Abstract
Many enzymes that hydrolyze insoluble crystalline polysaccharides such as cellulose and chitin guide detached single-polymer chains through long and deep active-site clefts, leading to processive (stepwise) degradation of the polysaccharide. We have studied the links between enzyme efficiency and processivity by analyzing the effects of mutating aromatic residues in the substrate-binding groove of a processive chitobiohydrolase, chitinase B from Serratia marcescens. Mutation of two tryptophan residues (Trp-97 and Trp-220) close to the catalytic center (subsites +1 and +2) led to reduced processivity and a reduced ability to degrade crystalline chitin, suggesting that these two properties are linked. Most remarkably, the loss of processivity in the W97A mutant was accompanied by a 29-fold increase in the degradation rate for single-polymer chains as present in the soluble chitin-derivative chitosan. The properties of the W220A mutant showed a similar trend, although mutational effects were less dramatic. Processivity is thought to contribute to the degradation of crystalline polysaccharides because detached single-polymer chains are kept from reassociating with the solid material. The present results show that this processivity comes at a large cost in terms of enzyme speed. Thus, in some cases, it might be better to focus strategies for enzymatic depolymerization of polysaccharide biomass on improving substrate accessibility for nonprocessive enzymes rather than on improving the properties of processive enzymes.
Keywords: cellulose, chitin, chitinase, chitosan, processivity
The enzymatic degradation of the closely related insoluble polysaccharides cellulose [β(1–4)-linked glucose] and chitin [β(1–4)-linked N-acetylglucosamine] is of large biological and economical importance. In recent review papers on the potential of biofuels, Ragauskas et al. (1) and Farrell et al. (2) emphasized the importance of improved cellulosic technologies and better cellulolytic enzymes for conversion of biomass to easily fermentable compounds such as glucose. Large efforts to improve cellulosic technologies are underway, stimulated by, among others, the U.S. Department of Energy (www1.eere.energy.gov/biomass). Chitin is the most important nonplant structural biopolymer, occurring in, e.g., the exoskeletons of invertebrates, fungal cell walls, and the digestive tracts of insects. Chitin is available in large quantities as an underutilized waste product (e.g., shrimp shells). It is used for production of glucosamine and chitosan and also could be converted to bioactive oligomeric compounds or building blocks for bioactive glycoconjugates if efficient enzyme technology were available (3). Chitin turnover plays a role in many important processes, including transmission of the malaria parasite (4), infectivity of insect viruses (5), plant defense responses (6), and modulation of immune responses and asthmatic inflammation in humans (7). Because chitin does not occur in humans, chitin metabolism is an interesting target area for development of drugs and pesticides.
Enzymes acting on cellulose or chitin face the challenges of associating with the insoluble substrate, disrupting crystal packing, and guiding a single-polymer chain into the catalytic center. In addition to their catalytic domains, cellulases and chitinases often contain one or multiple so-called carbohydrate-binding modules (CBMs) (8), which are beneficial for enzyme efficiency because they adhere to and sometimes disrupt the substrate (9–14). Recently, it has been shown that chitin-degrading microorganisms produce a separate noncatalytic protein whose function is to disrupt the crystallinity of the substrate, thus dramatically increasing the efficiency of hydrolysis by chitinases (15).
Another feature that overcomes the low accessibility of the substrate is the presence of long and deep, sometimes “tunnel-like,” substrate-binding clefts as revealed by the first crystal structures of cellulases (or cellobiohydrolases; refs. 16 and 17). These enzymes act processively, i.e., single-carbohydrate chains are threaded through the active-site cleft, while disaccharides are cleaved off at the catalytic center (Figs. 1 and 2; refs. 18 and 19). The general idea is that catalytic efficiency is improved by keeping the enzyme closely associated to the substrate in between subsequent hydrolytic reactions. In the case of crystalline substrates, a potential additional advantage of such processive enzymes is that they are capable of keeping once-detached single chains from reassociating with the insoluble material (19, 20).
Fig. 1.
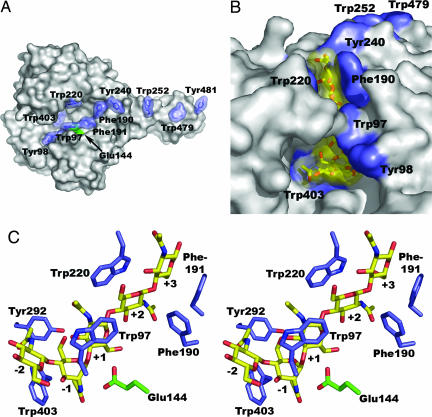
Enzyme–substrate interactions in ChiB. (A) Surface representation showing aromatic side chains lining the substrate-binding cleft and the binding surface of the chitin-binding domain (extending to the right; residues 479 and 481). The catalytic Glu-144 is colored green. (B) Surface representation of the E144Q mutant in complex with chitopentaose [(GlcNAc)5] bound to subsites −2 to +3 (27), showing that the substrate-binding cleft has a closed “roof” when substrate is bound. (GlcNAc)5 is shown with a yellow van der Waals surface. The surfaces of aromatic residues in the protein are blue. (C) Stereo picture showing (GlcNAc)5 and aromatic residues near the catalytic center. Individual sugars in the pentamer are labeled by the number of the enzyme subsite (from −2 to +3) to which they bind.
