Abstract
Eukaryotic mRNAs often recruit ribosomal subunits some distance upstream of the initiation codon; however, the mechanisms by which they reach the initiation codon remain to be fully elucidated. Although scanning is a widely accepted model, evidence for alternative mechanisms has accumulated. We previously suggested that this process may involve tethering of ribosomal complexes to the mRNA, in which the intervening mRNA is bypassed, or clustering, in which the initiation codon is reached by dynamic binding and release of ribosomal subunits at internal sites. The present studies tested the feasibility of these ideas by using model mRNAs and revealed that translation efficiency varied with the distance between the site of ribosomal recruitment and the initiation codon. The present studies also showed that translation could initiate efficiently at AUG codons located upstream of an internal site. These observations are consistent with ribosomal tethering at the cap structure and clustering at internal sites.
Keywords: 5′ leader, AUG codon, mRNA, ribosome
Translation initiation in eukaryotic cells is a key step in regulating protein expression. The first step in this process involves recruitment of a 40S ribosomal subunit, either at the m7G cap structure at the 5′-end of the mRNA or at an internal ribosome entry site (IRES) contained within the mRNA. In some cases, such as translation of the cricket paralysis virus RNA, this recruitment occurs at or near the initiation codon (1). For most mRNAs, however, ribosomal subunit recruitment occurs some distance upstream of the initiation codon. Thus, before protein synthesis can commence, the ribosomal subunits must reach the initiation codon. A variety of mechanisms, including linear scanning, leaky scanning, and shunting of the 5′ leaders of mRNAs, have been proposed to explain how this process occurs (2). Although linear scanning is a widely held model (3), observations in a number of instances are difficult to reconcile with this mechanism (see Discussion and refs. 4–15). For example, translation does not always initiate at the first AUG codon, even when it resides in a good context (7, 8). Indeed, up to 40% of human mRNAs have been reported to contain one or more AUG codons in their 5′ leader regions (14, 15). Although the authenticity of some of these 5′ leaders has been questioned, many others appear to be indisputable (16). Moreover, the number of upstream AUG (uAUG) codons in these mRNAs is related to the length of the 5′ leader, suggesting that there is no selective pressure to remove them. Data from several studies indicate that the relative utilization of two or more AUG codons is determined in large part by their distance from the ribosomal recruitment site (4, 9, 10). In addition, for several mRNAs it has been reported that as ribosomal subunits move to the initiation codon they appear to bypass or shunt segments of the 5′ leaders (5–8, 17). Moreover, our earlier studies showed that, by base-pairing to 18S rRNA, a short mRNA element from the 5′ leader of the Gtx homeodomain mRNA could facilitate ribosomal shunting (17–19).
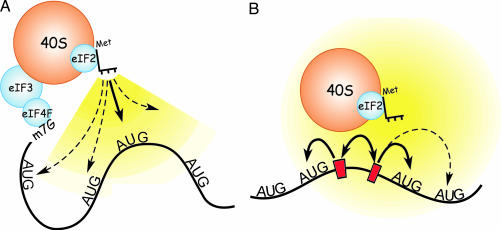
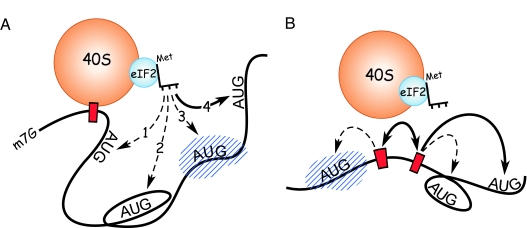
On the basis of these and other observations, we hypothesized that either tethering or clustering of the translation machinery could explain these results (17, 19). Tethering is a mechanism in which ribosomal subunits reach the initiation codon while bound to a fixed point in the mRNA (Fig. 1A). In such a tethered complex, the intervening mRNA sequences are bypassed when the initiator Met-tRNA base-pairs to the initiation codon, and this process may involve folding of the intervening sequences. In contrast, clustering does not require the ribosomal subunit to be tethered to the mRNA for it to reach the initiation codon (Fig. 1B). Rather, clustering is a dynamic process involving reversible binding of the ribosomal subunit to and detachment from various sites in the mRNA. This reversible binding at a number of sites would increase the local ribosomal subunit concentration and enhance the probability of binding of the initiator Met-tRNA to an AUG codon in the vicinity of such sites.
Fig. 1.
