniformly chosen number of base pairs between the parameters c1 and c2 is taken from both the right and left sides of the break. This loss is then replaced by copying from the other chromosome. The break may be resolved in a conversion or a cross-over event. The genetic material, including any mutations and the presence or absence of the hotspot motif, is transmitted as shown in Fig. 1. The hotspot motif is located a distance d to the right of the middle of the region. (We would like to emphasize that although we have decided to make the distance between the motif and the double-strand break deterministic and the amount of lost and copied DNA random, we could have made a similar model with the distance random and the amount deterministic or both the distance and the amount random. The important parameter is fh, the probability that a double-strand break due to the motif causes the loss of the motif in the subsequent loss and copying of DNA, which is a function of both the distance and the amount.)
We trace the genealogy of the original sample, and any copied regions due to the loss of an original sample's ancestral region, back to the most recent common ancestor. Note that different regions of the chromosome may have different most-recent common ancestors. A coalescent event decreases by one the number of pieces we have to track, whereas a recombination event increases this number by one. A recombination event may make an ancestor have the hotspot motif although its descendent does not, or vice versa. Once we have completed the genealogy, we then rain mutations down according to the infinite-site model (e.g., ref. 44): at each generation with probability m, there is a uniformly located mutation.
Next, we discuss the parameter values used in Table 1 in Results. The chromosomal region of interest is 100 kb. Breaks due to the hotspot occur at the middle of the region at position 50,000. The motif is d base pairs to the right of this position. The random amount of DNA lost to the right and left of a double-strand break is uniform between c1 = 100 and c2 = 200 base pairs. By varying the distance d, we vary the probability fh. For the entries in Table 1, d = 101 base pairs implies probability fh = 0.99; d = 110, fh = 0.90; and d = 190, fh = 0.10. The population size is N = 10,000 diploids; the sample size is s = 100 chromosomes. Per chromosome per generation, the mutation probability is m = 10−3, or 10−8 per base for the 100-kb region.Per chromosome per generation, the nonhotspot double-strand break probability is r = 5 × 10−4, or 5 × 10−9 per base; because there is a recombination event if either of two paired chromosomes suffers a break, the recombination probability is 10−3, or 10−8 per base. For chromosomes harboring the motif, there is an additional per generation break probability of h = 10−3. As discussed previously, this can be interpreted as a hotspot with width 1 kb and recombination probability elevated h1 = 100 times above the genome average. The probability that a double-strand break is resolved as a cross-over, as opposed to a conversion, is one.
Acknowledgments
I thank Norman Arnheim and Simon Tavaré for useful discussions. This work was supported by National Human Genome Research Institute Center of Excellence in Genomic Science Grant P50 HG002790 (M. Waterman, principal investigator).
Footnotes
The author declares no conflict of interest.
This article is a PNAS direct submission.
References
- 1.Crawford D, Bhangale T, Li N, Hellenthal G, Rieder M, Nickerson D, Stephens M. Nat Genet. 2004;36:700–706. doi: 10.1038/ng1376. [DOI] [PubMed] [Google Scholar]
- 2.Myers S, Bottolo L, Freeman C, McVean G, Donnelly P. Science. 2005;310:321–324. doi: 10.1126/science.1117196. [DOI] [PubMed] [Google Scholar]
- 3.The International HapMap Consortium. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jeffreys A, Richie A, Neumann R. Hum Mol Genet. 2000;9:724–733. doi: 10.1093/hmg/9.5.725. [DOI] [PubMed] [Google Scholar]
- 5.Jeffreys A, Kauppi L, Neumann R. Nat Genet. 2001;29:217–222. doi: 10.1038/ng1001-217. [DOI] [PubMed] [Google Scholar]
- 6.Tiemann-Boege I, Calabrese P, Cochran D, Sokol R, Arnheim N. PLoS Genet. 2006;2:0020070. doi: 10.1371/journal.pgen.0020070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kauppi L, Jeffreys A, Keeney S. Nat Rev Genet. 2004;5:413–424. doi: 10.1038/nrg1346. [DOI] [PubMed] [Google Scholar]