Fig. 2.
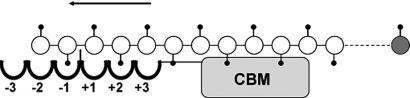
Schematic picture of ChiB in complex with a single chitin chain. The enzyme has six subsites, numbered −3 to +3. CBM, carbohydrate-binding module (8). The reducing end sugar is colored gray. A correctly positioned N-acetyl group (symbolized by small black balls on sticks) in the −1 subsite is essential for catalysis (which is “substrate-assisted”) to occur (25–27). Initial binding of the substrate will produce an odd- or even-numbered “overhang” leading to an odd- or even-numbered product (a trimer or dimer in case of exo activity). The scheme shows the situation during subsequent processive action when only dimers are produced. The arrow indicates the direction of the sliding of the substrate through the active-site cleft (31, 32). In the case of chitosan, complexes formed during processive action may be nonproductive because the sugar bound in the −1 subsite may lack the N-acetyl group. This leads to the production of longer even-numbered oligomers that is diagnostic for processivity (ref. 29; see text).
Generally, enzymatic degradation of crystalline polysaccharides is difficult to study because the insoluble substrate is not amenable to straightforward biochemical analysis and soluble intermediate oligosaccharide products are degraded fast and, therefore, difficult to detect. Thus, usually the only detectable products during degradation of chitin or cellulose are mono-, di-, and trisaccharides, which are typical end products (21). Processivity often is assessed by comparing the production of soluble and nonsoluble reducing ends, but this approach cannot discriminate between all possible modes of action (exo-acting enzymes will yield high soluble/nonsoluble ratios regardless of processivity, and so will endo-acting enzymes with a high degree of processivity; ref. 22). Processivity also may be assessed roughly by studying the ratio between produced dimers and monomers (ref. 23; see below).
Because the successive sugar units in chitin (and cellulose) are rotated by 180°, sliding of such polymers through the enzyme's active site will result in productive binding only for every second sugar, and the products of processive degradation are disaccharides (Fig. 2). This is particularly obvious in the case of family 18 chitinases, where catalysis is substrate-assisted and depends critically on a correctly positioned N-acetyl group on the sugar positioned in the −1 subsite (24) of the enzyme (see Fig. 2; refs. 25–28). Partial deacetylation of chitin produces a soluble polymer, chitosan, that, because of the lack of catalytically crucial N-acetyl groups, can bind both productively and nonproductively to chitinases. As explained in Fig. 2 legend and below, the combination of the soluble chitosan substrate with the substrate-assisted mechanism of a family 18 chitinase provides a unique model system for studying complex aspects of the enzymatic degradation of insoluble polysaccharides, such as processivity (21, 29).
In the present study, we have exploited this model system in the study of chitinase B from Serratia marcescens (ChiB). ChiB is a processive family 18 chitinase with a deep substrate-binding cleft (refs. 27 and 29–31; Fig. 1). Because ChiB sharpens the ends of chitin microfibrilles, it has been suggested that ChiB is an exochitinase (ref. 32; by analogy to the cellulase nomenclature, this would make ChiB a chitobiohydrolase). However, recent studies with chitosan have shown that ChiB tends to bind this substrate in an endo fashion. This indicates that the apparent exo-activity on chitin may be due to better accessibility of chain ends rather than to structural features of the enzyme (ref. 22; see also Fig. 4 and Supporting Materials and Methods, which are published on the PNAS web site). Initial endo binding is not commonly observed for processive tunnel-like cellobiohydrolases but has been suggested in a few cases (33, 34).
The substrate-binding clefts of processive cellulases and chitinases are lined with aromatic residues that are thought to facilitate substrate binding and, most importantly, sliding of the polymer chain through the cleft during a processive mode of action (Fig. 1; refs. 35 and 36). Indeed, there are a few examples showing that mutation of such residues impairs enzyme performance (37–41), but links to processivity are not clear. In this study we have mutated a series of aromatic residues in the substrate-binding cleft of ChiB. Analysis of mutational effects by using standard substrates (chitin and chito-oligosaccharides) revealed two particularly interesting mutants (W97A and W220A), which were analyzed in more detail by using the chitosan model substrate. This led to the discovery of previously undescribed aspects of the contribution of processivity to enzyme efficiency that are of crucial importance for the future development of more efficient enzyme technology for biomass turnover.