Schematic representations of ribosomal tethering and clustering. (A) Representation of the hypothesized ribosomal tethering at the cap structure. The 40S ribosomal subunit is shown tethered to the 5′ end of the mRNA at the cap structure (m7G), via the eIF4F complex of initiation factors and initiation factor eIF3. The 40S subunit interacts with an AUG initiation codon by means of the initiator Met-tRNA of the ternary complex, which also contains eIF2 and GTP, which are not shown. In both A and B, the range and probability of interaction with the various AUG codons is indicated schematically by the shaded yellow regions; brighter yellow indicates a higher probability of binding to an AUG codon in the vicinity (bold arrow). Pale yellow and dashed arrows indicate lower probabilities of binding to an AUG codon. (B) Schematic model of translation initiation by means of dynamic ribosomal clustering. Clustering of the translation machinery (indicated by the double-headed arrow) involves binding to and detaching from sites in the mRNA (indicated by red bars), increasing the probability of binding to an AUG codon in the vicinity.
Two key predictions of the tethering and clustering hypotheses are that (i) translation efficiency will be determined in large part by the distance between the ribosomal recruitment site and the initiation codon and that (ii) translation can initiate at AUG codons located both upstream and downstream of IRESs. These predictions are consistent with various observations from other laboratories (4, 9, 10, 13), but they have not yet been addressed systematically. Here we test the operation of these hypothesized mechanisms by using model mRNAs that contain either the cap structure or Gtx elements as ribosomal subunit recruitment sites.
Results
Evidence for Ribosomal Tethering at the Cap Structure.
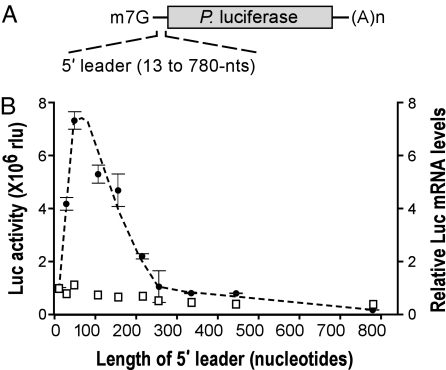
To determine whether translation efficiency is affected by distance between the cap structure and the initiation codon, we generated monocistronic Photinus luciferase reporter mRNAs (Fig. 2A). These mRNAs contained 5′ leaders that ranged in length from 13 to 780 nt and were composed of segments of, or multiple copies of, the 52-nt β-globin 5′ leader. The β-globin 5′ leader was selected because it efficiently mediates cap-dependent translation and does not appear to contain any sequence elements or RNA secondary structures that might significantly affect translation (20, 21). In addition, none of the 5′ leaders indicated in Fig. 2A, including up to 30 nt of the coding region, are predicted to form RNA structures more stable than the single β-globin 5′ leader (−9.5 kcal/mol; 1 cal = 4.18 J) as determined by analysis using the M-Fold algorithm (22).
Fig. 2.
Translation is affected by the distance between the cap structure and the initiation codon. (A) Schematic representation of Photinus luciferase reporter mRNAs, with the m7G cap structure and poly(A) tail [(A)n]. The 5′ leaders were derived from β-globin 5′ leader sequences and ranged in length from 13 to 780 nt (see Table 1). (B) Luciferase (Luc) activities from constructs transiently transfected into N2a cells are plotted as black dots with SEM indicated. Relative luciferase mRNA levels determined by RNase protection assays are indicated as open squares and are normalized with a value of 1.0 for the mRNA with the shortest (13 nt) 5′ leader.
Data obtained with these constructs in transiently transfected neuroblastoma N2a cells showed that luciferase activities varied with the length of the 5′ leader (Fig. 2B); maximal activity was observed for the mRNA with a single copy of the 52-nt β-globin 5′ leader and declined progressively with either increasingly longer or shorter 5′ leader sequences. Although ribonuclease protection assays revealed that the most efficiently translated mRNAs were present at somewhat higher levels than the mRNAs that were least efficiently translated, these differences in mRNA levels could not account for the differences in activity (Fig. 2B). For example, the luciferase activities obtained with the mRNAs containing 13-nt and 52-nt 5′ leaders differed by 7.8-fold, but their mRNA levels differed by only 15%. Results similar to those in Fig. 2B were obtained using a Renilla luciferase cistron, suggesting that these results were not due to features in the coding sequence (data not shown).
The possibility that the rapid decline in translation observed with the 5′ leader length increasing between 52 and 260 nt was due to reiterated secondary structures appears unlikely because translation levels of mRNAs with even longer 5′ leaders (for example, between 260 and 442 nt) were of similar magnitude. These effects of 5′ leader length on translation (Fig. 2B) are not easily explained by the notion of ribosomal scanning and are more consistent with a model of translation initiation that involves tethering of the ribosomal complex to the cap structure, a notion that is developed further in Discussion.
