The extraordinary diversity of sex-determination mechanisms has long been noted. Many species resort to environmental cues for the determination of sex. A well known example is temperature-dependent sex determination in many (although not all) reptiles. Yet, many other species use genetic mechanisms, i.e., sex chromosomes, for the determination of sex. The best-studied systems of genetic sex determination include the XX:XY system that arose independently in mammals and Drosophila, the XX:XO system in Caenorhabditis elegans, and the ZZ:ZW system in birds. It is generally accepted that environmental sex determination is the ancestral state and that genetic sex determination evolved as a derived condition. It is also recognized that genetic sex determination is evolutionarily highly labile, having evolved into existence on many independent occasions across diverse taxa. A case in point is sex-determination mechanisms in amniotes (a clade encompassing reptiles, birds, and mammals). The ancestral state in amniotes is likely temperature-dependent sex determination, which is still found in many extant reptilian species, such as crocodilians and some turtles and lizards (1). From this ancestral state, genetic sex determination evolved in birds, which utilize the ZZ:ZW system, and also independently in mammals, which use the XX:XY system. The ZZ:ZW system is also found in all snake species. The split of the mammalian lineage from the rest of amniotes occurred ≈315 million years ago, whereas the split between Lepidosauria (including snakes and lizards) and Archosauromorpha (encompassing crocodilians, birds, and possibly turtles) occurred ≈260 million years ago (1). In this phylogenetic context, the sharing of the ZZ:ZW system by both birds and snakes can be explained by either of two scenarios. One is that the ZZ:ZW system arose during the 55 million years after the splitting away of the mammal lineage but before the divergence between Lepidosauria and Archosauromorpha, with subsequent reversion to temperature-dependent sex determination in some Lepidosauria/Archosauromorpha species. The other scenario is that birds and snakes evolved the ZZ:ZW system independently. In this issue of PNAS, Matsubara et al. (2) provide definitive evidence for the latter scenario. As such, their results extend our understanding of the lability of sex-chromosome evolution in amniotes.
Matsubara et al. (2) used FISH to map the chromosomal locations of 109 cDNA clones in snakes. They found that genes present on snake sex chromosomes mapped to autosomes in both mammals and birds. Additionally, genes from bird sex chromosomes mapped to an autosome in the snake. These results demonstrated unambiguously a separate origin of snake sex chromosomes compared with the sex chromosomes in birds or mammals. Thus, the ZZ:ZW system has arisen at least twice during amniote evolution, once in the ancestors of birds and once in the ancestors of snakes. Furthermore, the autosome being converted to sex chromosomes in birds is different from that in snakes (Fig. 1).
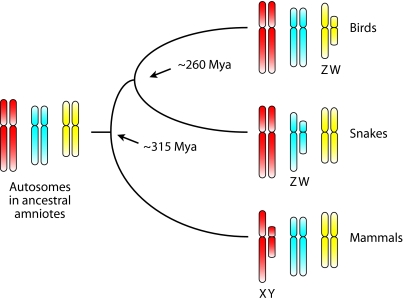
Fig. 1.
Independent origins of sex chromosomes in birds, snakes, and mammals. In ancestral amniotes, which presumably used temperature-dependent sex determination, there were no sex chromosomes. Sex chromosomes then evolved from autosomes on three independent occasions in birds, snakes, and mammals. A different autosome was converted to sex chromosomes in each of these three lineages. The ZZ:ZW system emerged twice (once in birds and once in snakes), whereas the XX:XY system emerged once in mammals.
One popular model of sex-chromosome evolution postulates that the sex-determining locus first arises when an autosomal gene involved in environmental sex determination acquires a new mutation that consistently gives rise to either male (in the case of the XX:XY system) or female (in the case of the ZZ:ZW system) development. In mammals, it is believed this transition occurred when a mutation turned the SOX3 gene into the male-determining SRY gene on the Y chromosome (3, 4). In birds, a mutation in the DMRT1 gene is hypothesized to have originated the sex-determining locus (5). For snakes, however, Matsubara et al. (2) show that both SOX3 and DMRT1 are located on autosomes, implicating another as-yet-unidentified gene as the sex-determining locus.