Results
As explained in Fig. 2 legend, initial productive binding of chitin may produce odd- or even-numbered oligomers, whereas all further cleavages resulting from the same initial enzyme-substrate association event (i.e., processive action) will produce dimers. All odd-numbered products eventually are converted to monomers and dimers (21). The monomer fraction thus is indicative for the number of odd-numbered products, which again relates to the number of initial binding events. Thus, dimer/monomer ratios in end-product mixtures give an impression of the degree of processivity (23).
Previous studies on the role of aromatic residues in chitinases mostly addressed residues that are located more remote from the catalytic center and did not address processivity (refs. 39–41; see also below). In the present study, we focused on aromatic residues close to the catalytic center, namely Trp-97, Phe-190, Phe-191, Trp-220, Tyr-240, and Trp-403 (Fig. 1), which were mutated to alanine. When degrading chitin, all mutants (except 403, for which no protein could be produced) displayed reduced dimer/monomer ratios (results not shown), indicative of reduced processivity. The most pronounced effects were found for W97A and W220A, where the ratio was reduced from 14 in the wild-type to four and five, respectively. The ratio was also four for a W97A-W220A double mutant.
These ratios do not provide direct quantitative measurements of processivity because they are affected by the enzyme's preferences for initial binding of the polymer (see Fig. 2; ref. 21). This may be illustrated by looking at the degradation of hexamers, which may be converted to two trimers (which ChiB will convert to two dimers and two monomers) or three dimers, depending on the subsite structure and binding preferences of the enzyme. In this particular case, it is relevant to note that the wild-type enzyme produces more trimers from hexamers than W97A (see Fig. 3 and below). Although binding preferences for the hexameric substrate and chitin may differ, the results with the hexamer do suggest that W97A has a higher, processivity-independent tendency to produce dimers than the wild type. Thus, the difference in processivity between the two enzymes may be larger than what is suggested by the difference in dimer/monomer ratios.
Fig. 3.
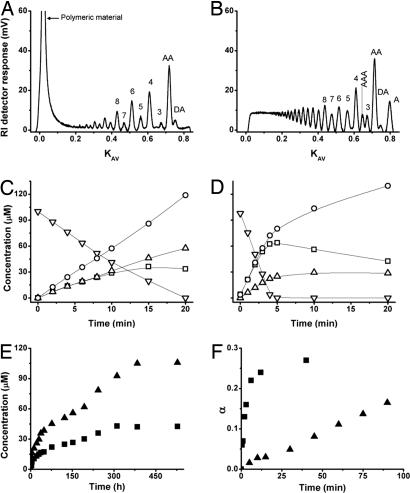
Characteristics of wild-type ChiB and the W97A mutant. (A and B) Size exclusion chromatography of products formed during degradation of chitosan (65% acetylated water-soluble chitin with random distribution of acetylated units) with ChiB (A) and W97A (B), after cleavage of 14% of the glycosidic bonds (i.e., α = 0.14). The products are labeled by chain length or, for the shortest products, by sequence (A, acetylated unit; D, deacetylated unit). (C and D) Degradation of chitohexaose with ChiB (C) and W97A (D). ▿, (GlcNAc)6; □, (GlcNAc)4; ▵, (GlcNAc)3; ○, (GlcNAc)2. (E) Degradation of chitin with ChiB (▴) and W97A (■). (F) Time curve for chitosan degradation with ChiB (▴) and W97A (■). α, fraction of cleaved glycosidic bonds (complete conversion of the substrate to exclusively dimers would yield α = 0.50).
Degradation experiments with the soluble polymeric substrate chitosan showed that wild-type ChiB predominantly produces even-numbered oligomers, whereas the polymer fraction disappears slowly (Fig. 3A). The occurrence of longer primarily even-numbered oligomeric products during the initial phase of the reaction proofs processivity, as previously discussed in refs. 21 and 29. If association of the enzyme with the chitosan chain leads to a nonproductive complex (e.g., because the −1 sugar lacks the N-acetyl group), the substrate either may dissociate and rebind in a different fashion or slide through the active-site cleft until a productive complex is formed (processive action). Because the enzyme has a limited number of subsites, the former solution would lead to a random distribution of odd-numbered and even-numbered products (for products longer than trimers, i.e., products that, when bound to the enzyme, extend beyond the substrate-binding cleft). If the enzyme acts processively, the initial product may be odd- or even-numbered, whereas all further products resulting from the same initial association event will be even-numbered. It is interesting to note that the initial formation of longer even-numbered oligomers shows that processive movement does not depend on the hydrolytic reaction to occur (29).
Fig. 3B shows that W97A has a dramatically different product profile. This mutant has lost most of its processive properties and the size distribution of the products looks like those obtained for nonprocessive endo enzymes, such as ChiC from S. marcescens (21) (i.e., no dominance of even-numbered products and rapid disappearance of the polymer fraction). The loss of processivity in W97A could also be shown by other, less reliable methods, such as simultaneous measurements of substrate viscosity and reducing end production (see Fig. 4 and Supporting Materials and Methods) and studies with oligomeric substrates (see below).