Dynamic Local Clustering of Ribosomal Subunits.
Although the above studies are consistent with the notion of ribosomal tethering at the cap structure, our earlier studies using an mRNA element from the Gtx mRNA, a ribosomal recruitment site that functions both as an IRES (23) and as a translational enhancer (18), suggested that another mechanism of ribosomal movement that involves the dynamic local clustering of ribosomal subunits (17) can be used. In that study, we showed that an uAUG codon or stable hairpin structure was efficiently shunted by a mechanism that required the base-pairing of the Gtx elements to 18S rRNA. Inasmuch as 18S rRNA contains only one binding site for the Gtx element and efficient shunting was observed only when the obstacles were flanked by Gtx elements, we postulated that the shunting required ribosomal subunits recruited at an upstream Gtx element to dissociate from the upstream site and reassociate with a Gtx element located downstream of the obstacle (17, 24).
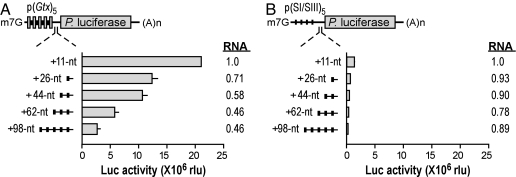
To investigate in the present study how translation might be affected by the distance between the initiation codon and the Gtx element, we performed transfection experiments using monocistronic reporter mRNAs containing five linked copies of this element in their 5′ leaders (Fig. 3A). The activities from these reporter mRNAs were compared with size-matched control mRNAs lacking these elements (Fig. 3B). As in our earlier studies (18, 23, 24), five linked Gtx elements were used to increase the signal-to-noise ratio seen with fewer copies. This group of elements was placed at various distances ranging from 11 to 98 nt upstream of the luciferase initiation codon using a spacer sequence (SI/SIII) that we previously showed does not enhance translation (23). Application of the M-Fold algorithm (22) indicated that these SI/SII spacer sequences were relatively unstructured and that none of them were predicted to form RNA structures more stable than −5.4 kcal/mol.
Fig. 3.
Translation is affected by distance between an IRES element and the initiation codon. (A) Schematic representation of Photinus luciferase (Luc) reporter mRNAs with five copies of the Gtx element in the 5′ leader, spaced 11–98 nt upstream of the initiation codon (see Table 1). Gtx elements are indicated as gray vertical bars, SI spacer sequences are indicated as thick black bars, and SIII spacer sequences are indicated as thin lines. The histogram shows luciferase activities in transiently transfected N2a cells. Relative luciferase mRNA levels are shown to the right and are normalized in reference to a value of 1.0 for the sample with the shortest (11 nt) 5′ leader. (B) Size-matched controls in which SIII spacer sequences replace Gtx elements.
Maximal luciferase activity was observed with the mRNA in which the Gtx elements were located closest to the initiation codon (11 nt); activity decreased as these elements were spaced progressively further upstream. As in Fig. 2, the most efficiently translated mRNAs were present at somewhat higher levels than the mRNAs that were least efficiently translated, but these differences could not account for the differences in activity.
Size-matched control mRNAs, which contained only the cap structure as a ribosomal recruitment site, yielded luciferase activities that were ≈13% or less than those obtained with mRNAs containing the Gtx elements (Fig. 3B). Translation of the control mRNAs decreased with increasing 5′ leader lengths. The 5′ leader lengths of these control constructs were all >90 nt, and the observed trend in activity was similar to that observed with the constructs in Fig. 2B for mRNAs with comparable 5′ leader lengths. The similar patterns of activity obtained by increasing the lengths of two different 5′ leader sequences suggested that these results were not due to peculiarities of the individual mRNA sequences.
The results obtained in this experiment are consistent with a dynamic clustering mechanism in which the local concentration of ribosomal subunits increases the probability of initiating translation in the immediate vicinity of the Gtx elements.
Translation Can Initiate Upstream of a Recruitment Site.