- 8.Carrington M, Cullen M. Trends Genet. 2004;20:196–205. doi: 10.1016/j.tig.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 9.Wiuf C, Posada D. Genetics. 2003;164:407–417. doi: 10.1093/genetics/164.1.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li N, Stephens M. Genetics. 2003;165:2213–2233. doi: 10.1093/genetics/165.4.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McVean G, Myers S, Hunt S, Deloukas P, Bentley D, Donnelly P. Science. 2004;304:581–584. doi: 10.1126/science.1092500. [DOI] [PubMed] [Google Scholar]
- 12.Hudson R. Theor Popul Biol. 1983;23:183–201. doi: 10.1016/0040-5809(83)90013-8. [DOI] [PubMed] [Google Scholar]
- 13.Griffiths R, Marjoram P. J Comp Biol. 1996;3:479–502. doi: 10.1089/cmb.1996.3.479. [DOI] [PubMed] [Google Scholar]
- 14.Griffiths R, Marjoram P. In: Progress in Population Genetics and Human Evolution. Donnelly P, Tavaré S, editors. New York: Springer; 1997. pp. 257–270. [Google Scholar]
- 15.Simonsen K, Churchill G. Theor Popul Biol. 1997;52:43–59. doi: 10.1006/tpbi.1997.1307. [DOI] [PubMed] [Google Scholar]
- 16.Wiuf C, Hein J. Theor Popul Biol. 1999;55:248–259. doi: 10.1006/tpbi.1998.1403. [DOI] [PubMed] [Google Scholar]
- 17.Neumann R, Jeffreys A. Hum Mol Genet. 2006;15:1401–1411. doi: 10.1093/hmg/ddl063. [DOI] [PubMed] [Google Scholar]
- 18.Winckler W, Myers S, Richter D, Onofrio R, McDonald G, Bontrop R, McVean G, Gabriel S, Reich D, Donnelly P, Altshuler D. Science. 2005;308:107–111. doi: 10.1126/science.1105322. [DOI] [PubMed] [Google Scholar]
- 19.Ptak S, Hinds D, Koehler K, Nickel B, Patil N, Ballinger D, Przeworski M, Frazer K, Paabo S. Nat Genet. 2005;37:429–434. doi: 10.1038/ng1529. [DOI] [PubMed] [Google Scholar]
- 20.Szostak J, Orr-Weaver T, Rothstein R, Stahl F. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- 21.Petes T. Nat Rev Genet. 2001;2:360–369. doi: 10.1038/35072078. [DOI] [PubMed] [Google Scholar]
- 22.Boulton A, Myers R, Redfield R. Proc Natl Acad Sci USA. 1997;94:8058–8063. doi: 10.1073/pnas.94.15.8058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pineda-Krch M, Redfield R. Genetics. 2005;169:2319–2333. doi: 10.1534/genetics.104.034363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Steiner W, Smith G. Mol Cell Biol. 2005;25:9054–9062. doi: 10.1128/MCB.25.20.9054-9062.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Steiner W, Schreckhise R, Smith G. Mol Cell. 2002;9:847–855. doi: 10.1016/s1097-2765(02)00489-6. [DOI] [PubMed] [Google Scholar]
- 26.Jeffreys A, May C. Nat Genet. 2004;36:151–156. doi: 10.1038/ng1287. [DOI] [PubMed] [Google Scholar]
- 27.Smith G, Amundsen S, Chaudhury A, Cheng K, Ponticelli A, Roberts C, Schultz D, Taylor A. Cold Spring Harb Symp Quant Biol. 1984;49:485–495. doi: 10.1101/sqb.1984.049.01.055. [DOI] [PubMed] [Google Scholar]
- 28.Myers R, Stahl F. Ann Rev Genet. 1994;28:49–70. doi: 10.1146/annurev.ge.28.120194.000405. [DOI] [PubMed] [Google Scholar]
- 29.Fox M, Yamada T, Ohta K, Smith G. Genetics. 2000;156:59–68. doi: 10.1093/genetics/156.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Myers S, Spencer C, Auton A, Bottolo L, Freeman C, Donnelly P, McVean G. Biochem Soc Trans. 2006;34:526–530. doi: 10.1042/BST0340526. [DOI] [PubMed] [Google Scholar]
- 31.Jeffreys A, Neumann R. Nat Genet. 2002;31:267–271. doi: 10.1038/ng910. [DOI] [PubMed] [Google Scholar]
- 32.Jeffreys A, Neumann R. Hum Mol Genet. 2005;14:2277–2287. doi: 10.1093/hmg/ddi232. [DOI] [PubMed] [Google Scholar]
- 33.Clark A. Nat Genet. 2005;37:563–564. doi: 10.1038/ng0605-563. [DOI] [PubMed] [Google Scholar]
- 34.Nordborg M. In: Handbook of Statistical Genetics. Balding D, Bishop M, Cannings C, editors. New York: Wiley; 2001. pp. 179–212. [Google Scholar]
- 35.Nagylaki T. Proc Natl Acad Sci USA. 1983;80:5941–5945. doi: 10.1073/pnas.80.19.5941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Naglyaki T. Proc Natl Acad Sci USA. 1983;80:6278–6281. doi: 10.1073/pnas.80.20.6278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wiuf C, Hein J. Genetics. 2000;155:451–462. doi: 10.1093/genetics/155.1.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wiuf C. Theor Popul Biol. 2000;57:357–367. doi: 10.1006/tpbi.2000.1462. [DOI] [PubMed] [Google Scholar]
- 39.Morton N. Outline of Genetic Epidemiology. Basel: Karger; 1982. [Google Scholar]
- 40.The Chimpanzee Sequencing Analysis Consortium. Nature. 2005;437:69–87. [Google Scholar]
- 41.Templeton A. Nature. 2002;416:45–51. doi: 10.1038/416045a. [DOI] [PubMed] [Google Scholar]
- 42.Karlin S, Taylor H. A Second Course in Stochastic Processes. New York: Academic; 1981. [Google Scholar]
- 43.Ewens W. Mathematical Population Genetics. New York: Springer; 2004. [Google Scholar]
- 44.Hudson R. Oxford Surv Evol Biol. 1990;7:1–44. [Google Scholar]
- 45.Kaplan N, Darden T, Hudson R. Genetics. 1988;120:819–829. doi: 10.1093/genetics/120.3.819. [DOI] [PMC free article] [PubMed] [Google Scholar]