Studies with soluble oligosaccharide substrates showed that the W97A mutant hydrolyzes (GlcNAc)4 (data not shown) and (GlcNAc)6 (Figs. 3 C and D) ≈4-fold faster than the wild-type enzyme, the initial specific activities for (GlcNAc)6 degradation being 33 s−1 and 131 s−1 for ChiB wild-type and W97A, respectively. Note that the wild-type enzyme initially produced more dimers than tetramers as expected for a processive enzyme, whereas the mutant produced equal amounts of both oligosaccharides, as expected for a nonprocessive enzyme.
Experiments with insoluble chitin showed that W97A is less efficient than the wild-type enzyme, both in terms of rate and yield (Fig. 3E).
Analysis of the kinetics of chitosan degradation revealed that the effect of the W97A mutation is indeed remarkable (Fig. 3F), the mutant showing a dramatic 29-fold increase in the hydrolysis rate. The initial specific activities for the wild-type and mutant enzyme were 16 s−1 and 466 s−1, respectively.
The effects of the W220A mutation (data not shown) were similar, but less extreme: W220A showed a higher degree of remaining processivity than W97A and was only four times faster than the wild-type toward chitosan. As expected, W220A also showed reduced activity toward chitin compared with the wild type.
As a control, we also mutated Asp-316, whose side chain makes a strong hydrogen bond to Trp-97, which is crucial for closing the “roof” of the active-site cleft upon substrate-binding (ref. 27; Fig. 1B). The D316A mutant showed wild-type-like processivity and activity toward chitin and no increase in efficiency toward soluble substrates. This shows that the effect of the W97A mutation is not due to disruption of the enzyme's ability to close the active-site cleft but rather to the removal of the aromatic side chain of residue 97.
Discussion
There are several studies in the literature showing that mutation of aromatic residues in the substrate-binding grooves of chitinases and cellulases reduces activity toward crystalline substrates, while not affecting, or in some cases increasing, the activity toward soluble or amorphous substrates (37–40). Katouno et al. (40) studied the role of aromatic residues in ChiB that are more remote from the catalytic center than the residues described in this report (Fig. 1). They mutated five residues to alanine and found that mutation of residues 240, 252, 479, and 481 (but not of 190) reduces binding to and hydrolysis of crystalline chitin while not reducing activity toward soluble substrates (processivity was not addressed in this study). Interestingly, Watanabe et al. (39) have mutated the equivalents of Trp-97 and Trp-220 in chitinase A1 from Bacillus circulans to alanine and noted that, besides reduced activity toward crystalline chitin, their mutants displayed an “unexpected” increase in activity (2- to 3-fold) toward chitopentaose. We show here that the gain in enzyme efficiency is in fact much higher when assessed with a polymeric substrate (Fig. 3F). Most importantly, we show that this gain is accompanied by strongly reduced processivity (Figs. 3 A and B). Our results provide a clear illustration of the link between enzyme efficiency and processivity and show that processivity, although being beneficial for hydrolyzing insoluble substrates, may drastically reduce enzyme efficiency toward more accessible substrates.
Interestingly, literature on cellulases contains a few examples that, when interpreted in the light of the present results, seem to confirm the link between processivity, efficiency toward crystalline substrates, and enzyme speed. Von Ossowski et al. (20) showed that deletion of a tunnel-forming loop in a processive cellobiohydrolase led to a 2-fold loss in efficiency toward crystalline cellulose, a 1.7-fold gain in activity toward amorphous cellulose, and a reduction in the cellobiose/glucose ratio from 23 to 14 (suggesting reduced processivity). Zhou et al. (42) have described a series of point mutations in a processive endoglucanase, including the mutation of a tyrosine in subsite −3 to Phe or Ala. Interestingly, both these mutations led to reduced processivity (assessed by measuring soluble versus nonsoluble reducing ends), increased activity on carboxymethylcellulose and reduced activity on crystalline cellulose. The results of these studies on processive cellulases are in accordance with the conclusions of the present study.
As explained above, the studies with chitosan show that the substrate remains associated with ChiB and slides through the active-site cleft, even if catalysis does not occur (hence, the formation of longer even-numbered oligomers visible in Fig. 3A). It is conceivable that the necessarily loose association of the polymeric substrate is due to an enzyme-based restriction of diffusion that disfavors full dissociation of the substrate (i.e., release to the solvent). Tunnel-like active-site clefts (18) or closure of a “roof” in the active-site cleft upon substrate binding (Fig. 1) may provide one limiting factor to diffusability (20), but the results obtained with the D316A control mutant suggest that this is not a dominant factor in ChiB. Structural work on a cellobiohydrolase has shown that tryptophans provide a flexible hydrophobic sheath that contributes to the necessary fluid binding of the polysaccharide ligand (36). On crystalline substrates, tight but fluid binding of the substrate is favorable because it keeps single-polysaccharide chains detached from the insoluble material, while permitting the necessary movement of the detached sugar chain through the enzyme active-site cleft. However, in kinetic terms, this “stickiness” of the enzyme means a low off rate, koff (43), which will slow down catalytic efficiency for easily diffusible soluble substrates. Increased off rates may explain why the W97A mutant and, to a lesser extent, the W220A mutant, displayed increased activities toward soluble substrates.