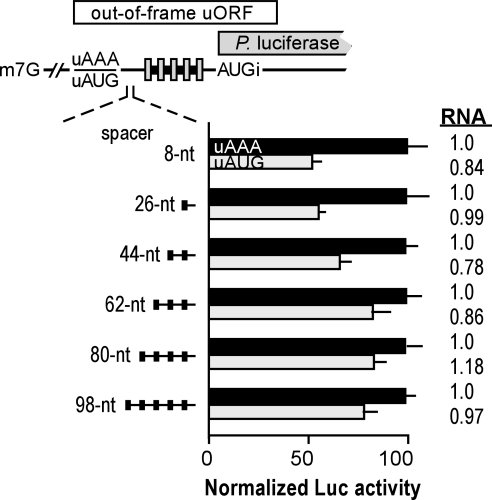
Dynamic clustering of the translation machinery at an internal mRNA element (Fig. 1B) also predicts that ribosomal subunits can move upstream or downstream after binding to such a site (17). We tested this notion by using luciferase reporter mRNAs, each of which contained an AUG codon located various distances upstream of both the authentic luciferase AUG codon and the Gtx elements (Fig. 4). These constructs were designed such that translation initiating at the uAUG codons, which were out of frame with the luciferase cistron and overlapped it in part, would inhibit translation of luciferase by diverting ribosomes from the luciferase initiation codon. Under these conditions, the extent to which luciferase translation is inhibited provides an indication of the amount of translation initiating at the uAUG codon.
Fig. 4.
Translation can initiate at AUG codons located upstream of multiple Gtx elements. The upper diagram is a schematic representation of mRNA reporter constructs. The uAUG codons in one set of constructs generates a coding region, indicated as a large horizontal white bar, that overlaps the luciferase cistron in a different reading frame, indicated as a large horizontal gray bar. The uAUG was spaced various distances upstream of the Gtx elements by using SI/SIII spacer sequences. Size-matched control constructs contained AAA in place of the uAUG (see Table 1). The histogram shows normalized luciferase activities in transiently transfected N2a cells. In each set of constructs, the relative luciferase activities are normalized by using a value of 100 for the activity of the uAAA construct. Black and gray bars indicate data from upstream AAA (uAAA)- and uAUG-containing constructs, respectively. Relative luciferase mRNA levels are shown to the right and are normalized by using a value of 1.0 for the upstream AAA construct having the shortest spacer sequence (8 nt).
The Gtx elements in these constructs were spaced 11 nt upstream of the luciferase AUG initiation codon. The uAUG codons, which resided in an optimal context (ACCAUGGA; the uAUG is italicized, and key flanking nucleotides are bold), were spaced various distances upstream of the Gtx elements by using repeats of the SI/SIII sequence. Inasmuch as the distances between the uAUG and the cap (150 nt) and between the Gtx elements and the luciferase cistron (11 nt) were the same in all constructs, any differences in uAUG utilization would be attributable to the spacing between the uAUG codon and the Gtx elements.
After transfection into N2a cells, the luciferase activities of constructs containing the uAUG codons were compared with those obtained with size-matched control mRNAs, which lacked the uAUG and contained the triplet AAA in its place. The results indicated that the uAUG codons were used to different extents depending on their distance from the Gtx elements (Fig. 4). Maximal utilization occurred when the AUG codon was spaced at <26 nt upstream of the Gtx elements, resulting in an ≈50% inhibition of luciferase activity (8 and 26 nt). This result suggested that up to half of the translation initiation events occurred upstream of the ribosomal recruitment site. However, the translation of the luciferase cistron may have been further decreased by ribosomes engaged in elongation of the peptide encoded by the upstream ORF, which may have masked the Gtx elements and the luciferase initiation codon.
As the distance between the uAUG codons and the Gtx elements increased, the uAUG codons were used progressively less, inhibiting luciferase activity by only ≈20% when the distance was >62 nt. The residual inhibition was most likely due to initiation at the uAUG by ribosomal subunits recruited at the cap structure. Ribonuclease protection assays revealed no significant differences in mRNA levels for the size-matched constructs (Fig. 4).
Discussion
In eukaryotes, the initiation of translation of various different cellular and viral mRNAs does not appear to occur by a single mechanism but rather by a multiplicity of mechanisms (1–3, 5, 6). Experiments using model mRNAs provide an opportunity to investigate factors influencing these mechanisms. In the present studies, we found that translation initiated most efficiently when the AUG codon was spaced ≈50 nt downstream of the cap structure (Fig. 2B). In addition, we found that translation decreased with distance from an internal ribosomal recruitment site (Fig. 3A) and that it could initiate translation efficiently at AUG codons located either upstream or downstream of such a recruitment site (Fig. 4).