After the emergence of the sex-determining locus, the pair of autosomes bearing the locus become sex chromosomes. Initially, the two sex chromosomes are essentially identical except at the sex-determining locus. However, the two sex chromosomes are destined for an evolutionary trajectory whereby they become progressively differentiated from each other (6). The homogametic sex chromosome (X or Z) more or less maintains its initial size and gene content, although it does acquire some of its own characteristics, such as increased rate of gene trafficking to and away from the chromosome and a mechanism of dosage compensation (6–8). The heterogametic sex chromosome (Y or W), on the other hand, tends to wither away slowly, losing both gene content and size (6). This genetic decay occurs as recombination is suppressed between the sex chromosomes and the heterogametic chromosome accumulates deleterious mutations and deletions without the ability to remove them (6). The process of Y degeneration remains incomplete in mammals, as evidenced by the presence of gametologs (X-Y homologous genes) on the two sex chromosomes and the persistence of pseudoautosomal regions where recombination between X and Y still occurs in male meioses. In birds, the W chromosome remains even more intact, although still diminutive compared with the Z.
Genetic sex determination is evolutionarily highly labile.
The work by Matsubara et al. (2) shows that the phylogenetic depth of snake lineages can be leveraged to take informative snapshots of the W degeneration process (2). In the Burmese python, the most ancient of the snakes studied, the Z and W chromosomes remain largely undifferentiated from each other based on cytogenetic criteria. At a more intermediate level, the W chromosome of the Japanese four-striped rat snake has lost both size and genes during evolution, whereas its cytogenetic banding patterns have clearly diverged from the Z chromosome. By the time the evolutionarily recent habu arose, nearly all genes under study and half the size had vanished from the W chromosome, leaving it clearly impoverished compared with its partner, the Z chromosome.
Despite our current appreciation of the origins and evolutionary fates of the sex chromosomes, it remains unclear how and why the path from environmental to genetic sex determination should have occurred in so many different fashions and at so many different times. The knowledge of another independent origin of sex chromosomes in snakes offers yet more data to those seeking to understand these questions. Indeed, given their phylogenetic diversity, snakes may prove to be a most informative system for the study of sex-chromosome evolution.
Footnotes
The authors declare no conflict of interest.
See companion article on page 18190.
References
- 1.Smith CA, Sinclair AH. BioEssays. 2004;26:120–132. doi: 10.1002/bies.10400. [DOI] [PubMed] [Google Scholar]
- 2.Matsubara K, Tarui H, Toriba M, Yamada K, Nishida-Umehara C, Agata K, Matsuda Y. Proc Natl Acad Sci USA. 2006;103:18190–18195. doi: 10.1073/pnas.0605274103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Foster JW, Graves JA. Proc Natl Acad Sci USA. 1994;91:1927–1931. doi: 10.1073/pnas.91.5.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lahn BT, Pearson NM, Jegalian K. Nat Rev Genet. 2001;2:207–216. doi: 10.1038/35056058. [DOI] [PubMed] [Google Scholar]
- 5.Smith CA, Katz M, Sinclair AH. Biol Reprod. 2003;68:560–570. doi: 10.1095/biolreprod.102.007294. [DOI] [PubMed] [Google Scholar]
- 6.Vallender EJ, Lahn BT. BioEssays. 2004;26:159–169. doi: 10.1002/bies.10393. [DOI] [PubMed] [Google Scholar]
- 7.Wang PJ, McCarrey JR, Yang F, Page DC. Nat Genet. 2001;27:422–426. doi: 10.1038/86927. [DOI] [PubMed] [Google Scholar]
- 8.Emerson JJ, Kaessmann H, Betran E, Long M. Science. 2004;303:537–540. doi: 10.1126/science.1090042. [DOI] [PubMed] [Google Scholar]