Processive enzymes are abundant among natural chitinases and cellulases (18, 19, 44). The present results show that processivity may drastically reduce enzyme efficiency for certain substrates, meaning that processive enzymes not necessarily are the most effective starting point for development of industrial biocatalysts for biomass conversion. Fortunately, nature does provide other means for improving enzymatic degradation of polysaccharides, e.g., in the form of nonprocessive enzymes that contain one or more CBMs. For example, the three-domain endochitinase ChiC from S. marcescens has an open substrate-binding groove and is not processive but is more effective toward crystalline chitin than ChiB (11, 15, 21). In this case, the efficiency toward the crystalline substrate is not due to processivity but to the presence of the additional substrate-binding and substrate-disrupting domains (10, 11). CBMs also have been shown to improve the efficiency of cellulases (12). However, of the naturally occurring CBM-containing cellulases, processive enzymes are the most effective for degradation of crystalline cellulose (19).
In conclusion, we have shown that strategies toward improved enzymatic turnover of chitin and, possibly, other insoluble crystalline polysaccharides, should include use of substrate-disrupting CBMs (8–14, 45, 46) or proteins (15, 47), or chemical methods (48, 49), to increase substrate accessibility and alleviate the need for processivity and low off rates. In some cases, focusing on substrate accessibility and nonprocessive enzymes (natural or engineered) containing optimal CBMs may be a better strategy for reducing the costs of biomass turnover than using processive enzymes.
Materials and Methods
Chemicals.
Squid pen β-chitin was purchased from France Chitin (Marseille, France). Chitosan, with a degree of N-acetylation of 65% and a number-average degree of polymerization of 700 (DPn = 700), was prepared by homogeneous N-deacetylation of milled (1.0-mm sieve) shrimp shell chitin (50) and was converted to the chitosan hydrochloride salt (51), which is readily soluble in water. This procedure results in a chitosan with a random distribution of N-acetylated and de-N-acetylated units (52). Chito-oligosaccharides and all other chemicals were purchased from Sigma (St Louis, MO).
Enzymes.
The chitinase gene chib from S. marcescens strain BJL200 was expressed in Escherichia coli DH5α (Life Technologies, Rockville, MD) under control of its own promoter (30). Site-directed mutations were introduced by using the QuikChange kit from Stratagene (La Jolla, CA) essentially as described by the manufacturer by using cloning and DNA sequencing procedures that have been described in refs. 27 and 28. ChiB and its mutants were purified from periplasmic extracts of early stationary phase cultures as described in ref. 53.
Enzyme purity was verified by SDS/PAGE and estimated to be >95% in all cases. Protein concentrations were determined by using the Bio-Rad Protein Assay (Bio-Rad, Hercules, CA) with BSA as the standard.
Enzymatic Degradation of Chitin, Chito-Oligosaccharides, and Chitosan.
β-chitin (0.1 mg/ml) was degraded with 50 nM enzyme as described in ref. 15. Samples (30 μl) were taken regularly, and the reaction was stopped by adding 90 μl of acetonitrile.
Hydrolysis of (GlcNAc)4 and (GlcNAc)6 was carried out in 50 mM sodium acetate with 50 μg/ml BSA at pH 6.1 and 37°C. The enzyme concentration was 3.0 nM, and the total volume of the reaction mixture was 1 ml. Reaction samples (100 μl) were withdrawn at regular time intervals, and the enzyme was inactivated by adding 5 μl of 2.5 M HCl.
For studies of chitosan degradation, 10 mg of chitosan was dissolved in 1.0 ml H2O followed by the addition of 1.0 ml buffer (0.08 M NaAc/0.2 M NaCl, pH 5.5) and 0.2 mg BSA. Hydrolysis was carried out at 37.0°C in a shaking water bath with 5 μg of enzyme. Degradation was allowed to proceed for increasing time intervals, and reactions were stopped by lowering the pH to 2.5 by the addition of 1.0 M HCl and immersing the samples in boiling water for 2 min.
Chromatography of Oligosaccharides.