A variety of studies using both natural and model mRNAs have revealed the effects of altering the distance between the ribosomal recruitment site and AUG codons on translation (4, 9, 11, 12). For example, in the case of the yeast MOD5 mRNA (9), the relative utilization of two AUG codons could be dramatically altered when the distance between these codons and the cap structure was altered. The first AUG codon was used ≈5–10% of the time when spaced 10 nt from the 5′ end of the mRNA; when the spacing of this AUG codon was increased up to 65 nt, it was used exclusively. Comparable results were obtained in a second example using model mRNAs containing two AUG codons (31). In this study, the relative utilization of the first AUG codon increased progressively from ≈40% to almost 100% when its spacing was increased incrementally from 3 to 32 nt from the 5′ end of the mRNA. These results are consistent with our results in Fig. 2.
The effects of distance on translation were also seen in a study of the turnip yellow mosaic virus RNA (10). In this RNA, ≈65% of translation was initiated at the first of two closely spaced AUG codons. However, when the second AUG codon was spaced farther downstream, its utilization decreased dramatically, whereas that of the uAUG codon increased, so that almost all translation initiated at this codon. The ability of the position of the second AUG codon to affect the utilization of the uAUG codon suggests that the two codons competed for the translation machinery and that this competition was affected by their distances from the cap structure.
Experiments using model mRNAs showed that translation initiated preferentially at an AUG codon located closest to the ribosomal recruitment site, either the cap structure or poly(A) tail (11). When an mRNA designated BIGCAT was capped but not polyadenylated, translation initiated at the first of two AUG codons (see Fig. 2 in ref. 11). When this mRNA was both capped and polyadenylated, translation initiated at both AUGs but predominantly at the uAUG codon. However, when this mRNA was polyadenylated but lacked a cap structure, translation initiated predominantly at the downstream AUG codon.
Effects on translation of the distance between an IRES and AUG codons also have been observed. For example, in the encephalomyocarditis virus mRNA, translation normally initiates at AUG-11, which is the second AUG codon located downstream of an IRES in this RNA (12). However, the relative utilization of different AUG codons could be switched by altering their distance from the IRES. For example, shortening the distance between the IRES and AUG-11 resulted in almost exclusive utilization of AUG-12, albeit at a reduced efficiency. Lengthening the distance increased utilization of AUG-10 and reduced utilization of AUG-11.
All of these examples involve AUG codons located downstream of ribosomal recruitment sites. A natural example of translation that initiates upstream of a ribosomal recruitment site is provided by the HIV-2 RNA. In this RNA, it was shown that translation of a Gag protein initiates at an AUG codon located 50 nt upstream of an IRES that is contained entirely within the coding sequence (13).
Can ribosomal scanning explain the results in the studies discussed above and in the present studies? The inefficient translation observed when an AUG codon was spaced very close to the ribosomal recruitment site (Fig. 2B) (12) could be consistent with the notion of scanning if a minimum distance from the ribosomal recruitment site were required to load ribosomal subunits onto the mRNA (discussed in ref. 25). A scanning mechanism does not, however, readily explain why translation at AUG codons located farther from the ribosomal recruitment site becomes less and less efficient with increasing distance (Fig. 2B and 3B) (4, 9, 10, 12) unless, for example, one invokes a diminishing rate of ribosomal scanning as 5′ leader lengths increase or if scanning ribosomal subunits are released from the mRNA at a very high rate such that fewer scanning subunits reach the initiation codon as 5′ leader length increases. Although such mechanisms appear feasible, studies showing that translation can initiate at AUG codons located upstream of a ribosomal recruitment site (Fig. 4) (13) are not easily explained by ribosomal scanning. Extensive “backward” scanning (of >60 nt) would be required to account for the translation initiation we observed at AUG codons located upstream of ribosomal recruitment sites. A recent study has provided the suggestion that backward movement can occur but only to a maximum of 15 nt (10). Nevertheless, the scanning hypothesis does not appear generally to account for this type of movement (26).
We suggest that these various observations and our own data are more readily explained by mechanisms that involve tethering of the ribosomal complex to the mRNA or clustering of ribosomal subunits at internal sites (Fig. 1). For example, results showing that translation initiates most efficiently when the AUG codon is located a specific distance downstream of the cap structure or an IRES would be predicted to occur if the ribosomal subunit was tethered to the mRNA and recognition of the AUG codon occurred by direct binding of the initiator Met-tRNA. At suboptimal distances, translation would be inefficient because of steric effects, i.e., the mRNA tether would be too short to enable the translation machinery to reorient to allow optimal binding of the initiator Met-tRNA to the initiation codon. When the 5′ leader is sufficiently long (≈50-nt in Fig. 2B), translation would be more efficient because steric effects are minimal, and the likelihood of the initiator Met-tRNA binding to the initiation codon is increased. With increasingly longer 5′ leaders, however, translation becomes less efficient because the probability of locating the initiation codon is diminished.