Chito-oligosaccharides were analyzed by HPLC with a Tosoh TSK Amide 80 column (0.46 × 25 cm) with an Amide 80 guard column (Tosoh, Tokyo, Japan). A 10-μl (degradation of GlcNAc4 and GlcNAc6) or 50-μl (degradation of chitin) sample was injected on the column, and the oligosaccharides were eluted isocratically at 0.7 ml/min with 70% acetonitrile at room temperature. The chito-oligosaccharides were monitored by measuring absorbance at 210 nm, and the amounts were quantified by measuring peak areas. Peak areas were compared with peak areas obtained with standard samples with known concentrations of chito-oligosaccharides. Using these standard samples, it was established that there was a linear correlation between peak area and oligosaccharide concentration within the concentration range used in this study for each of the oligomers that were analyzed.
Oligomers produced by enzymatic depolymerization of chitosan were separated on three XK 26 columns, packed with Superdex 30, from Amersham Pharmacia Biotech (Uppsala, Sweden), with an overall dimension of 2.60 × 180 cm. The mobile phase was 0.15 M ammonium acetate (pH 4.50), and the flow rate was 0.80 ml/min. The relative amounts of oligomers were monitored with an online refractive index (RI) detector (Shimadzu RID 6A; Duisburg, Germany), and the data were logged with a CR 510 Basic Data logger, from Campbell Scientific (Logan, UT). This method and its performance have been described in detail in ref. 29. It has been shown that this method allows the separation of mixtures of partially N-acetylated oligomers according to size [degree of polymerization (DP)], regardless of chemical composition, in the separation range between DP = 4 and a DP of ≈20. Within the monomer-trimer range, fully N-acetylated oligomers were separated from partially N-acetylated oligomers of the same DP. Studies with standard samples have shown that there is a linear relationship between peak areas and the amount (mass) of injected oligomer, irrespective of DP and degree of N-acetylation (29).
1H-NMR Spectroscopy.
1H-NMR Spectroscopy was used to sequence shorter oligomers and to calculate the number-average degree of polymerization (DPn) in the reaction mixtures as described in ref. 52. The extent of chitosan degradation is given as the degree of scission, α (= 1/DPn), which represents the fraction of glycosidic linkages that has been cleaved. Complete conversion of the polymer to dimers (DP = 2) would yield an α of 0.50.
Supplementary Material
Acknowledgments
The authors thank Audun Sørbotten, Morten Melgård, Bjørge Westereng, Sigrid Gåseidnes, and Inger Marie Krokeide for their help in some experiments. This work was supported by Norwegian Research Council Grants 140497/420 and 164653/V40.
Abbreviations
- CBM
carbohydrate-binding module
- ChiB
chitinase B from Serratia marcescens
- DP
degree of polymerization
Footnotes
The authors declare no conflict of interest.
References
- 1.Ragauskas AJ, Williams CK, Davison BH, Britovsek G, Cairney J, Eckert CA, Frederick WJ, Hallett JP, Leak DJ, Liotta CL, et al. Science. 2006;311:484–489. doi: 10.1126/science.1114736. [DOI] [PubMed] [Google Scholar]
- 2.Farrell AE, Plevin RJ, Turner BT, Jones AD, O'Hare M, Kammen DM. Science. 2006;311:5006–5008. doi: 10.1126/science.1121416. [DOI] [PubMed] [Google Scholar]
- 3.Peter MG. In: Biopolymers, Vol. 6: Polysaccharides II. De Baets S, Vandamme EJ, Steinbüchel A, editors. Weinheim, Germany: Wiley; 2002. pp. 481–574. [Google Scholar]
- 4.Langer RC, Vinetz JM. Trends Parasitol. 2001;17:269–272. doi: 10.1016/s1471-4922(01)01918-3. [DOI] [PubMed] [Google Scholar]
- 5.Hawtin RE, Zarkowska T, Arnold K, Thomas CJ, Gooday GW, King LA, Kuzio JA, Possee RD. Virology. 1997;238:243. doi: 10.1006/viro.1997.8816. [DOI] [PubMed] [Google Scholar]