The notion of ribosomal clustering, which involves the binding and release of ribosomal complexes at specific mRNA elements, was supported by the results of our earlier studies (17). This mechanism can explain why translation decreases progressively at AUG codons located at increasing distances upstream or downstream from the Gtx elements (Figs. 3 and 4). For the present experiments, however, we cannot completely rule out that some translation initiation events occurred while the ribosomal subunits were tethered to Gtx elements.
Ribosomal tethering or clustering can account for numerous other observations, including ribosomal shunting (17). Moreover, these mechanisms can explain why translation does not always initiate at the first AUG codon, even if it resides in a good context. The operation of these mechanisms may also explain why individual mRNA secondary structures are or are not inhibitory to translation. An RNA secondary structure may affect translation mediated by tethered or clustered ribosomal complexes by altering the flexibility of the mRNA or by masking mRNA elements, including ribosomal recruitment sites as well as the initiation codon (Figs. 5). Inasmuch as the 5′ leaders of different mRNAs are likely to have different conformations and to interact with different factors, the optimal distance from the ribosomal recruitment site for translation initiation is likely to vary for each mRNA.
Fig. 5.
Schematic representation of factors affecting translation efficiency mediated by ribosomal tethering or clustering. (A) The 40S subunit is shown tethered directly to an internal site in the mRNA (indicated by a red box). Tethering may also occur through intermediary factors or via the cap structure (see Fig. 1A). Dashed arrows represent inefficient initiation due to steric effects (dashed arrow 1), masking of an AUG codon by mRNA secondary structure (enclosing an AUG) (dashed arrow 2), and masking of an AUG codon by an RNA-binding protein (indicated as a hatched blue object) (dashed arrow 3). Arrow 4 shows an AUG recognized during tethering as an initiation codon because it is accessible to the initiator Met-tRNA. (B) Factors affecting translation efficiency from a complex undergoing dynamic clustering will be similar to those affecting a tethered complex, except that the ribosomal complex will be less constrained.
The present hypotheses are premised on the notion that scanning alone cannot explain a variety of data, which prompts us to ask whether the scanning model is valid in any case, and, if so, whether it is exclusive. Approximately 30 years of investigation have failed to elucidate the mechanistic details that underlie scanning or to visualize scanning ribosomes directly (27). The processive mode of scanning prompts a question concerning its energy requirements. In contrast to other biological examples of processive molecular motors, such as kinesin movement on microtubules, which utilizes one ATP per step (28), there appears to be no energy requirement for ribosomal movement to the initiation codon on unstructured RNAs (29). This finding suggests that the operation of ribosomal scanning can proceed without an energy source. For structured mRNAs, ribosomal complex formation at the initiation codon does appear to require ATP (29), which is presumably required for the helicase activity of eIF4A. However, this helicase is not processive (30) and is unlikely to drive scanning ribosomes.
We suggest that, for some mRNAs, tethering or clustering are testable alternatives to scanning. They accommodate the known mechanisms of ribosomal recruitment at the cap structure or at internal mRNA sequences and can explain various observations that are not easily explained by ribosomal scanning. In addition, these models are consistent with the idea that multiple mechanisms can be used to facilitate and control translation initiation.
Methods
DNA Constructs.
The 5′ leader sequences used in this study are presented in Table 1, which is published as supporting information on the PNAS web site, and were generated by PCR amplification of oligonucleotide templates using Pfu DNA polymerase (Stratagene, La Jolla CA). The 5′ leaders were cloned into the pGL3c 5′ multiple cloning site (MCS) reporter construct used in our previous studies that encodes Photinus (firefly) luciferase (18).
Cloning of inserts corresponding to increasing numbers of repeats of the β-globin 5′ leader sequences required the introduction of an AatII restriction site into the putative transcription start site of pGL3c 5′ MCS by using site-directed mutagenesis. PCR-amplified inserts were then introduced into the modified pGL3c 5′ MCS by using AatII and NcoI restriction sites, placing them between the putative transcription start site of the reporter gene and the initiation codon.
Constructs in which five linked copies of the 8-nt Gtx translational enhancer elements, or a size-matched control sequence, were spaced at increasing distance upstream of the Photinus luciferase initiation codon were generated by cloning two PCR-amplified inserts into pGL3c 5′ MCS. The first inserts corresponded to either the 8-nt Gtx translational enhancer elements or to a size-matched control sequence and were cloned into pGL3c 5′ MCS by using SpeI and NcoI restriction sites. A second set of inserts corresponding to increasing numbers of repeats of the poly(A)/β-globin (SI/SIII) spacer sequence were subsequently cloned downstream of the first insert by using AflII and NcoI restriction sites.
Constructs in which the 8-nt Gtx translational enhancer elements are placed downstream of an upstream initiation codon in excellent context (ACCATGGA) were generated by cloning two inserts into pGL3c 5′ MCS. The first insert corresponds to five linked copies of the 8-nt Gtx translational enhancer elements and was cloned into pGL3c 5′ MCS by using SpeI and NcoI restriction sites. A second set of inserts corresponding to increasing numbers of repeats of the poly(A)/β-globin (SI/SIII) spacer sequence containing the uAUG codon in excellent context was subsequently cloned upstream of the first insert in the resulting plasmid by using SpeI and EcoRI restriction sites. An NcoI fragment was isolated from these constructs and cloned into the NcoI site of the construct containing three tandem repeats of the β-globin 5′ leader sequence, which were included to minimize the effects of ribosomal recruitment at the cap structure. The latter manipulation altered the context of the upstream initiation codon (TCCATGGA), which was subsequently restored with site-directed mutagenesis. Finally, size-matched control constructs were generated by site-directed mutagenesis of the upstream initiation codons (ACCATGGA to ACCAAAGA).
Site-Directed Mutagenesis.
Site-directed mutagenesis was performed in reactions containing 125-ng forward and reverse oligonucleotide primers containing mutated nucleotides (presented in Table 2, which is published as supporting information on the PNAS web site), 600 μM dNTPs, and 1 unit of Phusion DNA polymerase (New England Biolabs, Ipswich, MA) with 25 cycles of amplification for 1.5 min/kb extension times. Eight microliters was used to transform DH5α subcloning efficiency cells (Invitrogen, Carlsbad, CA), which were then plated onto ampicillin agar plates.
Analyses of Reporter Gene Activity.
Transfection of mouse N2a cells, reporter gene assays, and total RNA isolations were performed as described in our previous studies (18). Reporter mRNA levels in mammalian cells were determined by using ribonuclease protection assays (RPAIII kit; Ambion, Austin, TX) with 1 μg of DNase-treated total RNA. Protected fragments were size-fractionated on 6% polyacrylamide-urea gels, visualized on a Storm 860 PhosphorImager (Molecular Dynamics, Sunnyvale, CA) and quantified by using AlphaEaseFC Stand Alone software (Alpha Innotech, San Leandro, CA).
Supplementary Material
Acknowledgments
We thank Luke Burman for excellent technical assistance and Drs. Bruce Cunningham and Joseph Gally for valuable comments and critical readings of the manuscript. This work was supported by National Institutes of Health Grant GM61725 (to V.P.M.), the G. Harold and Leila Y. Mathers Charitable Foundation (V.P.M.), and the Skaggs Institute for Chemical Biology (S.A.C.).
Abbreviations
- uAUG
upstream AUG
- IRES
internal ribosome entry site
Note Added in Proof.
Recently, Junemann et al. (32) showed that translation can initiate at a cistron located upstream of picornavirus IRESs. These results would require the ribosomal subunits to initiate translation up to ≈900 nt upstream of the IRES. These findings appear to be inconsistent with backward scanning and are more consistent with a nonlinear mechanism of ribosomal movement that involves either tethering or clustering of the ribosomal subunits.
Footnotes
The authors declare no conflict of interest.