- 6.Chisholm ST, Coaker G, Day B, Staskawicz BJ. Cell. 2006;124:803–814. doi: 10.1016/j.cell.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 7.Zhu Z, Zheng T, Homer RJ, Kim YK, Chen NY, Cohn L, Hamid Q, Elias JA. Science. 2004;304:1678–1682. doi: 10.1126/science.1095336. [DOI] [PubMed] [Google Scholar]
- 8.Boraston AB, Bolan DN, Gilbert HJ, Davies GJ. Biochem J. 2004;382:769–781. doi: 10.1042/BJ20040892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Din N, Damude HG, Gilkes NR, Miller RC, Warren RAJ, Kilburn DG. Proc Natl Acad Sci USA. 1994;91:11383–11387. doi: 10.1073/pnas.91.24.11383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Watanabe T, Ito Y, Yamada T, Hashimoto M, Sekine S, Tanaka H. J Bacteriol. 1994;176:4465–44472. doi: 10.1128/jb.176.15.4465-4472.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suzuki K, Taiyoji M, Sugawara N, Nikaidou N, Henrissat B, Watanabe T. Biochem J. 1999;343:587–596. [PMC free article] [PubMed] [Google Scholar]
- 12.Carrard G, Koivula A, Söderlund H, Beguin P. Proc Natl Acad Sci USA. 2000;97:10342–10347. doi: 10.1073/pnas.160216697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lehtio J, Sugiyama J, Gustavsson M, Fransson L, Linder M, Teeri TT. Proc Natl Acad Sci USA. 2003;100:484–489. doi: 10.1073/pnas.212651999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCartney L, Gilbert HJ, Bolam DN, Boraston AB, Knox JP. Anal Biochem. 2004;326:49–54. doi: 10.1016/j.ab.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 15.Vaaje-Kolstad G, Horn SJ, Van Aalten DMF, Synstad B, Eijsink VGH. J Biol Chem. 2005;280:28492–28497. doi: 10.1074/jbc.M504468200. [DOI] [PubMed] [Google Scholar]
- 16.Rouvinen J, Bergfors T, Teeri T, Knowles JKC, Jones TA. Science. 1990;249:380–386. doi: 10.1126/science.2377893. [DOI] [PubMed] [Google Scholar]
- 17.Divne C, Stahlberg J, Reinikainen T, Ruohonen L, Pettersson G, Knowles JKC, Teeri TT, Jones TA. Science. 1994;265:524–528. doi: 10.1126/science.8036495. [DOI] [PubMed] [Google Scholar]
- 18.Davies G, Henrissat B. Structure (London) 1995;3:853–859. doi: 10.1016/S0969-2126(01)00220-9. [DOI] [PubMed] [Google Scholar]
- 19.Teeri T. Trends Biotechnol. 1997;15:160–167. [Google Scholar]
- 20.Von Ossowski I, Ståhlberg J, Koivula A, Piens K, Becker D, Boer H, Harle R, Harris M, Divne C, Mahdi S., et al. J Mol Biol. 2003;333:817–829. doi: 10.1016/s0022-2836(03)00881-7. [DOI] [PubMed] [Google Scholar]
- 21.Horn SJ, Sørbotten A, Synstad B, Sikorski P, Sørlie M, Vårum KM, Eijsink VGH. FEBS J. 2006;273:491–503. doi: 10.1111/j.1742-4658.2005.05079.x. [DOI] [PubMed] [Google Scholar]
- 22.Sikorski P, Sørbotten A, Horn SJ, Eijsink VGH, Vårum KM. Biochemistry. 2006;45:9566–9574. doi: 10.1021/bi060370l. [DOI] [PubMed] [Google Scholar]
- 23.Teeri TT, Koivula A, Linder M, Wohlfahrt G, Divne C, Jones TA. Biochem Soc Trans. 1998;26:173–178. doi: 10.1042/bst0260173. [DOI] [PubMed] [Google Scholar]
- 24.Davies GJ, Wilson KS, Henrissat B. Biochem J. 1997;321:557–559. doi: 10.1042/bj3210557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Van Scheltinga ACT, Armand S, Kalk KH, Isogai A, Henrissat B, Dijkstra BW. Biochemistry. 1995;34:15619–15623. doi: 10.1021/bi00048a003. [DOI] [PubMed] [Google Scholar]
- 26.Tews I, van Scheltinga ACT, Perrakis A, Wilson KS, Dijkstra BW. J Am Chem Soc. 1997;119:7954–7959. [Google Scholar]
- 27.van Aalten DMF, Komander D, Synstad B, Gåseidnes S, Peter MG, Eijsink VGH. Proc Natl Acad Sci USA. 2001;98:8979–8984. doi: 10.1073/pnas.151103798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Synstad B, Gåseidnes S, van Aalten DMF, Vriend G, Nielsen JE, Eijsink VGH. Eur J Biochem. 2004;271:253–262. doi: 10.1046/j.1432-1033.2003.03923.x. [DOI] [PubMed] [Google Scholar]
- 29.Sørbotten A, Horn SJ, Eijsink VGH, Vårum KM. FEBS J. 2005;272:538–549. doi: 10.1111/j.1742-4658.2004.04495.x. [DOI] [PubMed] [Google Scholar]
- 30.Brurberg MB, Eijsink VGH, Haandrikman AJ, Venema G, Nes IF. Microbiology. 1995;141:123–131. doi: 10.1099/00221287-141-1-123. [DOI] [PubMed] [Google Scholar]
- 31.van Aalten DMF, Synstad B, Brurberg MB, Hough E, Riise BW, Eijsink VGH, Wierenga RK. Proc Natl Acad Sci USA. 2000;97:5842–5847. doi: 10.1073/pnas.97.11.5842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hult EL, Katouno F, Uchiyama T, Watanabe T, Sugiyama J. Biochem J. 2005;388:851–856. doi: 10.1042/BJ20050090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ståhlberg J, Johansson G, Pettersson G. Biochim Biophys Acta. 1993;1157:107–113. doi: 10.1016/0304-4165(93)90085-m. [DOI] [PubMed] [Google Scholar]
- 34.Boisset C, Fraschini C, Schülein M, Henrissat B, Chanzy H. Appl Environ Microbiol. 2000;66:1444–1452. doi: 10.1128/aem.66.4.1444-1452.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Divne C, Ståhlberg J, Teeri TT, Jones TA. J Mol Biol. 1998;275:309–325. doi: 10.1006/jmbi.1997.1437. [DOI] [PubMed] [Google Scholar]
- 36.Varrot A, Frandsen TP, von Ossowski I, Boyer V, Cottaz S, Driguez H, Schulein M, Davies GJ. Structure (London) 2003;11:855–864. doi: 10.1016/s0969-2126(03)00124-2. [DOI] [PubMed] [Google Scholar]
- 37.Koivula A, Kinnari T, Harjunpaa V, Ruohonen L, Teleman A, Drakenberg T, Rouvinen J, Jones TA, Teeri TT. FEBS Lett. 1998;429:341–346. doi: 10.1016/s0014-5793(98)00596-1. [DOI] [PubMed] [Google Scholar]
- 38.Zhang S, Irwin DC, Wilson DB. Eur J Biochem. 2000;267:3101–3115. doi: 10.1046/j.1432-1327.2000.01315.x. [DOI] [PubMed] [Google Scholar]
- 39.Watanabe T, Ariga Y, Sato U, Toratani T, Hashimoto M, Nikaidou N, Kezuka Y, Nonaka T, Sugiyama J. Biochem J. 2003;376:237–244. doi: 10.1042/BJ20030419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Katouno F, Taguchi M, Sakurai K, Uchiyama T, Nikaidou N, Nonaka T, Sugiyama J, Watanabe T. J Biochem. 2004;136:163–168. doi: 10.1093/jb/mvh105. [DOI] [PubMed] [Google Scholar]
- 41.Uchiyama T, Katouno F, Nikaidou N, Nonaka T, Sugiyama J, Watanabe T. J Biol Chem. 2001;276:41343–41349. doi: 10.1074/jbc.M103610200. [DOI] [PubMed] [Google Scholar]
- 42.Zhou W, Irwin DC, Escovar-Kousen J, Wilson DB. Biochemistry. 2004;43:9655–9663. doi: 10.1021/bi049394n. [DOI] [PubMed] [Google Scholar]
- 43.Harjunpää V, Teleman A, Koivula A, Ruohonen L, Teeri TT, Teleman O, Drakenberg T. Eur J Biochem. 1996;240:584–591. doi: 10.1111/j.1432-1033.1996.0584h.x. [DOI] [PubMed] [Google Scholar]
- 44.Davies GJ, Gloster TM, Henrissat B. Curr Opin Struct Biol. 2005;15:637–645. doi: 10.1016/j.sbi.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 45.Irwin D, Shin DH, Zhang S, Barr BK, Sakon J, Karplus PA, Wilson DB. J Bacteriol. 1998;180:1709–1714. doi: 10.1128/jb.180.7.1709-1714.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Limon MC, Margolles-Clark E, Benitez T, Penttila M. FEMS Microbiol Lett. 2001;198:57–63. doi: 10.1111/j.1574-6968.2001.tb10619.x. [DOI] [PubMed] [Google Scholar]
- 47.Saloheimo M, Paloheimo M, Hakola S, Pere J, Swanson B, Nyyssonen E, Bhatia A, Ward M, Penttila M. Eur J Biochem. 2002;269:4202–4211. doi: 10.1046/j.1432-1033.2002.03095.x. [DOI] [PubMed] [Google Scholar]
- 48.Sun Y, Cheng JY. Bioresour Technol. 2002;83:1–11. doi: 10.1016/s0960-8524(01)00212-7. [DOI] [PubMed] [Google Scholar]
- 49.Mosier N, Wyman C, Dale B, Elander R, Lee YY, Holtzapple M, Ladisch M. Bioresour Technol. 2005;96:673–686. doi: 10.1016/j.biortech.2004.06.025. [DOI] [PubMed] [Google Scholar]
- 50.Sannan T, Kurita K, Ogura K, Iwakura Y. Polymer. 1978;19:458–459. [Google Scholar]
- 51.Draget KI, Vårum KM, Moen E, Gynnild H, Smidsrød O. Biomaterials. 1992;13:635–638. doi: 10.1016/0142-9612(92)90032-j. [DOI] [PubMed] [Google Scholar]
- 52.Vårum KM, Anthonsen MW, Grasdalen H, Smidsrød O. Carbohydr Res. 1991;211:17–23. doi: 10.1016/0008-6215(91)84142-2. [DOI] [PubMed] [Google Scholar]
- 53.Brurberg MB, Nes IF, Eijsink VGH. Microbiology. 1996;142:1581–1589. doi: 10.1099/13500872-142-7-1581. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.