References
- 1.Pisarev AV, Shirokikh NE, Hellen CU. C R Biol. 2005;328:589–605. doi: 10.1016/j.crvi.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 2.Jackson RJ. In: Translational Control of Gene Expression. Sonenberg N, Hershey JWB, Mathews MB, editors. Woodbury, NY: Cold Spring Harbor Lab Press; 2000. pp. 127–183. [Google Scholar]
- 3.Kozak M. Gene. 2002;299:1–34. doi: 10.1016/S0378-1119(02)01056-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sedman SA, Gelembiuk GW, Mertz JE. J Virol. 1990;64:453–457. doi: 10.1128/jvi.64.1.453-457.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yueh A, Schneider RJ. Genes Dev. 2000;14:414–421. [PMC free article] [PubMed] [Google Scholar]
- 6.Ryabova LA, Pooggin MM, Hohn T. Prog Nucleic Acid Res Mol Biol. 2002;72:1–39. doi: 10.1016/S0079-6603(02)72066-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rogers GW, Jr, Edelman GM, Mauro VP. Proc Natl Acad Sci USA. 2004;101:2794–2799. doi: 10.1073/pnas.0308576101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lammich S, Schobel S, Zimmer AK, Lichtenthaler SF, Haass C. EMBO Rep. 2004;5:620–625. doi: 10.1038/sj.embor.7400166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Slusher LB, Gillman EC, Martin NC, Hopper AK. Proc Natl Acad Sci USA. 1991;88:9789–9793. doi: 10.1073/pnas.88.21.9789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matsuda D, Dreher TW. RNA. 2006;12:1338–1349. doi: 10.1261/rna.67906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Preiss T, Hentze MW. Nature. 1998;392:516–520. doi: 10.1038/33192. [DOI] [PubMed] [Google Scholar]
- 12.Kaminski A, Belsham GJ, Jackson RJ. EMBO J. 1994;13:1673–1681. doi: 10.1002/j.1460-2075.1994.tb06431.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herbreteau CH, Weill L, Decimo D, Prevot D, Darlix JL, Sargueil B, Ohlmann T. Nat Struct Mol Biol. 2005;12:1001–1007. doi: 10.1038/nsmb1011. [DOI] [PubMed] [Google Scholar]
- 14.Suzuki Y, Ishihara D, Sasaki M, Nakagawa H, Hata H, Tsunoda T, Watanabe M, Komatsu T, Ota T, Isogai T, et al. Genomics. 2000;64:286–297. doi: 10.1006/geno.2000.6076. [DOI] [PubMed] [Google Scholar]
- 15.Peri S, Pandey A. Trends Genet. 2001;17:685–687. doi: 10.1016/s0168-9525(01)02493-3. [DOI] [PubMed] [Google Scholar]
- 16.Kozak M. Genomics. 2000;70:396–406. doi: 10.1006/geno.2000.6412. [DOI] [PubMed] [Google Scholar]
- 17.Chappell SA, Dresios J, Edelman GM, Mauro VP. Proc Natl Acad Sci USA. 2006;103:9488–9493. doi: 10.1073/pnas.0603597103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chappell SA, Edelman GM, Mauro VP. Proc Natl Acad Sci USA. 2004;101:9590–9594. doi: 10.1073/pnas.0308759101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mauro VP, Edelman GM. Proc Natl Acad Sci USA. 2002;99:12031–12036. doi: 10.1073/pnas.192442499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kozak M. J Mol Biol. 1994;235:95–110. doi: 10.1016/s0022-2836(05)80019-1. [DOI] [PubMed] [Google Scholar]
- 21.Krowczynska A, Brawerman G. Proc Natl Acad Sci USA. 1986;83:902–906. doi: 10.1073/pnas.83.4.902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zuker M, Mathews DH, Turner DH. In: RNA Biochemistry and Biotechnology. Barciszewski J, Clark BFC, editors. The Netherlands: Kluwer, Dordrecht; 1999. pp. 11–43. [Google Scholar]
- 23.Chappell SA, Edelman GM, Mauro VP. Proc Natl Acad Sci USA. 2000;97:1536–1541. doi: 10.1073/pnas.97.4.1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dresios J, Chappell SA, Zhou W, Mauro VP. Nat Struct Mol Biol. 2006;13:30–34. doi: 10.1038/nsmb1031. [DOI] [PubMed] [Google Scholar]
- 25.Sonenberg N. Trends Genet. 1991;7:105–106. doi: 10.1016/0168-9525(91)90440-2. [DOI] [PubMed] [Google Scholar]
- 26.Kozak M. Proc Natl Acad Sci USA. 1995;92:7134. doi: 10.1073/pnas.92.15.7134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Merrick WC. Gene. 2004;332:1–11. doi: 10.1016/j.gene.2004.02.051. [DOI] [PubMed] [Google Scholar]
- 28.Carter NJ, Cross RA. Curr Opin Cell Biol. 2006;18:61–67. doi: 10.1016/j.ceb.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 29.Pestova TV, Kolupaeva VG. Genes Dev. 2002;16:2906–2922. doi: 10.1101/gad.1020902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rogers GW, Jr, Komar AA, Merrick WC. Prog Nucleic Acid Res Mol Biol. 2002;72:307–331. doi: 10.1016/s0079-6603(02)72073-4. [DOI] [PubMed] [Google Scholar]
- 31.Kozak M. Gene Expression. 1991;1:111–115. [PMC free article] [PubMed] [Google Scholar]
- 32.Junemann C, Song Y, Bassili G, Goergen D, Henke J, Niepmann M. J Biol Chem. 2006 Nov 8; doi: 10.1074/jbc.M608750200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.